Addressing the Reciprocal Crosstalk between the AR and the PI3K/AKT/mTOR Signaling Pathways for Prostate Cancer Treatment
Abstract
1. Introduction
2. AR Signaling in Prostate Cancer
2.1. AR Pathway Overview
2.2. AR Reprogramming in Prostate Cancer
2.3. Neuroendocrine Prostate Cancer
3. PI3K/AKT/mTOR Signaling in Prostate Cancer
3.1. PI3K/AKT/mTOR Pathway Overview
3.2. PI3K Alterations in Prostate Cancer
3.3. AKT Alterations in Prostate Cancer
3.4. mTOR Complex Alterations in Prostate Cancer
3.5. FOXO Alterations in Prostate Cancer
3.6. PTEN Loss in Prostate Cancer
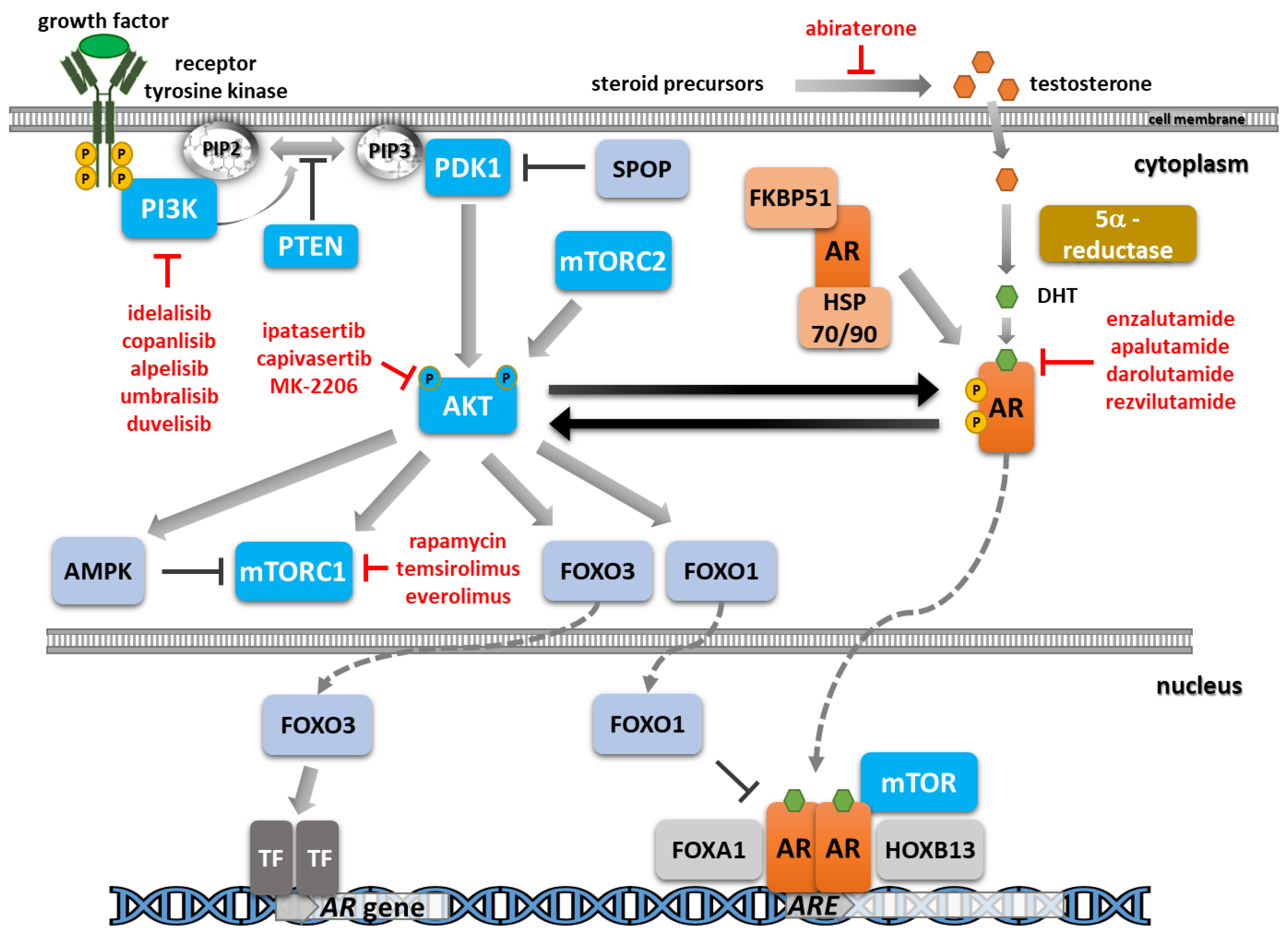
4. Crosstalk between PI3K/AKT/mTOR and AR Signaling
4.1. AR Signaling Impacts the PI3K/AK/mTOR Pathway
4.2. The PI3K/AKT/mTOR Pathway Impacts AR Signaling
5. AR Inhibitors
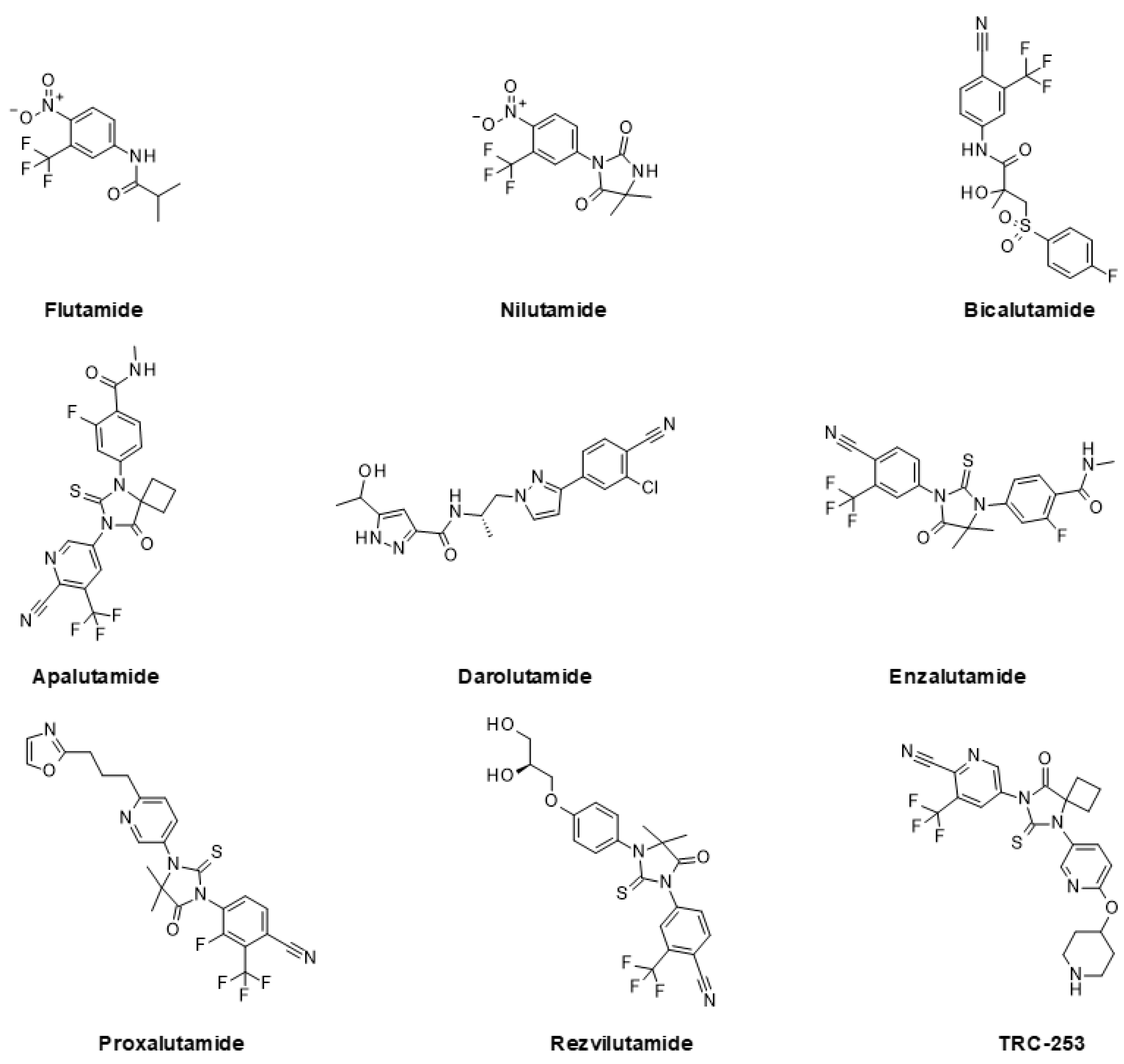
5.1. Approved and Clinically Advanced AR Inhibitors
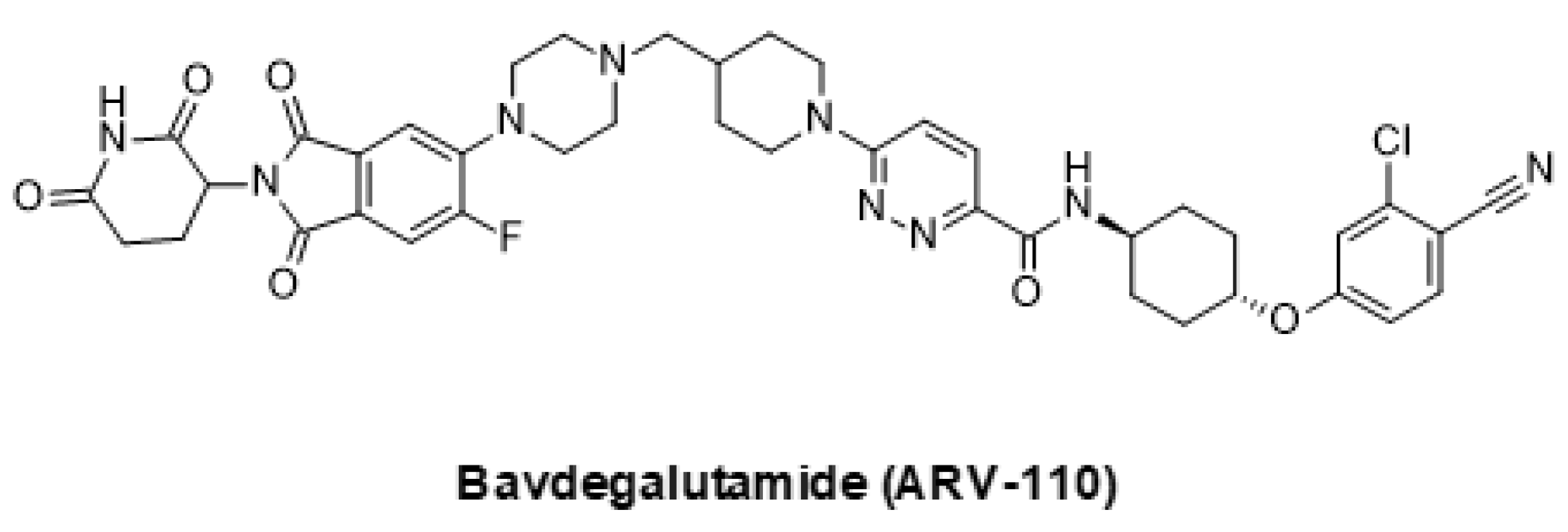
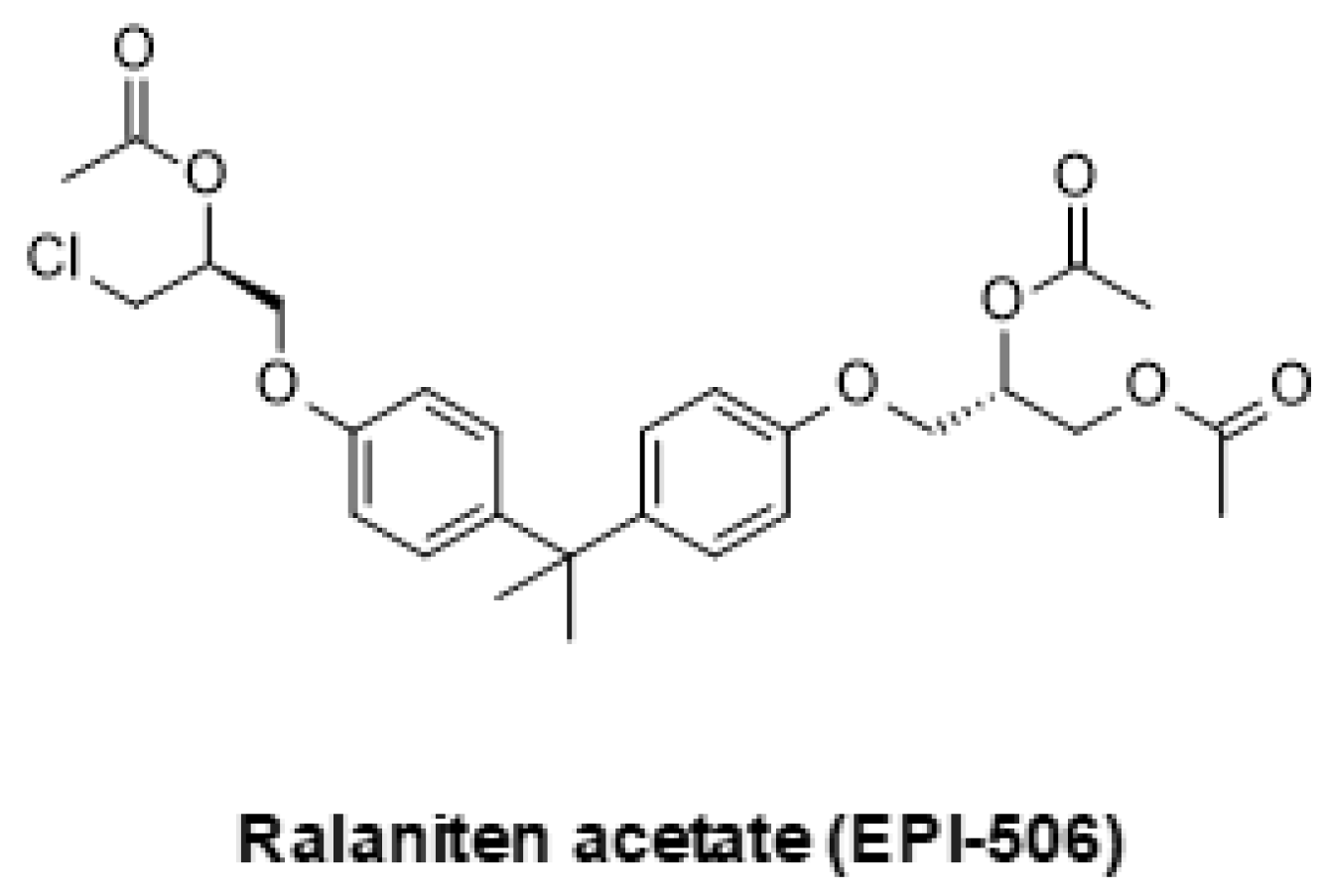
5.2. Emerging Strategies for Inhibition of AR Signaling
6. PI3K/AKT/mTOR Inhibitors
6.1. Clinically Advanced PI3K/AKT/mTOR Inhibitors
6.1.1. FDA-Approved and Clinically Advanced PI3K Inhibitors and Dual PI3K/mTOR Inhibitors
6.1.2. AKT Inhibitors
6.1.3. mTOR Inhibitors
6.2. Emerging Strategies for PI3K/AKT/mTOR Inhibition
7. Recent Clinical Studies Combining Inhibitors of the PI3K/AKT/mTOR and AR Pathways for Prostate Cancer Treatment
8. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Cronin, K.A.; Scott, S.; Firth, A.U.; Sung, H.; Henley, S.J.; Sherman, R.L.; Siegel, R.L.; Anderson, R.N.; Kohler, B.A.; Benard, V.B.; et al. Annual report to the nation on the status of cancer, part 1: National cancer statistics. Cancer 2022, 128, 4251–4284. [Google Scholar] [CrossRef] [PubMed]
- Bancroft, E.K.; Raghallaigh, H.N.; Page, E.C.; Eeles, R.A. Updates in prostate cancer research and screening in men at genetically higher risk. Curr. Genet. Med. Rep. 2021, 9, 47–58. [Google Scholar] [CrossRef]
- Nevedomskaya, E.; Haendler, B. From omics to multi-omics approaches for in-depth analysis of the molecular mechanisms of prostate cancer. Int. J. Mol. Sci. 2022, 23, 6281. [Google Scholar] [CrossRef] [PubMed]
- Wengner, A.M.; Scholz, A.; Haendler, B. Targeting DNA damage response in prostate and breast cancer. Int. J. Mol. Sci. 2020, 21, 8273. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, S.; Moore, C.M.; Chiong, E.; Beltran, H.; Bristow, R.G.; Williams, S.G. Prostate cancer. Lancet 2021, 398, 1075–1090. [Google Scholar] [CrossRef]
- King, L.; Bernaitis, N.; Christie, D.; Chess-Williams, R.; Sellers, D.; McDermott, C.; Dare, W.; Anoopkumar-Dukie, S. Drivers of radioresistance in prostate cancer. J. Clin. Med. 2022, 11, 5637. [Google Scholar] [CrossRef] [PubMed]
- Nevedomskaya, E.; Baumgart, S.J.; Haendler, B. Recent advances in prostate cancer treatment and drug discovery. Int. J. Mol. Sci. 2018, 19, 1359. [Google Scholar] [CrossRef] [PubMed]
- Turco, F.; Gillessen, S.; Cathomas, R.; Buttigliero, C.; Vogl, U.M. Treatment landscape for patients with castration-resistant prostate cancer: Patient selection and unmet clinical needs. Res. Rep. Urol. 2022, 14, 339–350. [Google Scholar] [CrossRef]
- Liu, S.; Alabi, B.R.; Yin, Q.; Stoyanova, T. Molecular mechanisms underlying the development of neuroendocrine prostate cancer. Semin. Cancer Biol. 2022, 86, 57–68. [Google Scholar] [CrossRef]
- Merkens, L.; Sailer, V.; Lessel, D.; Janzen, E.; Greimeier, S.; Kirfel, J.; Perner, S.; Pantel, K.; Werner, S.; von Amsberg, G. Aggressive variants of prostate cancer: Underlying mechanisms of neuroendocrine transdifferentiation. J. Exp. Clin. Cancer Res. 2022, 41, 46. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Ning, S.; Hu, J. Molecular mechanisms of neuroendocrine differentiation in prostate cancer progression. J. Cancer Res. Clin. Oncol. 2022, 148, 1813–1823. [Google Scholar] [CrossRef]
- Tang, F.; Xu, D.; Wang, S.; Wong, C.K.; Martinez-Fundichely, A.; Lee, C.J.; Cohen, S.; Park, J.; Hill, C.E.; Eng, K.; et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science 2022, 376, eabe1505. [Google Scholar] [CrossRef] [PubMed]
- Coleman, I.M.; DeSarkar, N.; Morrissey, C.; Xin, L.; Roudier, M.P.; Sayar, E.; Li, D.; Corey, E.; Haffner, M.C.; Nelson, P.S. Therapeutic implications for intrinsic phenotype classification of metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2022, 28, 3127–3140. [Google Scholar] [CrossRef] [PubMed]
- Feng, E.; Rydzewski, N.R.; Zhang, M.; Lundberg, A.; Bootsma, M.; Helzer, K.T.; Lang, J.M.; Aggarwal, R.; Small, E.J.; Quigley, D.A.; et al. Intrinsic molecular subtypes of metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2022, 28, 5396–5404. [Google Scholar] [CrossRef]
- Hatano, K.; Nonomura, N. Genomic profiling of prostate cancer: An updated review. World J. Mens Health 2022, 40, 368–379. [Google Scholar] [CrossRef]
- Zhou, C.K.; Young, D.; Yeboah, E.D.; Coburn, S.B.; Tettey, Y.; Biritwum, R.B.; Adjei, A.A.; Tay, E.; Niwa, S.; Truelove, A.; et al. TMPRSS2:ERG gene fusions in prostate cancer of West African men and a meta-analysis of racial differences. Am. J. Epidemiol. 2017, 186, 1352–1361. [Google Scholar] [CrossRef]
- Jamaspishvili, T.; Berman, D.M.; Ross, A.E.; Scher, H.I.; De Marzo, A.M.; Squire, J.A.; Lotan, T.L. Clinical implications of PTEN loss in prostate cancer. Nat. Rev. Urol. 2018, 15, 222–234. [Google Scholar] [CrossRef]
- Shorning, B.Y.; Dass, M.S.; Smalley, M.J.; Pearson, H.B. The PI3K-AKT-mTOR pathway and prostate cancer: At the crossroads of AR, MAPK, and WNT signaling. Int. J. Mol. Sci. 2020, 21, 4507. [Google Scholar] [CrossRef]
- Blanco-Aparicio, C.; Renner, O.; Leal, J.F.; Carnero, A. PTEN, more than the AKT pathway. Carcinogenesis 2007, 28, 1379–1386. [Google Scholar] [CrossRef]
- Pungsrinont, T.; Kallenbach, J.; Baniahmad, A. Role of PI3K-AKT-mTOR pathway as a pro-survival signaling and resistance-mediating mechanism to therapy of prostate cancer. Int. J. Mol. Sci. 2021, 22, 11088. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, X.; Yu, H.; Liu, X.; Sun, H.; Shao, C. Alternations of gene expression in PI3K and AR pathways and DNA methylation features contribute to metastasis of prostate cancer. Cell Mol. Life Sci. 2022, 79, 436. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, A.O. Molecular basis of androgen insensitivity. Mol. Cell Endocrinol. 2001, 179, 105–109. [Google Scholar] [CrossRef]
- Jin, J.M.; Yang, W.X. Molecular regulation of hypothalamus-pituitary-gonads axis in males. Gene 2014, 551, 15–25. [Google Scholar] [CrossRef]
- Gerald, T.; Raj, G. Testosterone and the androgen receptor. Urol. Clin. N. Am. 2022, 49, 603–614. [Google Scholar] [CrossRef] [PubMed]
- McPhaul, M.J. Factors that mediate and modulate androgen action. J. Investig. Dermatol. Symp. Proc. 2003, 8, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Klocker, H.; Culig, Z.; Eder, I.E.; Nessler-Menardi, C.; Hobisch, A.; Putz, T.; Bartsch, G.; Peterziel, H.; Cato, A.C. Mechanism of androgen receptor activation and possible implications for chemoprevention trials. Eur. Urol. 1999, 35, 413–419. [Google Scholar] [CrossRef]
- Claessens, F.; Denayer, S.; Van Tilborgh, N.; Kerkhofs, S.; Helsen, C.; Haelens, A. Diverse roles of androgen receptor (AR) domains in AR-mediated signaling. Nucl. Recept. Signal. 2008, 6, e008. [Google Scholar] [CrossRef] [PubMed]
- Cano, L.Q.; Lavery, D.N.; Bevan, C.L. Mini-review: Foldosome regulation of androgen receptor action in prostate cancer. Mol. Cell Endocrinol. 2013, 369, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, C.; Malik, A.; Abeysinghe, P.; Clements, J.; Batra, J. SWATH-MS based proteomic profiling of prostate cancer cells reveals adaptive molecular mechanisms in response to anti-androgen therapy. Cancers 2021, 13, 715. [Google Scholar] [CrossRef]
- Nevedomskaya, E.; Sugawara, T.; Baumgart, S.J.; Lesche, R.; Hahne, H.; Mumberg, D.; Haendler, B. Comparative proteomic and transcriptomic analysis of the impact of androgen stimulation and darolutamide inhibition. Cancers 2022, 15, 2. [Google Scholar] [CrossRef] [PubMed]
- Sadeesh, N.; Scaravilli, M.; Latonen, L. Proteomic landscape of prostate cancer: The view provided by quantitative proteomics, integrative analyses, and protein interactomes. Cancers 2021, 13, 4829. [Google Scholar] [CrossRef]
- Chauhan, G.; Heemers, H.V. Somatic alterations impact AR transcriptional activity and efficacy of AR-targeting therapies in prostate cancer. Cancers 2021, 13, 3947. [Google Scholar] [CrossRef] [PubMed]
- Messner, E.A.; Steele, T.M.; Tsamouri, M.M.; Hejazi, N.; Gao, A.C.; Mudryj, M.; Ghosh, P.M. The androgen receptor in prostate cancer: Effect of structure, ligands and spliced variants on therapy. Biomedicines 2020, 8, 422. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Feng, Q. Androgen receptor signaling and spatial chromatin organization in castration-resistant prostate cancer. Front. Med. 2022, 9, 924087. [Google Scholar] [CrossRef]
- Centenera, M.M.; Harris, J.M.; Tilley, W.D.; Butler, L.M. The contribution of different androgen receptor domains to receptor dimerization and signaling. Mol. Endocrinol. 2008, 22, 2373–2382. [Google Scholar] [CrossRef] [PubMed]
- Mohler, M.L.; Sikdar, A.; Ponnusamy, S.; Hwang, D.J.; He, Y.; Miller, D.D.; Narayanan, R. An overview of next-generation androgen receptor-targeted therapeutics in development for the treatment of prostate cancer. Int. J. Mol. Sci. 2021, 22, 2124. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, Q.; Hankey, W.; Fang, X.; Yuan, F. Second generation androgen receptor antagonists and challenges in prostate cancer treatment. Cell Death Dis. 2022, 13, 632. [Google Scholar] [CrossRef] [PubMed]
- van Royen, M.E.; van Cappellen, W.A.; de Vos, C.; Houtsmuller, A.B.; Trapman, J. Stepwise androgen receptor dimerization. J. Cell Sci. 2012, 125, 1970–1979. [Google Scholar] [CrossRef]
- Faus, H.; Haendler, B. Androgen receptor acetylation sites differentially regulate gene control. J. Cell Biochem. 2008, 104, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Koryakina, Y.; Ta, H.Q.; Gioeli, D. Androgen receptor phosphorylation: Biological context and functional consequences. Endocr. Relat. Cancer 2014, 21, T131–T145. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Niu, Y.; Huang, H. Posttranslational regulation of androgen dependent and independent androgen receptor activities in prostate cancer. Asian J. Urol. 2020, 7, 203–218. [Google Scholar] [CrossRef]
- Jaiswal, B.; Agarwal, A.; Gupta, A. Lysine acetyltransferases and their role in AR signaling and prostate cancer. Front. Endocrinol. 2022, 13, 886594. [Google Scholar] [CrossRef] [PubMed]
- McEwan, I.J. Intrinsic disorder in the androgen receptor: Identification, characterisation and drugability. Mol. Biosyst. 2012, 8, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Sheikhhassani, V.; Scalvini, B.; Ng, J.; Heling, L.; Ayache, Y.; Evers, T.M.J.; Estebanez-Perpina, E.; McEwan, I.J.; Mashaghi, A. Topological dynamics of an intrinsically disordered N-terminal domain of the human androgen receptor. Protein Sci. 2022, 31, e4334. [Google Scholar] [CrossRef]
- Xie, J.; He, H.; Kong, W.; Li, Z.; Gao, Z.; Xie, D.; Sun, L.; Fan, X.; Jiang, X.; Zheng, Q.; et al. Targeting androgen receptor phase separation to overcome antiandrogen resistance. Nat. Chem Biol. 2022, 18, 1341–1350. [Google Scholar] [CrossRef]
- Takayama, K.I.; Inoue, S. Targeting phase separation on enhancers induced by transcription factor complex formations as a new strategy for treating drug-resistant cancers. Front. Oncol. 2022, 12, 1024600. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef]
- Baumgart, S.J.; Nevedomskaya, E.; Haendler, B. Dysregulated transcriptional control in prostate cancer. Int. J. Mol. Sci. 2019, 20, 2883. [Google Scholar] [CrossRef]
- Launonen, K.M.; Paakinaho, V.; Sigismondo, G.; Malinen, M.; Sironen, R.; Hartikainen, J.M.; Laakso, H.; Visakorpi, T.; Krijgsveld, J.; Niskanen, E.A.; et al. Chromatin-directed proteomics-identified network of endogenous androgen receptor in prostate cancer cells. Oncogene 2021, 40, 4567–4579. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Koul, H.K. Androgen receptor (AR) cistrome in prostate differentiation and cancer progression. Am. J. Clin. Exp. Urol. 2017, 5, 18–24. [Google Scholar]
- Shen, M.; Demers, L.K.; Bailey, S.D.; Labbe, D.P. To bind or not to bind: Cistromic reprogramming in prostate cancer. Front. Oncol. 2022, 12, 963007. [Google Scholar] [CrossRef]
- Severson, T.; Qiu, X.; Alshalalfa, M.; Sjostrom, M.; Quigley, D.; Bergman, A.; Long, H.; Feng, F.; Freedman, M.L.; Zwart, W.; et al. Androgen receptor reprogramming demarcates prognostic, context-dependent gene sets in primary and metastatic prostate cancer. Clin. Epigenetics 2022, 14, 60. [Google Scholar] [CrossRef]
- Stelloo, S.; Nevedomskaya, E.; van der Poel, H.G.; de Jong, J.; van Leenders, G.J.; Jenster, G.; Wessels, L.F.; Bergman, A.M.; Zwart, W. Androgen receptor profiling predicts prostate cancer outcome. EMBO Mol. Med. 2015, 7, 1450–1464. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, M.M.; Li, F.; Takeda, D.Y.; Lenci, R.; Chonkar, A.; Chabot, M.; Cejas, P.; Vazquez, F.; Cook, J.; Shivdasani, R.A.; et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet. 2015, 47, 1346–1351. [Google Scholar] [CrossRef]
- Pomerantz, M.M.; Qiu, X.; Zhu, Y.; Takeda, D.Y.; Pan, W.; Baca, S.C.; Gusev, A.; Korthauer, K.D.; Severson, T.M.; Ha, G.; et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat. Genet. 2020, 52, 790–799. [Google Scholar] [CrossRef] [PubMed]
- Sharp, A.; Coleman, I.; Yuan, W.; Sprenger, C.; Dolling, D.; Rodrigues, D.N.; Russo, J.W.; Figueiredo, I.; Bertan, C.; Seed, G.; et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J. Clin. Investig. 2019, 129, 192–208. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, D.; Thomas-Ahner, J.M.; Lu, C.; Zhao, P.; Zhang, Q.; Geraghty, C.; Yan, P.S.; Hankey, W.; Sunkel, B.; et al. Diverse AR-V7 cistromes in castration-resistant prostate cancer are governed by HoxB13. Proc. Natl. Acad. Sci. USA 2018, 115, 6810–6815. [Google Scholar] [CrossRef]
- Basil, P.; Robertson, M.J.; Bingman, W.E., 3rd; Dash, A.K.; Krause, W.C.; Shafi, A.A.; Piyarathna, B.; Coarfa, C.; Weigel, N.L. Cistrome and transcriptome analysis identifies unique androgen receptor (AR) and AR-V7 splice variant chromatin binding and transcriptional activities. Sci. Rep. 2022, 12, 5351. [Google Scholar] [CrossRef]
- Morova, T.; McNeill, D.R.; Lallous, N.; Gonen, M.; Dalal, K.; Wilson, D.M., 3rd; Gursoy, A.; Keskin, O.; Lack, N.A. Androgen receptor-binding sites are highly mutated in prostate cancer. Nat. Commun. 2020, 11, 832. [Google Scholar] [CrossRef]
- Davies, A.; Nouruzi, S.; Ganguli, D.; Namekawa, T.; Thaper, D.; Linder, S.; Karaoglanoglu, F.; Omur, M.E.; Kim, S.; Kobelev, M.; et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat. Cell Biol. 2021, 23, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Labaf, M.; Li, M.; Ting, L.; Karno, B.; Zhang, S.; Gao, S.; Patalano, S.; Macoska, J.A.; Zarringhalam, K.; Han, D.; et al. Increased AR expression in castration-resistant prostate cancer rapidly induces AR signaling reprogramming with the collaboration of EZH2. Front. Oncol. 2022, 12, 1021845. [Google Scholar] [CrossRef]
- He, Y.; Lu, J.; Ye, Z.; Hao, S.; Wang, L.; Kohli, M.; Tindall, D.J.; Li, B.; Zhu, R.; Wang, L.; et al. Androgen receptor splice variants bind to constitutively open chromatin and promote abiraterone-resistant growth of prostate cancer. Nucleic Acids Res. 2018, 46, 1895–1911. [Google Scholar] [CrossRef]
- Spratt, D.E.; Alshalalfa, M.; Fishbane, N.; Weiner, A.B.; Mehra, R.; Mahal, B.A.; Lehrer, J.; Liu, Y.; Zhao, S.G.; Speers, C.; et al. Transcriptomic heterogeneity of androgen receptor activity defines a de novo low AR-active subclass in treatment naive primary prostate cancer. Clin. Cancer Res. 2019, 25, 6721–6730. [Google Scholar] [CrossRef]
- Sharma, N.L.; Massie, C.E.; Ramos-Montoya, A.; Zecchini, V.; Scott, H.E.; Lamb, A.D.; MacArthur, S.; Stark, R.; Warren, A.Y.; Mills, I.G.; et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell 2013, 23, 35–47. [Google Scholar] [CrossRef]
- Alumkal, J.J.; Sun, D.; Lu, E.; Beer, T.M.; Thomas, G.V.; Latour, E.; Aggarwal, R.; Cetnar, J.; Ryan, C.J.; Tabatabaei, S.; et al. Transcriptional profiling identifies an androgen receptor activity-low, stemness program associated with enzalutamide resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 12315–12323. [Google Scholar] [CrossRef]
- Cato, L.; de Tribolet-Hardy, J.; Lee, I.; Rottenberg, J.T.; Coleman, I.; Melchers, D.; Houtman, R.; Xiao, T.; Li, W.; Uo, T.; et al. ARv7 represses tumor-suppressor genes in castration-resistant prostate cancer. Cancer Cell. 2019, 35, 401–413.e6. [Google Scholar] [CrossRef]
- Baca, S.C.; Takeda, D.Y.; Seo, J.H.; Hwang, J.; Ku, S.Y.; Arafeh, R.; Arnoff, T.; Agarwal, S.; Bell, C.; O’Connor, E.; et al. Reprogramming of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer. Nat. Commun. 2021, 12, 1979. [Google Scholar] [CrossRef]
- Storck, W.K.; May, A.M.; Westbrook, T.C.; Duan, Z.; Morrissey, C.; Yates, J.A.; Alumkal, J.J. The role of epigenetic change in therapy-induced neuroendocrine prostate cancer lineage plasticity. Front. Endocrinol. 2022, 13, 926585. [Google Scholar] [CrossRef]
- Turnham, D.J.; Bullock, N.; Dass, M.S.; Staffurth, J.N.; Pearson, H.B. The PTEN conundrum: How to target PTEN-deficient prostate cancer. Cells 2020, 9, 2342. [Google Scholar] [CrossRef]
- Mishra, R.; Patel, H.; Alanazi, S.; Kilroy, M.K.; Garrett, J.T. PI3K inhibitors in cancer: Clinical implications and adverse effects. Int. J. Mol. Sci. 2021, 22, 3464. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, Y.; Zhou, C.; Mei, W.; Zeng, C. PI3K/Akt/mTOR pathway and its role in cancer therapeutics: Are we making headway? Front. Oncol. 2022, 12, 819128. [Google Scholar] [CrossRef]
- Calleja, V.; Alcor, D.; Laguerre, M.; Park, J.; Vojnovic, B.; Hemmings, B.A.; Downward, J.; Parker, P.J.; Larijani, B. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 2007, 5, e95. [Google Scholar] [CrossRef]
- Degan, S.E.; Gelman, I.H. Emerging roles for AKT isoform preference in cancer progression pathways. Mol. Cancer Res. 2021, 19, 1251–1257. [Google Scholar] [CrossRef]
- Yan, Y.; Huang, H. Interplay among PI3K/AKT, PTEN/FOXO and AR signaling in prostate cancer. Adv. Exp. Med. Biol. 2019, 1210, 319–331. [Google Scholar]
- Giguere, V. DNA-PK, nuclear mTOR, and the androgen pathway in prostate cancer. Trends Cancer 2020, 6, 337–347. [Google Scholar] [CrossRef]
- Baffi, T.R.; Newton, A.C. mTOR regulation of AGC kinases: New twist to an old tail. Mol. Pharmacol. 2022, 101, 213–218. [Google Scholar] [CrossRef]
- Burke, J.E.; Triscott, J.; Emerling, B.M.; Hammond, G.R.V. Beyond PI3Ks: Targeting phosphoinositide kinases in disease. Nat. Rev. Drug Discov. 2022, 1–30. [Google Scholar] [CrossRef]
- Qiao, Y.; Choi, J.E.; Tien, J.C.; Simko, S.A.; Rajendiran, T.; Vo, J.N.; Delekta, A.D.; Wang, L.; Xiao, L.; Hodge, N.B.; et al. Autophagy inhibition by targeting PIKfyve potentiates response to immune checkpoint blockade in prostate cancer. Nat. Cancer 2021, 2, 978–993. [Google Scholar] [CrossRef]
- Karlsson, R.; Larsson, P.; Miftakhova, R.; Syed Khaja, A.S.; Sarwar, M.; Semenas, J.; Chen, S.; Hedblom, A.; Wang, T.; Ekstrom-Holka, K.; et al. Establishment of prostate tumor growth and metastasis is supported by bone marrow cells and is mediated by PIP5K1alpha lipid kinase. Cancers 2020, 12, 2719. [Google Scholar] [CrossRef]
- Wang, T.; Sarwar, M.; Whitchurch, J.B.; Collins, H.M.; Green, T.; Semenas, J.; Ali, A.; Roberts, C.J.; Morris, R.D.; Hubert, M.; et al. PIP5K1alpha is required for promoting tumor progression in castration-resistant prostate cancer. Front. Cell Dev. Biol. 2022, 10, 798590. [Google Scholar] [CrossRef]
- Hamila, S.A.; Ooms, L.M.; Rodgers, S.J.; Mitchell, C.A. The INPP4B paradox: Like PTEN, but different. Adv. Biol. Regul. 2021, 82, 100817. [Google Scholar] [CrossRef]
- Xue, C.; Corey, E.; Gujral, T.S. Proteomic and transcriptomic profiling reveals mitochondrial oxidative phosphorylation as therapeutic vulnerability in androgen receptor pathway active prostate tumors. Cancers 2022, 14, 1739. [Google Scholar] [CrossRef]
- Mossmann, D.; Park, S.; Hall, M.N. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 2018, 18, 744–757. [Google Scholar] [CrossRef]
- Stoykova, G.E.; Schlaepfer, I.R. Lipid metabolism and endocrine resistance in prostate cancer, and new opportunities for therapy. Int. J. Mol. Sci. 2019, 20, 2626. [Google Scholar] [CrossRef]
- Sena, L.A.; Denmeade, S.R. Fatty acid synthesis in prostate cancer: Vulnerability or epiphenomenon? Cancer Res. 2021, 81, 4385–4393. [Google Scholar] [CrossRef]
- Chetta, P.; Zadra, G. Metabolic reprogramming as an emerging mechanism of resistance to endocrine therapies in prostate cancer. Cancer Drug Resist. 2021, 4, 143–162. [Google Scholar] [CrossRef]
- Mao, N.; Zhang, Z.; Lee, Y.S.; Choi, D.; Rivera, A.A.; Li, D.; Lee, C.; Haywood, S.; Chen, X.; Chang, Q.; et al. Defining the therapeutic selective dependencies for distinct subtypes of PI3K pathway-altered prostate cancers. Nat. Commun. 2021, 12, 5053. [Google Scholar] [CrossRef]
- Arafeh, R.; Samuels, Y. PIK3CA in cancer: The past 30 years. Semin. Cancer Biol. 2019, 59, 36–49. [Google Scholar] [CrossRef]
- Jia, W.; Luo, S.; Guo, H.; Kong, D. Development of PI3Kalpha inhibitors for tumor therapy. J. Biomol. Struct. Dyn. 2022, 1–18. [Google Scholar] [CrossRef]
- Mak, B.; Lin, H.M.; Kwan, E.M.; Fettke, H.; Tran, B.; Davis, I.D.; Mahon, K.; Stockler, M.R.; Briscoe, K.; Marx, G.; et al. Combined impact of lipidomic and genetic aberrations on clinical outcomes in metastatic castration-resistant prostate cancer. BMC Med. 2022, 20, 112. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
- Pearson, H.B.; Li, J.; Meniel, V.S.; Fennell, C.M.; Waring, P.; Montgomery, K.G.; Rebello, R.J.; Macpherson, A.A.; Koushyar, S.; Furic, L.; et al. Identification of Pik3ca mutation as a genetic driver of prostate cancer that cooperates with Pten loss to accelerate progression and castration-resistant growth. Cancer Discov. 2018, 8, 764–779. [Google Scholar] [CrossRef]
- Zhang, M.; Jang, H.; Nussinov, R. The mechanism of PI3Kalpha activation at the atomic level. Chem. Sci. 2019, 10, 3671–3680. [Google Scholar] [CrossRef] [PubMed]
- Nussinov, R.; Zhang, M.; Tsai, C.J.; Jang, H. Phosphorylation and driver mutations in PI3Kalpha and PTEN autoinhibition. Mol. Cancer Res. 2021, 19, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, G.; Nandakumar, S.; Hirani, R.; Nguyen, B.; Stopsack, K.H.; Kreitzer, C.; Rajanala, S.H.; Ghale, R.; Mazzu, Y.Z.; Pillarsetty, N.V.K.; et al. The impact of PIK3R1 mutations and insulin-PI3K-glycolytic pathway regulation in prostate cancer. Clin. Cancer Res. 2022, 28, 3603–3617. [Google Scholar] [CrossRef]
- Herberts, C.; Murtha, A.J.; Fu, S.; Wang, G.; Schonlau, E.; Xue, H.; Lin, D.; Gleave, A.; Yip, S.; Angeles, A.; et al. Activating AKT1 and PIK3CA mutations in metastatic castration-resistant prostate cancer. Eur. Urol. 2020, 78, 834–844. [Google Scholar] [CrossRef]
- Jiang, Q.; Zheng, N.; Bu, L.; Zhang, X.; Zhang, X.; Wu, Y.; Su, Y.; Wang, L.; Zhang, X.; Ren, S.; et al. SPOP-mediated ubiquitination and degradation of PDK1 suppresses AKT kinase activity and oncogenic functions. Mol. Cancer 2021, 20, 100. [Google Scholar] [CrossRef]
- Blattner, M.; Liu, D.; Robinson, B.D.; Huang, D.; Poliakov, A.; Gao, D.; Nataraj, S.; Deonarine, L.D.; Augello, M.A.; Sailer, V.; et al. SPOP mutation drives prostate tumorigenesis In vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer Cell 2017, 31, 436–451. [Google Scholar] [CrossRef]
- Wu, C.; Huang, J. Phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin pathway is essential for neuroendocrine differentiation of prostate cancer. J. Biol. Chem. 2007, 282, 3571–3583. [Google Scholar] [CrossRef]
- Ciarlo, M.; Benelli, R.; Barbieri, O.; Minghelli, S.; Barboro, P.; Balbi, C.; Ferrari, N. Regulation of neuroendocrine differentiation by AKT/hnRNPK/AR/beta-catenin signaling in prostate cancer cells. Int. J. Cancer 2012, 131, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Kwon, O.J.; Zhang, L.; Jia, D.; Zhou, Z.; Li, Z.; Haffner, M.; Lee, J.K.; True, L.; Morrissey, C.; Xin, L. De novo induction of lineage plasticity from human prostate luminal epithelial cells by activated AKT1 and c-Myc. Oncogene 2020, 39, 7142–7151. [Google Scholar] [CrossRef] [PubMed]
- Wiesehofer, M.; Czyrnik, E.D.; Spahn, M.; Ting, S.; Reis, H.; Dankert, J.T.; Wennemuth, G. Increased expression of AKT3 in neuroendocrine differentiated prostate cancer cells alters the response towards anti-androgen treatment. Cancers 2021, 13, 578. [Google Scholar] [CrossRef] [PubMed]
- Alwhaibi, A.; Kolhe, R.; Gao, F.; Cobran, E.K.; Somanath, P.R. Genome atlas analysis based profiling of Akt pathway genes in the early and advanced human prostate cancer. Oncoscience 2019, 6, 317–336. [Google Scholar] [CrossRef]
- Dufour, C.R.; Scholtes, C.; Yan, M.; Chen, Y.; Han, L.; Li, T.; Xia, H.; Deng, Q.; Vernier, M.; Giguere, V. The mTOR chromatin-bound interactome in prostate cancer. Cell Rep. 2022, 38, 110534. [Google Scholar] [CrossRef]
- Gagne, L.M.; Morin, N.; Lavoie, N.; Bisson, N.; Lambert, J.P.; Mallette, F.A.; Huot, M.E. Tyrosine phosphorylation of DEPTOR functions as a molecular switch to activate mTOR signaling. J. Biol. Chem. 2021, 297, 101291. [Google Scholar] [CrossRef]
- Peterson, T.R.; Laplante, M.; Thoreen, C.C.; Sancak, Y.; Kang, S.A.; Kuehl, W.M.; Gray, N.S.; Sabatini, D.M. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009, 137, 873–886. [Google Scholar] [CrossRef]
- Vega, M.; Chen, Y.; Shi, Y.; Gera, J.; Lichtenstein, A. Turnover of the mTOR inhibitor, DEPTOR, and downstream AKT phosphorylation in multiple myeloma cells, is dependent on ERK1-mediated phosphorylation. J. Biol. Chem. 2022, 298, 101750. [Google Scholar] [CrossRef]
- Chen, X.; Xiong, X.; Cui, D.; Yang, F.; Wei, D.; Li, H.; Shu, J.; Bi, Y.; Dai, X.; Gong, L.; et al. DEPTOR is an in vivo tumor suppressor that inhibits prostate tumorigenesis via the inactivation of mTORC1/2 signals. Oncogene 2020, 39, 1557–1571. [Google Scholar] [CrossRef] [PubMed]
- Guertin, D.A.; Stevens, D.M.; Saitoh, M.; Kinkel, S.; Crosby, K.; Sheen, J.H.; Mullholland, D.J.; Magnuson, M.A.; Wu, H.; Sabatini, D.M. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell 2009, 15, 148–159. [Google Scholar] [CrossRef]
- Oneyama, C.; Kito, Y.; Asai, R.; Ikeda, J.; Yoshida, T.; Okuzaki, D.; Kokuda, R.; Kakumoto, K.; Takayama, K.; Inoue, S.; et al. MiR-424/503-mediated Rictor upregulation promotes tumor progression. PLoS ONE 2013, 8, e80300. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.L.; Chou, C.C.; Chuang, H.C.; Hsu, E.C.; Chiu, P.C.; Kulp, S.K.; Byrd, J.C.; Chen, C.S. Functional role of mTORC2 versus integrin-linked kinase in mediating Ser473-Akt phosphorylation in PTEN-negative prostate and breast cancer cell lines. PLoS ONE 2013, 8, e67149. [Google Scholar] [CrossRef] [PubMed]
- Gheghiani, L.; Shang, S.; Fu, Z. Targeting the PLK1-FOXO1 pathway as a novel therapeutic approach for treating advanced prostate cancer. Sci. Rep. 2020, 10, 12327. [Google Scholar] [CrossRef] [PubMed]
- Bohrer, L.R.; Liu, P.; Zhong, J.; Pan, Y.; Angstman, J.; Brand, L.J.; Dehm, S.M.; Huang, H. FOXO1 binds to the TAU5 motif and inhibits constitutively active androgen receptor splice variants. Prostate 2013, 73, 1017–1027. [Google Scholar] [CrossRef]
- Yang, Y.; Blee, A.M.; Wang, D.; An, J.; Pan, Y.; Yan, Y.; Ma, T.; He, Y.; Dugdale, J.; Hou, X.; et al. Loss of FOXO1 cooperates with TMPRSS2-ERG overexpression to promote prostate tumorigenesis and cell invasion. Cancer Res. 2017, 77, 6524–6537. [Google Scholar] [CrossRef]
- Su, B.; Zhang, L.; Zhuang, W.; Zhang, W.; Chen, X. Knockout of Akt1/2 suppresses the metastasis of human prostate cancer cells CWR22rv1 in vitro and in vivo. J. Cell Mol. Med. 2021, 25, 1546–1553. [Google Scholar] [CrossRef]
- Habrowska-Gorczynska, D.E.; Koziel, M.J.; Kowalska, K.; Piastowska-Ciesielska, A.W. FOXO3a and its regulators in prostate cancer. Int. J. Mol. Sci. 2021, 22, 12530. [Google Scholar] [CrossRef]
- Shukla, S.; Bhaskaran, N.; Maclennan, G.T.; Gupta, S. Deregulation of FoxO3a accelerates prostate cancer progression in TRAMP mice. Prostate 2013, 73, 1507–1517. [Google Scholar] [CrossRef]
- Gurbuz, V.; Sozen, S.; Bilen, C.Y.; Konac, E. miR-148a, miR-152 and miR-200b promote prostate cancer metastasis by targeting DNMT1 and PTEN expression. Oncol. Lett. 2021, 22, 805. [Google Scholar] [CrossRef]
- Guo, Y.; He, J.; Zhang, H.; Chen, R.; Li, L.; Liu, X.; Huang, C.; Qiang, Z.; Zhou, Z.; Wang, Y.; et al. Linear ubiquitination of PTEN impairs its function to promote prostate cancer progression. Oncogene 2022, 41, 4877–4892. [Google Scholar] [CrossRef]
- Hamid, A.A.; Gray, K.P.; Shaw, G.; MacConaill, L.E.; Evan, C.; Bernard, B.; Loda, M.; Corcoran, N.M.; Van Allen, E.M.; Choudhury, A.D.; et al. Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. Eur. Urol. 2019, 76, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Limberger, T.; Schlederer, M.; Trachtova, K.; Garces de Los Fayos Alonso, I.; Yang, J.; Hogler, S.; Sternberg, C.; Bystry, V.; Oppelt, J.; Tichy, B.; et al. KMT2C methyltransferase domain regulated INK4A expression suppresses prostate cancer metastasis. Mol. Cancer 2022, 21, 89. [Google Scholar] [CrossRef]
- Sjostrom, M.; Zhao, S.G.; Levy, S.; Zhang, M.; Ning, Y.; Shrestha, R.; Lundberg, A.; Herberts, C.; Foye, A.; Aggarwal, R.; et al. The 5-hydroxymethylcytosine landscape of prostate cancer. Cancer Res. 2022, 82, 3888–3902. [Google Scholar] [CrossRef]
- De Velasco, M.A.; Uemura, H. Preclinical remodeling of human prostate cancer through the PTEN/AKT pathway. Adv. Urol. 2012, 2012, 419348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kim, S.; Li, L.; Kemp, C.J.; Jiang, C.; Lu, J. Proteomic and transcriptomic profiling of Pten gene-knockout mouse model of prostate cancer. Prostate 2020, 80, 588–605. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, B.; Rowan, B.G.; Jazwinski, S.M.; Abdel-Mageed, A.B.; Steele, C.; Wang, A.R.; Sartor, O.; Niu, T.; Zhang, Q. A novel controlled PTEN-knockout mouse model for prostate cancer study. Front. Mol. Biosci. 2021, 8, 696537. [Google Scholar] [CrossRef]
- van Duijn, P.W.; Marques, R.B.; Ziel-van der Made, A.C.J.; van Zoggel, H.; Aghai, A.; Berrevoets, C.; Debets, R.; Jenster, G.; Trapman, J.; van Weerden, W.M. Tumor heterogeneity, aggressiveness, and immune cell composition in a novel syngeneic PSA-targeted Pten knockout mouse prostate cancer (MuCaP) model. Prostate 2018, 78, 1013–1023. [Google Scholar] [CrossRef]
- Jia, S.; Liu, Z.; Zhang, S.; Liu, P.; Zhang, L.; Lee, S.H.; Zhang, J.; Signoretti, S.; Loda, M.; Roberts, T.M.; et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature 2008, 454, 776–779. [Google Scholar] [CrossRef]
- Ni, J.; Liu, Q.; Xie, S.; Carlson, C.; Von, T.; Vogel, K.; Riddle, S.; Benes, C.; Eck, M.; Roberts, T.; et al. Functional characterization of an isoform-selective inhibitor of PI3K-p110beta as a potential anticancer agent. Cancer Discov. 2012, 2, 425–433. [Google Scholar] [CrossRef]
- Gao, X.; Wang, Y.; Ribeiro, C.F.; Manokaran, C.; Chang, H.; Von, T.; Rodrigues, S.; Cizmecioglu, O.; Jia, S.; Korpal, M.; et al. Blocking PI3K p110beta attenuates development of PTEN-deficient castration-resistant prostate cancer. Mol. Cancer Res. 2022, 20, 673–685. [Google Scholar] [CrossRef]
- Marques, R.B.; Aghai, A.; de Ridder, C.M.A.; Stuurman, D.; Hoeben, S.; Boer, A.; Ellston, R.P.; Barry, S.T.; Davies, B.R.; Trapman, J.; et al. High efficacy of combination therapy using PI3K/AKT inhibitors with androgen deprivation in prostate cancer preclinical models. Eur. Urol. 2015, 67, 1177–1185. [Google Scholar] [CrossRef]
- Crumbaker, M.; Khoja, L.; Joshua, A.M. AR signaling and the PI3K pathway in prostate cancer. Cancers 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A.D. PTEN-PI3K pathway alterations in advanced prostate cancer and clinical implications. Prostate 2022, 82 (Suppl. S1), S60–S72. [Google Scholar] [CrossRef] [PubMed]
- Jacob, A.; Raj, R.; Allison, D.B.; Myint, Z.W. Androgen receptor signaling in prostate cancer and therapeutic strategies. Cancers 2021, 13, 5417. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.T.; Huitema, A.D.R.; Chau, C.H.; Figg, W.D. Resistance to second-generation androgen receptor antagonists in prostate cancer. Nat. Rev. Urol. 2021, 18, 209–226. [Google Scholar] [CrossRef]
- Lee, S.H.; Johnson, D.; Luong, R.; Sun, Z. Crosstalking between androgen and PI3K/AKT signaling pathways in prostate cancer cells. J. Biol. Chem. 2015, 290, 2759–2768. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, M.C.; Shao, L.J.; Frolov, A.; Li, R.; Peterson, L.E.; Ayala, G.; Ittmann, M.M.; Weigel, N.L.; Agoulnik, I.U. Decreased expression and androgen regulation of the tumor suppressor gene INPP4B in prostate cancer. Cancer Res. 2011, 71, 572–582. [Google Scholar] [CrossRef]
- Carver, B.S.; Chapinski, C.; Wongvipat, J.; Hieronymus, H.; Chen, Y.; Chandarlapaty, S.; Arora, V.K.; Le, C.; Koutcher, J.; Scher, H.; et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011, 19, 575–586. [Google Scholar] [CrossRef]
- Qi, W.; Morales, C.; Cooke, L.S.; Johnson, B.; Somer, B.; Mahadevan, D. Reciprocal feedback inhibition of the androgen receptor and PI3K as a novel therapy for castrate-sensitive and -resistant prostate cancer. Oncotarget 2015, 6, 41976–41987. [Google Scholar] [CrossRef]
- Mulholland, D.J.; Tran, L.M.; Li, Y.; Cai, H.; Morim, A.; Wang, S.; Plaisier, S.; Garraway, I.P.; Huang, J.; Graeber, T.G.; et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 2011, 19, 792–804. [Google Scholar] [CrossRef]
- Shang, Z.; Yu, J.; Sun, L.; Tian, J.; Zhu, S.; Zhang, B.; Dong, Q.; Jiang, N.; Flores-Morales, A.; Chang, C.; et al. LncRNA PCAT1 activates AKT and NF-kappaB signaling in castration-resistant prostate cancer by regulating the PHLPP/FKBP51/IKKalpha complex. Nucleic Acids Res. 2019, 47, 4211–4225. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Habara, M.; Kawaguchi, M.; Matsumoto, H.; Hanaki, S.; Masaki, T.; Sato, Y.; Matsuyama, H.; Kunieda, K.; Nakagawa, H.; et al. FKBP51 and FKBP52 regulate androgen receptor dimerization and proliferation in prostate cancer cells. Mol. Oncol. 2022, 16, 940–956. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Xu, J.; Shen, Y.; Cahuzac, K.M.; Park, K.S.; Dale, B.; Liu, J.; Parsons, R.E.; Jin, J. Discovery of potent, selective, and in vivo efficacious AKT kinase protein degraders via structure-activity relationship studies. J. Med. Chem. 2022, 65, 3644–3666. [Google Scholar] [CrossRef] [PubMed]
- Audet-Walsh, E.; Vernier, M.; Yee, T.; Laflamme, C.; Li, S.; Chen, Y.; Giguere, V. SREBF1 activity is regulated by an AR/mTOR nuclear axis in prostate cancer. Mol. Cancer Res. 2018, 16, 1396–1405. [Google Scholar] [CrossRef]
- Audet-Walsh, E.; Dufour, C.R.; Yee, T.; Zouanat, F.Z.; Yan, M.; Kalloghlian, G.; Vernier, M.; Caron, M.; Bourque, G.; Scarlata, E.; et al. Nuclear mTOR acts as a transcriptional integrator of the androgen signaling pathway in prostate cancer. Genes Dev. 2017, 31, 1228–1242. [Google Scholar] [CrossRef]
- Martin, P.L.; Yin, J.J.; Seng, V.; Casey, O.; Corey, E.; Morrissey, C.; Simpson, R.M.; Kelly, K. Androgen deprivation leads to increased carbohydrate metabolism and hexokinase 2-mediated survival in Pten/Tp53-deficient prostate cancer. Oncogene 2017, 36, 525–533. [Google Scholar] [CrossRef]
- Butler, L.M.; Mah, C.Y.; Machiels, J.; Vincent, A.D.; Irani, S.; Mutuku, S.M.; Spotbeen, X.; Bagadi, M.; Waltregny, D.; Moldovan, M.; et al. Lipidomic profiling of clinical prostate cancer reveals targetable alterations in membrane lipid composition. Cancer Res. 2021, 81, 4981–4993. [Google Scholar] [CrossRef]
- Roudsari, N.M.; Lashgari, N.A.; Momtaz, S.; Abaft, S.; Jamali, F.; Safaiepour, P.; Narimisa, K.; Jackson, G.; Bishayee, A.; Rezaei, N.; et al. Inhibitors of the PI3K/Akt/mTOR pathway in prostate cancer chemoprevention and intervention. Pharmaceutics 2021, 13, 1195. [Google Scholar] [CrossRef]
- Park, H.; Kim, Y.; Sul, J.W.; Jeong, I.G.; Yi, H.J.; Ahn, J.B.; Kang, J.S.; Yun, J.; Hwang, J.J.; Kim, C.S. Synergistic anticancer efficacy of MEK inhibition and dual PI3K/mTOR inhibition in castration-resistant prostate cancer. Prostate 2015, 75, 1747–1759. [Google Scholar] [CrossRef]
- Toren, P.; Kim, S.; Johnson, F.; Zoubeidi, A. Combined AKT and MEK pathway blockade in pre-clinical models of enzalutamide-resistant prostate cancer. PLoS ONE 2016, 11, e0152861. [Google Scholar] [CrossRef] [PubMed]
- Adelaiye-Ogala, R.; Gryder, B.E.; Nguyen, Y.T.M.; Alilin, A.N.; Grayson, A.R.; Bajwa, W.; Jansson, K.H.; Beshiri, M.L.; Agarwal, S.; Rodriguez-Nieves, J.A.; et al. Targeting the PI3K/AKT pathway overcomes enzalutamide resistance by inhibiting induction of the glucocorticoid receptor. Mol. Cancer Ther. 2020, 19, 1436–1447. [Google Scholar] [CrossRef] [PubMed]
- Politz, O.; Siegel, F.; Barfacker, L.; Bomer, U.; Hagebarth, A.; Scott, W.J.; Michels, M.; Ince, S.; Neuhaus, R.; Meyer, K.; et al. BAY 1125976, a selective allosteric AKT1/2 inhibitor, exhibits high efficacy on AKT signaling-dependent tumor growth in mouse models. Int. J. Cancer 2017, 140, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Lamoureux, F.; Crafter, C.; Davies, B.R.; Beraldi, E.; Fazli, L.; Kim, S.; Thaper, D.; Gleave, M.E.; Zoubeidi, A. Synergistic targeting of PI3K/AKT pathway and androgen receptor axis significantly delays castration-resistant prostate cancer progression in vivo. Mol. Cancer Ther. 2013, 12, 2342–2355. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, T.; Baumgart, S.J.; Nevedomskaya, E.; Reichert, K.; Steuber, H.; Lejeune, P.; Mumberg, D.; Haendler, B. Darolutamide is a potent androgen receptor antagonist with strong efficacy in prostate cancer models. Int. J. Cancer. 2019, 145, 1382–1394. [Google Scholar] [CrossRef]
- Wang, W.; Shen, T.; Dong, B.; Creighton, C.J.; Meng, Y.; Zhou, W.; Shi, Q.; Zhou, H.; Zhang, Y.; Moore, D.D.; et al. MAPK4 overexpression promotes tumor progression via noncanonical activation of AKT/mTOR signaling. J. Clin. Investig. 2019, 129, 1015–1029. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Wang, W.; Zhou, W.; Coleman, I.; Cai, Q.; Dong, B.; Ittmann, M.M.; Creighton, C.J.; Bian, Y.; Meng, Y.; et al. MAPK4 promotes prostate cancer by concerted activation of androgen receptor and AKT. J. Clin. Investig. 2021, 131, e135465. [Google Scholar] [CrossRef]
- Yan, Y.; An, J.; Yang, Y.; Wu, D.; Bai, Y.; Cao, W.; Ma, L.; Chen, J.; Yu, Z.; He, Y.; et al. Dual inhibition of AKT-mTOR and AR signaling by targeting HDAC3 in PTEN- or SPOP-mutated prostate cancer. EMBO Mol. Med. 2018, 10, e8478. [Google Scholar] [CrossRef]
- Chen, H.; Li, H.; Chen, Q. INPP4B overexpression suppresses migration, invasion and angiogenesis of human prostate cancer cells. Clin. Exp. Pharmacol. Physiol. 2017, 44, 700–708. [Google Scholar] [CrossRef]
- Zhang, M.; Suarez, E.; Vasquez, J.L.; Nathanson, L.; Peterson, L.E.; Rajapakshe, K.; Basil, P.; Weigel, N.L.; Coarfa, C.; Agoulnik, I.U. Inositol polyphosphate 4-phosphatase type II regulation of androgen receptor activity. Oncogene 2019, 38, 1121–1135. [Google Scholar] [CrossRef]
- Gupta, S.; Halabi, S.; Kemeny, G.; Anand, M.; Giannakakou, P.; Nanus, D.M.; George, D.J.; Gregory, S.G.; Armstrong, A.J. Circulating tumor cell genomic evolution and hormone therapy outcomes in men with metastatic castration-resistant prostate cancer. Mol. Cancer Res. 2021, 19, 1040–1050. [Google Scholar] [CrossRef]
- Fu, X.; Zhang, Z.; Liu, M.; Li, J.; Fu, L.; Huang, C.; Dong, J.T. AR imposes different effects on ZFHX3 transcription depending on androgen status in prostate cancer cells. J. Cell Mol. Med. 2022, 26, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xing, C.; Fu, X.; Li, J.; Zhang, B.; Frierson, H.F., Jr.; Dong, J.T. Additive effect of Zfhx3/Atbf1 and Pten deletion on mouse prostatic tumorigenesis. J. Genet. Genomics 2015, 42, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Gharibpoor, F.; Kamali Zonouzi, S.; Razi, S.; Rezaei, N. AMPK’s double-faced role in advanced stages of prostate cancer. Clin. Transl. Oncol. 2022, 24, 2064–2073. [Google Scholar] [CrossRef] [PubMed]
- Lemos, C.; Schulze, V.K.; Baumgart, S.J.; Nevedomskaya, E.; Heinrich, T.; Lefranc, J.; Bader, B.; Christ, C.D.; Briem, H.; Kuhnke, L.P.; et al. The potent AMPK inhibitor BAY-3827 shows strong efficacy in androgen-dependent prostate cancer models. Cell Oncol. 2021, 44, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Brave, M.; Weinstock, C.; Brewer, J.R.; Chi, D.C.; Suzman, D.L.; Cheng, J.; Zhang, L.; Sridhara, R.; Ibrahim, A.; Kluetz, P.G.; et al. An FDA review of drug development in nonmetastatic castration-resistant prostate cancer. Clin. Cancer Res. 2020, 26, 4717–4722. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.M.; Brave, M.; Maher, V.E.; Zhang, L.; Tang, S.; Sridhara, R.; Kim, G.; Ibrahim, A.; Pazdur, R.U.S. Food and Drug Administration approval summary: Enzalutamide for the treatment of patients with chemotherapy-naive metastatic castration-resistant prostate cancer. Oncologist 2015, 20, 960–966. [Google Scholar] [CrossRef]
- Gul, A.; Garcia, J.A.; Barata, P.C. Treatment of non-metastatic castration-resistant prostate cancer: Focus on apalutamide. Cancer Manag. Res. 2019, 11, 7253–7262. [Google Scholar] [CrossRef]
- Fizazi, K.; Shore, N.; Tammela, T.L.; Ulys, A.; Vjaters, E.; Polyakov, S.; Jievaltas, M.; Luz, M.; Alekseev, B.; Kuss, I.; et al. Darolutamide in nonmetastatic, castration-resistant prostate cancer. N. Engl. J. Med. 2019, 380, 1235–1246. [Google Scholar] [CrossRef]
- Fizazi, K.; Shore, N.; Tammela, T.L.; Ulys, A.; Vjaters, E.; Polyakov, S.; Jievaltas, M.; Luz, M.; Alekseev, B.; Kuss, I.; et al. Nonmetastatic, castration-resistant prostate cancer and survival with darolutamide. N. Engl. J. Med. 2020, 383, 1040–1049. [Google Scholar] [CrossRef]
- Smith, M.R.; Hussain, M.; Saad, F.; Fizazi, K.; Sternberg, C.N.; Crawford, E.D.; Kopyltsov, E.; Park, C.H.; Alekseev, B.; Montesa-Pino, A.; et al. Darolutamide and survival in metastatic, hormone-sensitive prostate cancer. N. Engl. J. Med. 2022, 386, 1132–1142. [Google Scholar] [CrossRef]
- Keam, S.J. Rezvilutamide: First approval. Drugs 2023. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Xu, W.; Zhang, W.; Sun, Y.; Yan, H.; Gao, X.; Wang, F.; Zhou, Q.; Hou, J.; Ren, S.; et al. Preclinical profile and phase I clinical trial of a novel androgen receptor antagonist GT0918 in castration-resistant prostate cancer. Eur. J. Cancer 2020, 134, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Rathkopf, D.E.; Saleh, M.; Tsai, F.Y.; Rosen, L.S.; Adams, B.J.; Liu, L.; Theuer, C.; Freddo, J.L.; Agarwal, N. An open-label phase 1/2a study to evaluate the safety, pharmacokinetics, pharmacodynamics, and preliminary efficacy of TRC253, an androgen receptor antagonist, in patients with metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2018, 36, TPS403. [Google Scholar] [CrossRef]
- Bekes, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef]
- Petrylak, D.P.; Gao, X.; Vogelzang, N.J.; Garfield, M.H.; Taylor, I.; Moore, M.D.; Peck, R.A.; Burris, H.A. First-in-human phase I study of ARV-110, an androgen receptor PROTAC degrader in patients with metastatic castrate-resistant prostate cancer following enzalutamide and/or abiraterone. J. Clin. Oncol. 2020, 38, 3500. [Google Scholar] [CrossRef]
- Kregel, S.; Wang, C.; Han, X.; Xiao, L.; Fernandez-Salas, E.; Bawa, P.; McCollum, B.L.; Wilder-Romans, K.; Apel, I.J.; Cao, X.; et al. Androgen receptor degraders overcome common resistance mechanisms developed during prostate cancer treatment. Neoplasia 2020, 22, 111–119. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Chandhasin, C.; Osbourne, E.; Luo, J.; Sadar, M.D.; Perabo, F. Targeting the N-terminal domain of the androgen receptor: A new approach for the treatment of advanced prostate cancer. Oncologist 2016, 21, 1427–1435. [Google Scholar] [CrossRef]
- Maurice-Dror, C.; Le Moigne, R.; Vaishampayan, U.; Montgomery, R.B.; Gordon, M.S.; Hong, N.H.; DiMascio, L.; Perabo, F.; Chi, K.N. A phase 1 study to assess the safety, pharmacokinetics, and anti-tumor activity of the androgen receptor n-terminal domain inhibitor epi-506 in patients with metastatic castration-resistant prostate cancer. Investig. New Drugs 2022, 40, 322–329. [Google Scholar] [CrossRef]
- Sadar, M.D. Discovery of drugs that directly target the intrinsically disordered region of the androgen receptor. Expert. Opin. Drug Discov. 2020, 15, 551–560. [Google Scholar] [CrossRef]
- Hong, N.H.; Biannic, B.; Virsik, P.; Zhou, H.J.; Le Moigne, R. Advances in the development of a targeted N-terminal domain androgen receptor (AR) degrader (ANITAC) for the treatment of prostate cancer. Eur. J. Cancer 2022, 174 (Suppl. S1), S37. [Google Scholar] [CrossRef]
- Narayanan, R. Androgen receptor (AR) N-terminus-domain-binding small molecule degraders for the treatment of AR splice variant-positive castration-resistant prostate cancer. Mol. Cancer Ther. 2021, 20, LBA016. [Google Scholar] [CrossRef]
- Bird, S.T.; Tian, F.; Flowers, N.; Przepiorka, D.; Wang, R.; Jung, T.H.; Kessler, Z.; Woods, C.; Kim, B.; Miller, B.W.; et al. Idelalisib for treatment of relapsed follicular lymphoma and chronic lymphocytic leukemia: A comparison of treatment outcomes in clinical trial participants vs. medicare beneficiaries. JAMA Oncol. 2020, 6, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.W.; Przepiorka, D.; de Claro, R.A.; Lee, K.; Nie, L.; Simpson, N.; Gudi, R.; Saber, H.; Shord, S.; Bullock, J.; et al. FDA approval: Idelalisib monotherapy for the treatment of patients with follicular lymphoma and small lymphocytic lymphoma. Clin. Cancer Res. 2015, 21, 1525–1529. [Google Scholar] [CrossRef]
- Markham, A. Copanlisib: First global approval. Drugs 2017, 77, 2057–2062. [Google Scholar] [CrossRef]
- de Souza, A.; Aggarwal, R.; Safran, H.; Golijanin, D.; Wood, R.; Olszewski, A.; El-Deiry, W.S.; Mega, A.; Carneiro, B.A. BrUOG360: A phase Ib/II study of copanlisib combined with rucaparib in patients with metastatic castration-resistant prostate cancer (mCRPC). Cancer Res. 2021, 81, 128. [Google Scholar] [CrossRef]
- Damodaran, S.; Zhao, F.; Deming, D.A.; Mitchell, E.P.; Wright, J.J.; Gray, R.J.; Wang, V.; McShane, L.M.; Rubinstein, L.V.; Patton, D.R.; et al. Phase II study of copanlisib in patients with tumors with PIK3CA mutations: Results from the NCI-MATCH ECOG-ACRIN trial (EAY131) subprotocol Z1F. J. Clin. Oncol. 2022, 40, 1552–1561. [Google Scholar] [CrossRef] [PubMed]
- Narayan, P.; Prowell, T.M.; Gao, J.J.; Fernandes, L.L.; Li, E.; Jiang, X.; Qiu, J.; Fan, J.; Song, P.; Yu, J.; et al. FDA approval summary: Alpelisib plus fulvestrant for patients with HR-positive, HER2-negative, PIK3CA-mutated, advanced or metastatic breast cancer. Clin. Cancer Res. 2021, 27, 1842–1849. [Google Scholar] [CrossRef] [PubMed]
- Hus, I.; Pula, B.; Robak, T. PI3K inhibitors for the treatment of chronic lymphocytic leukemia: Current status and future perspectives. Cancers 2022, 14, 1571. [Google Scholar] [CrossRef]
- Deng, C.; Lipstein, M.R.; Scotto, L.; Jirau Serrano, X.O.; Mangone, M.A.; Li, S.; Vendome, J.; Hao, Y.; Xu, X.; Deng, S.X.; et al. Silencing c-Myc translation as a therapeutic strategy through targeting PI3Kdelta and CK1epsilon in hematological malignancies. Blood 2017, 129, 88–99. [Google Scholar] [CrossRef]
- Aschenbrenner, D.S. FDA evaluating possible serious risks from umbralisib. Am. J. Nurs. 2022, 122, 23. [Google Scholar] [CrossRef]
- Vanhaesebroeck, B.; Perry, M.W.D.; Brown, J.R.; Andre, F.; Okkenhaug, K. PI3K inhibitors are finally coming of age. Nat. Rev. Drug Discov. 2021, 20, 741–769. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Zhang, J.; Liu, H.; Wu, L.; Jiang, H.; Wu, Q.; Liu, T.; Lou, M.; Wu, H. The dual PI3K/mTOR inhibitor dactolisib elicits anti-tumor activity in vitro and in vivo. Oncotarget 2018, 9, 706–717. [Google Scholar] [CrossRef]
- Wise-Draper, T.M.; Moorthy, G.; Salkeni, M.A.; Karim, N.A.; Thomas, H.E.; Mercer, C.A.; Beg, M.S.; O’Gara, S.; Olowokure, O.; Fathallah, H.; et al. A phase Ib study of the dual PI3K/mTOR inhibitor dactolisib (BEZ235) combined with everolimus in patients with advanced solid malignancies. Target. Oncol. 2017, 12, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Carlo, M.I.; Molina, A.M.; Lakhman, Y.; Patil, S.; Woo, K.; DeLuca, J.; Lee, C.H.; Hsieh, J.J.; Feldman, D.R.; Motzer, R.J.; et al. A phase Ib study of BEZ235, a dual inhibitor of phosphatidylinositol 3-kinase (PI3K) and mammalian target of rapamycin (mTOR), in patients with advanced renal cell carcinoma. Oncologist 2016, 21, 787–788. [Google Scholar] [CrossRef]
- Shor, R.E.; Dai, J.; Lee, S.Y.; Pisarsky, L.; Matei, I.; Lucotti, S.; Lyden, D.; Bissell, M.J.; Ghajar, C.M. The PI3K/mTOR inhibitor gedatolisib eliminates dormant breast cancer cells in organotypic culture, but fails to prevent metastasis in preclinical settings. Mol. Oncol. 2022, 16, 130–147. [Google Scholar] [CrossRef] [PubMed]
- Layman, R.; Wesolowski, R.; Han, Y.; Specht, J.M.; Stringer-Reasor, E.M.; Dees, E.C.; Kabos, P.; Mayer, I.A.; Vaishampayan, U.; Lu, J.; et al. Phase 1b expansion study of gedatolisib in combination with palbociclib and endocrine therapy in women with ER-positive metastatic breast cancer. Cancer Res. 2022, 82, PD13-02. [Google Scholar] [CrossRef]
- Curigliano, G.; Shapiro, G.I.; Kristeleit, R.S.; Abdul Razak, A.R.; Leong, S.; Alsina, M.; Giordano, A.; Gelmon, K.A.; Stringer-Reasor, E.; Vaishampayan, U.N.; et al. A Phase 1B open-label study of gedatolisib (PF-05212384) in combination with other anti-tumour agents for patients with advanced solid tumours and triple-negative breast cancer. Br. J. Cancer 2023, 128, 30–41. [Google Scholar] [CrossRef]
- Heffron, T.P.; Ndubaku, C.O.; Salphati, L.; Alicke, B.; Cheong, J.; Drobnick, J.; Edgar, K.; Gould, S.E.; Lee, L.B.; Lesnick, J.D.; et al. Discovery of clinical development candidate GDC-0084, a brain penetrant inhibitor of PI3K and mTOR. ACS Med. Chem. Lett. 2016, 7, 351–356. [Google Scholar] [CrossRef]
- Wen, P.Y.; Cloughesy, T.F.; Olivero, A.G.; Morrissey, K.M.; Wilson, T.R.; Lu, X.; Mueller, L.U.; Coimbra, A.F.; Ellingson, B.M.; Gerstner, E.; et al. First-in-human phase I study to evaluate the brain-penetrant PI3K/mTOR inhibitor GDC-0084 in patients with progressive or recurrent high-grade glioma. Clin. Cancer Res. 2020, 26, 1820–1828. [Google Scholar] [CrossRef]
- Szklener, K.; Mazurek, M.; Wieteska, M.; Waclawska, M.; Bilski, M.; Mandziuk, S. New directions in the therapy of glioblastoma. Cancers 2022, 14, 5377. [Google Scholar] [CrossRef]
- Piha-Paul, S.A.; Taylor, M.H.; Spitz, D.; Schwartzberg, L.; Beck, J.T.; Bauer, T.M.; Meric-Bernstam, F.; Purkayastha, D.; Karpiak, L.; Szpakowski, S.; et al. Efficacy and safety of buparlisib, a PI3K inhibitor, in patients with malignancies harboring a PI3K pathway activation: A phase 2, open-label, single-arm study. Oncotarget 2019, 10, 6526–6535. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Yang, J.; Gu, Y.; Yi, J. Research update on the anticancer effects of buparlisib. Oncol. Lett. 2021, 21, 266. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Lu, H.; Zhou, X.; Wang, Y. The efficacy and safety of neoadjuvant buparlisib for breast cancer: A meta-analysis of randomized controlled studies. Medicine 2019, 98, e17614. [Google Scholar] [CrossRef]
- Armstrong, A.J.; Halabi, S.; Healy, P.; Alumkal, J.J.; Winters, C.; Kephart, J.; Bitting, R.L.; Hobbs, C.; Soleau, C.F.; Beer, T.M.; et al. Phase II trial of the PI3 kinase inhibitor buparlisib (BKM-120) with or without enzalutamide in men with metastatic castration resistant prostate cancer. Eur. J. Cancer 2017, 81, 228–236. [Google Scholar] [CrossRef]
- Bendell, J.C.; Patel, M.R.; Moore, K.N.; Chua, C.C.; Arkenau, H.T.; Dukart, G.; Harrow, K.; Liang, C. Phase I, first-in-human, dose-escalation study to evaluate the safety, tolerability, and pharmacokinetics of vorolanib in patients with advanced solid tumors. Oncologist 2019, 24, 455-e121. [Google Scholar] [CrossRef]
- Sweeney, C.J.; Percent, I.J.; Babu, S.; Cultrera, J.L.; Mehlhaff, B.A.; Goodman, O.B.; Morris, D.S.; Schnadig, I.D.; Albany, C.; Shore, N.D.; et al. Phase Ib/II study of enzalutamide with samotolisib (LY3023414) or placebo in patients with metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2022, 28, 2237–2247. [Google Scholar] [CrossRef] [PubMed]
- Hoegenauer, K.; Soldermann, N.; Zecri, F.; Strang, R.S.; Graveleau, N.; Wolf, R.M.; Cooke, N.G.; Smith, A.B.; Hollingworth, G.J.; Blanz, J.; et al. Discovery of CDZ173 (Leniolisib), representing a structurally novel class of PI3K delta-selective inhibitors. ACS Med. Chem. Lett. 2017, 8, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.K.; Webster, S.; Sediva, A.; Plebani, A.; Schuetz, C.; Shcherbina, A.; Conlon, N.; Coulter, T.I.; Dalm, V.A.; Trizzino, A.; et al. Randomized, placebo-controlled, phase 3 trial of PI3Kdelta inhibitor leniolisib for activated PI3Kdelta syndrome. Blood, 2022; in press. [Google Scholar]
- Pagel, J.M.; Soumerai, J.D.; Reddy, N.; Jagadeesh, D.; Stathis, A.; Asch, A.; Salman, H.; Kenkre, V.P.; Iasonos, A.; Llorin-Sangalang, J.; et al. Zandelisib with continuous or intermittent dosing as monotherapy or in combination with rituximab in patients with relapsed or refractory B-cell malignancy: A multicentre, first-in-patient, dose-escalation and dose-expansion, phase 1b trial. Lancet Oncol. 2022, 23, 1021–1030. [Google Scholar] [CrossRef]
- Xu, Y.; Afify, S.M.; Du, J.; Liu, B.; Hassan, G.; Wang, Q.; Li, H.; Liu, Y.; Fu, X.; Zhu, Z.; et al. The efficacy of PI3Kgamma and EGFR inhibitors on the suppression of the characteristics of cancer stem cells. Sci. Rep. 2022, 12, 347. [Google Scholar] [CrossRef]
- Zhang, Z.; Richmond, A. The role of PI3K inhibition in the treatment of breast cancer, alone or combined with immune checkpoint inhibitors. Front. Mol. Biosci. 2021, 8, 648663. [Google Scholar] [CrossRef]
- Mateo, J.; Ganji, G.; Lemech, C.; Burris, H.A.; Han, S.W.; Swales, K.; Decordova, S.; DeYoung, M.P.; Smith, D.A.; Kalyana-Sundaram, S.; et al. A first-time-in-human study of GSK2636771, a phosphoinositide 3 kinase beta-selective inhibitor, in patients with advanced solid tumors. Clin. Cancer Res. 2017, 23, 5981–5992. [Google Scholar] [CrossRef] [PubMed]
- Sarker, D.; Dawson, N.A.; Aparicio, A.M.; Dorff, T.B.; Pantuck, A.J.; Vaishampayan, U.N.; Henson, L.; Vasist, L.; Roy-Ghanta, S.; Gorczyca, M.; et al. A phase I, open-label, dose-finding study of GSK2636771, a PI3Kbeta inhibitor, administered with enzalutamide in patients with metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2021, 27, 5248–5257. [Google Scholar] [CrossRef] [PubMed]
- Hanan, E.J.; Braun, M.G.; Heald, R.A.; MacLeod, C.; Chan, C.; Clausen, S.; Edgar, K.A.; Eigenbrot, C.; Elliott, R.; Endres, N.; et al. Discovery of GDC-0077 (Inavolisib), a highly selective inhibitor and degrader of mutant PI3Kalpha. J. Med. Chem. 2022, 65, 16589–16621. [Google Scholar] [CrossRef] [PubMed]
- Chien, A.J.; Tripathy, D.; Albain, K.S.; Symmans, W.F.; Rugo, H.S.; Melisko, M.E.; Wallace, A.M.; Schwab, R.; Helsten, T.; Forero-Torres, A.; et al. MK-2206 and standard neoadjuvant chemotherapy Improves response in patients with human epidermal growth factor receptor 2-positive and/or hormone receptor-negative breast cancers in the I-SPY 2 trial. J. Clin. Oncol. 2020, 38, 1059–1069. [Google Scholar] [CrossRef]
- Ma, C.X.; Suman, V.; Goetz, M.P.; Northfelt, D.; Burkard, M.E.; Ademuyiwa, F.; Naughton, M.; Margenthaler, J.; Aft, R.; Gray, R.; et al. A Phase II trial of neoadjuvant MK-2206, an AKT inhibitor, with anastrozole in clinical stage II or III PIK3CA-mutant ER-positive and HER2-negative breast cancer. Clin. Cancer Res. 2017, 23, 6823–6832. [Google Scholar] [CrossRef]
- Xing, Y.; Lin, N.U.; Maurer, M.A.; Chen, H.; Mahvash, A.; Sahin, A.; Akcakanat, A.; Li, Y.; Abramson, V.; Litton, J.; et al. Phase II trial of AKT inhibitor MK-2206 in patients with advanced breast cancer who have tumors with PIK3CA or AKT mutations, and/or PTEN loss/PTEN mutation. Breast Cancer Res. 2019, 21, 78. [Google Scholar] [CrossRef]
- Myers, A.P.; Konstantinopoulos, P.A.; Barry, W.T.; Luo, W.; Broaddus, R.R.; Makker, V.; Drapkin, R.; Liu, J.; Doyle, A.; Horowitz, N.S.; et al. Phase II, 2-stage, 2-arm, PIK3CA mutation stratified trial of MK-2206 in recurrent endometrial cancer. Int. J. Cancer 2020, 147, 413–422. [Google Scholar] [CrossRef]
- Stover, E.H.; Xiong, N.; Myers, A.P.; Tayob, N.; Engvold, V.; Polak, M.; Broaddus, R.R.; Makker, V.; Drapkin, R.; Liu, J.F.; et al. A phase II study of MK-2206, an AKT inhibitor, in uterine serous carcinoma. Gynecol. Oncol. Rep. 2022, 40, 100974. [Google Scholar] [CrossRef]
- Lee, J.B.; Jung, M.; Beom, S.H.; Kim, G.M.; Kim, H.R.; Choi, H.J.; Sohn, J.H.; Ahn, J.B.; Rha, S.Y.; Chung, H.C. Phase 2 study of TAS-117, an allosteric akt inhibitor in advanced solid tumors harboring phosphatidylinositol 3-kinase/v-akt murine thymoma viral oncogene homolog gene mutations. Investig. New Drugs. 2021, 39, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Rodon, J.; Funchain, P.; Laetsch, T.W.; Arkenau, H.T.; Hervieu, A.; Singer, C.F.; Murciano-Goroff, Y.R.; Chawla, S.P.; Anthony, K.; Yamamiya, I.; et al. A phase II study of TAS-117 in patients with advanced solid tumors harboring germline PTEN-inactivating mutations. Future Oncol. 2022, 18, 3377–3387. [Google Scholar] [CrossRef]
- Yamaji, M.; Ota, A.; Wahiduzzaman, M.; Karnan, S.; Hyodo, T.; Konishi, H.; Tsuzuki, S.; Hosokawa, Y.; Haniuda, M. Novel ATP-competitive Akt inhibitor afuresertib suppresses the proliferation of malignant pleural mesothelioma cells. Cancer Med. 2017, 6, 2646–2659. [Google Scholar] [CrossRef] [PubMed]
- Blagden, S.P.; Hamilton, A.L.; Mileshkin, L.; Wong, S.; Michael, A.; Hall, M.; Goh, J.C.; Lisyanskaya, A.S.; DeSilvio, M.; Frangou, E.; et al. Phase IB dose escalation and expansion study of AKT inhibitor afuresertib with carboplatin and paclitaxel in recurrent platinum-resistant ovarian cancer. Clin. Cancer Res. 2019, 25, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.I.; Paul, H.; Le, L.W.; Wei, E.N.; Snitzler, S.; Wang, T.; Levina, O.; Kakar, S.; Lau, A.; Queau, M.; et al. A phase 2 study of ofatumumab (Arzerra) in combination with a pan-AKT inhibitor (afuresertib) in previously treated patients with chronic lymphocytic leukemia (CLL). Leuk. Lymphoma 2019, 60, 92–100. [Google Scholar] [CrossRef]
- Crabb, S.J.; Griffiths, G.; Dunkley, D.; Downs, N.; Ellis, M.; Radford, M.; Light, M.; Northey, J.; Whitehead, A.; Wilding, S.; et al. Overall survival update for patients with metastatic castration-resistant prostate cancer treated with capivasertib and docetaxel in the phase 2 ProCAID clinical trial. Eur. Urol. 2022, 82, 512–515. [Google Scholar] [CrossRef] [PubMed]
- Kolinsky, M.P.; Rescigno, P.; Bianchini, D.; Zafeiriou, Z.; Mehra, N.; Mateo, J.; Michalarea, V.; Riisnaes, R.; Crespo, M.; Figueiredo, I.; et al. A phase I dose-escalation study of enzalutamide in combination with the AKT inhibitor AZD5363 (capivasertib) in patients with metastatic castration-resistant prostate cancer. Ann. Oncol. 2020, 31, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Howell, S.J.; Casbard, A.; Carucci, M.; Ingarfield, K.; Butler, R.; Morgan, S.; Meissner, M.; Bale, C.; Bezecny, P.; Moon, S.; et al. Fulvestrant plus capivasertib versus placebo after relapse or progression on an aromatase inhibitor in metastatic, oestrogen receptor-positive, HER2-negative breast cancer (FAKTION): Overall survival, updated progression-free survival, and expanded biomarker analysis from a randomised, phase 2 trial. Lancet Oncol. 2022, 23, 851–864. [Google Scholar]
- Westin, S.N.; Labrie, M.; Litton, J.K.; Blucher, A.; Fang, Y.; Vellano, C.P.; Marszalek, J.R.; Feng, N.; Ma, X.; Creason, A.; et al. Phase Ib dose expansion and translational analyses of olaparib in combination with capivasertib in recurrent endometrial, triple-negative breast, and ovarian cancer. Clin. Cancer Res. 2021, 27, 6354–6365. [Google Scholar] [CrossRef]
- Blake, J.F.; Xu, R.; Bencsik, J.R.; Xiao, D.; Kallan, N.C.; Schlachter, S.; Mitchell, I.S.; Spencer, K.L.; Banka, A.L.; Wallace, E.M.; et al. Discovery and preclinical pharmacology of a selective ATP-competitive Akt inhibitor (GDC-0068) for the treatment of human tumors. J. Med. Chem. 2012, 55, 8110–8127. [Google Scholar] [CrossRef]
- Sweeney, C.; Bracarda, S.; Sternberg, C.N.; Chi, K.N.; Olmos, D.; Sandhu, S.; Massard, C.; Matsubara, N.; Alekseev, B.; Parnis, F.; et al. Ipatasertib plus abiraterone and prednisolone in metastatic castration-resistant prostate cancer (IPATential150): A multicentre, randomised, double-blind, phase 3 trial. Lancet 2021, 398, 131–142. [Google Scholar] [CrossRef]
- Kotani, N.; Wilkins, J.J.; Wade, J.R.; Dang, S.; Sutaria, D.S.; Yoshida, K.; Sundrani, S.; Ding, H.; Garcia, J.; Hinton, H.; et al. Characterization of exposure-response relationships of ipatasertib in patients with metastatic castration-resistant prostate cancer in the IPATential150 study. Cancer Chemother. Pharmacol. 2022, 90, 511–521. [Google Scholar] [CrossRef]
- Sutaria, D.S.; Rasuo, G.; Harris, A.; Johnson, R.; Miles, D.; Gallo, J.D.; Sane, R. Drug-drug interaction study to evaluate the pharmacokinetics, safety, and tolerability of ipatasertib in combination with darolutamide in patients with advanced prostate cancer. Pharmaceutics 2022, 14, 2101. [Google Scholar] [CrossRef]
- Bose, S.; Kalinsky, K. Durable clinical activity to the AKT inhibitor ipatasertib in a heavily pretreated patient with an AKT1 E17K mutant metastatic breast cancer. Clin. Breast Cancer 2021, 21, e150–e153. [Google Scholar] [CrossRef] [PubMed]
- Dent, R.; Oliveira, M.; Isakoff, S.J.; Im, S.A.; Espie, M.; Blau, S.; Tan, A.R.; Saura, C.; Wongchenko, M.J.; Xu, N.; et al. Final results of the double-blind placebo-controlled randomized phase 2 LOTUS trial of first-line ipatasertib plus paclitaxel for inoperable locally advanced/metastatic triple-negative breast cancer. Breast Cancer Res. Treat. 2021, 189, 377–386. [Google Scholar] [CrossRef]
- Turner, N.; Dent, R.A.; O’Shaughnessy, J.; Kim, S.B.; Isakoff, S.J.; Barrios, C.; Saji, S.; Bondarenko, I.; Nowecki, Z.; Lian, Q.; et al. Ipatasertib plus paclitaxel for PIK3CA/AKT1/PTEN-altered hormone receptor-positive HER2-negative advanced breast cancer: Primary results from cohort B of the IPATunity130 randomized phase 3 trial. Breast Cancer Res. Treat. 2022, 191, 565–576. [Google Scholar] [CrossRef]
- Kajiwara, M.; Masuda, S. Role of mTOR inhibitors in kidney disease. Int. J. Mol. Sci. 2016, 17, 975. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wu, Y.; Zhou, X.; Qian, J.; Zhu, W.; Shu, Y.; Liu, P. Clinical efficacy of mTOR inhibitors in solid tumors: A systematic review. Future Oncol. 2015, 11, 1687–1699. [Google Scholar] [CrossRef] [PubMed]
- Mao, B.; Zhang, Q.; Ma, L.; Zhao, D.S.; Zhao, P.; Yan, P. Overview of research into mTOR inhibitors. Molecules 2022, 27, 5295. [Google Scholar] [CrossRef]
- Occhiuzzi, M.A.; Lico, G.; Ioele, G.; De Luca, M.; Garofalo, A.; Grande, F. Recent advances in PI3K/PKB/mTOR inhibitors as new anticancer agents. Eur. J. Med. Chem. 2023, 246, 114971. [Google Scholar] [CrossRef]
- Klippel, A.; Wang, R.; Puca, L.; Faber, A.L.; Shen, W.; Bhagwat, S.V.; Karukurichi, K.; Zhang, F.F.; Perez, C.; Rama, R.; et al. Preclinical characterization of LOXO-22783, a highly potent, mutant-selective and brain-penetrant allosteric PI3Ka H1047R inhibitor. Mol. Cancer Ther. 2021, 20, P142. [Google Scholar] [CrossRef]
- Perez, C.A.; Henry, J.T.; Varkaris, A.; Subbiah, V.; Spira, A.I.; Schmidt, O.; Shen, J.; Guo, W.; Hunter, T.; Jen, K.Y.; et al. First-in-human global multi-center study of RLY-2608, a pan-mutant and isoform-selective PI3Ka inhibitor, as a single agent in patients with advanced solid tumors and in combination with fulvestrant in patients with advanced breast cancer. J. Clin. Oncol. 2022, 40, TPS1124. [Google Scholar] [CrossRef]
- Buckbinder, L.; St Jean, D.J.; Tieu, T.; Wang, W.; Kryukov, G.; Jonsson, P.; Alltucker, J.; Manimala, S.; Ladd, B.; Guzman-Perez, A.; et al. Discovery and characterization of a PI3KaH1047X inhibitor with a best-in-class profile. Cancer Res. 2022, 82, LB194. [Google Scholar] [CrossRef]
- Wang, H.; Li, C.; Liu, X.; Ma, M. Design, synthesis and activity study of a novel PI3K degradation by hijacking VHL E3 ubiquitin ligase. Bioorg. Med. Chem. 2022, 61, 116707. [Google Scholar] [CrossRef]
- You, I.; Erickson, E.C.; Donovan, K.A.; Eleuteri, N.A.; Fischer, E.S.; Gray, N.S.; Toker, A. Discovery of an AKT degrader with prolonged inhibition of downstream signaling. Cell Chem. Biol. 2020, 27, 66–73.e7. [Google Scholar] [CrossRef]
- Yu, X.; Xu, J.; Xie, L.; Wang, L.; Shen, Y.; Cahuzac, K.M.; Chen, X.; Liu, J.; Parsons, R.E.; Jin, J. Design, synthesis, and evaluation of potent, selective, and bioavailable AKT kinase degraders. J. Med. Chem. 2021, 64, 18054–18081. [Google Scholar] [CrossRef]
- Song, K.W.; Edgar, K.A.; Hanan, E.J.; Hafner, M.; Oeh, J.; Merchant, M.; Sampath, D.; Nannini, M.A.; Hong, R.; Phu, L.; et al. RTK-dependent inducible degradation of mutant PI3Kalpha drives GDC-0077 (inavolisib) efficacy. Cancer Discov. 2022, 12, 204–219. [Google Scholar] [CrossRef]
- Cham, J.; Venkateswaran, A.R.; Bhangoo, M. Targeting the PI3K-AKT-mTOR pathway in castration resistant prostate cancer: A review article. Clin. Genitourin. Cancer 2021, 19, 563.e1–563.e7. [Google Scholar] [CrossRef]
- Gasmi, A.; Roubaud, G.; Dariane, C.; Barret, E.; Beauval, J.B.; Brureau, L.; Crehange, G.; Fiard, G.; Fromont, G.; Gauthe, M.; et al. Overview of the development and use of Akt inhibitors in prostate cancer. J. Clin. Med. 2021, 11, 160. [Google Scholar] [CrossRef]
- He, Y.; Xu, W.; Xiao, Y.T.; Huang, H.; Gu, D.; Ren, S. Targeting signaling pathways in prostate cancer: Mechanisms and clinical trials. Signal. Transduct. Target. Ther. 2022, 7, 198. [Google Scholar] [CrossRef]
- Vanhaesebroeck, B.; Burke, J.E.; Madsen, R.R. Precision targeting of mutant PI3Kalpha in cancer by selective degradation. Cancer Discov. 2022, 12, 20–22. [Google Scholar] [CrossRef]
- Li, W.; Gao, C.; Zhao, L.; Yuan, Z.; Chen, Y.; Jiang, Y. Phthalimide conjugations for the degradation of oncogenic PI3K. Eur. J. Med. Chem. 2018, 151, 237–247. [Google Scholar] [CrossRef]
- Zhu, C.L.; Luo, X.; Tian, T.; Rao, Z.; Wang, H.; Zhou, Z.; Mi, T.; Chen, D.; Xu, Y.; Wu, Y.; et al. Structure-based rational design enables efficient discovery of a new selective and potent AKT PROTAC degrader. Eur. J. Med. Chem. 2022, 238, 114459. [Google Scholar] [CrossRef]
- Harmon, S.A.; Patel, P.G.; Sanford, T.H.; Caven, I.; Iseman, R.; Vidotto, T.; Picanco, C.; Squire, J.A.; Masoudi, S.; Mehralivand, S.; et al. High throughput assessment of biomarkers in tissue microarrays using artificial intelligence: PTEN loss as a proof-of-principle in multi-center prostate cancer cohorts. Mod. Pathol. 2021, 34, 478–489. [Google Scholar] [CrossRef]
- Herberts, C.; Annala, M.; Sipola, J.; Ng, S.W.S.; Chen, X.E.; Nurminen, A.; Korhonen, O.V.; Munzur, A.D.; Beja, K.; Schonlau, E.; et al. Deep whole-genome ctDNA chronology of treatment-resistant prostate cancer. Nature 2022, 608, 199–208. [Google Scholar] [CrossRef]

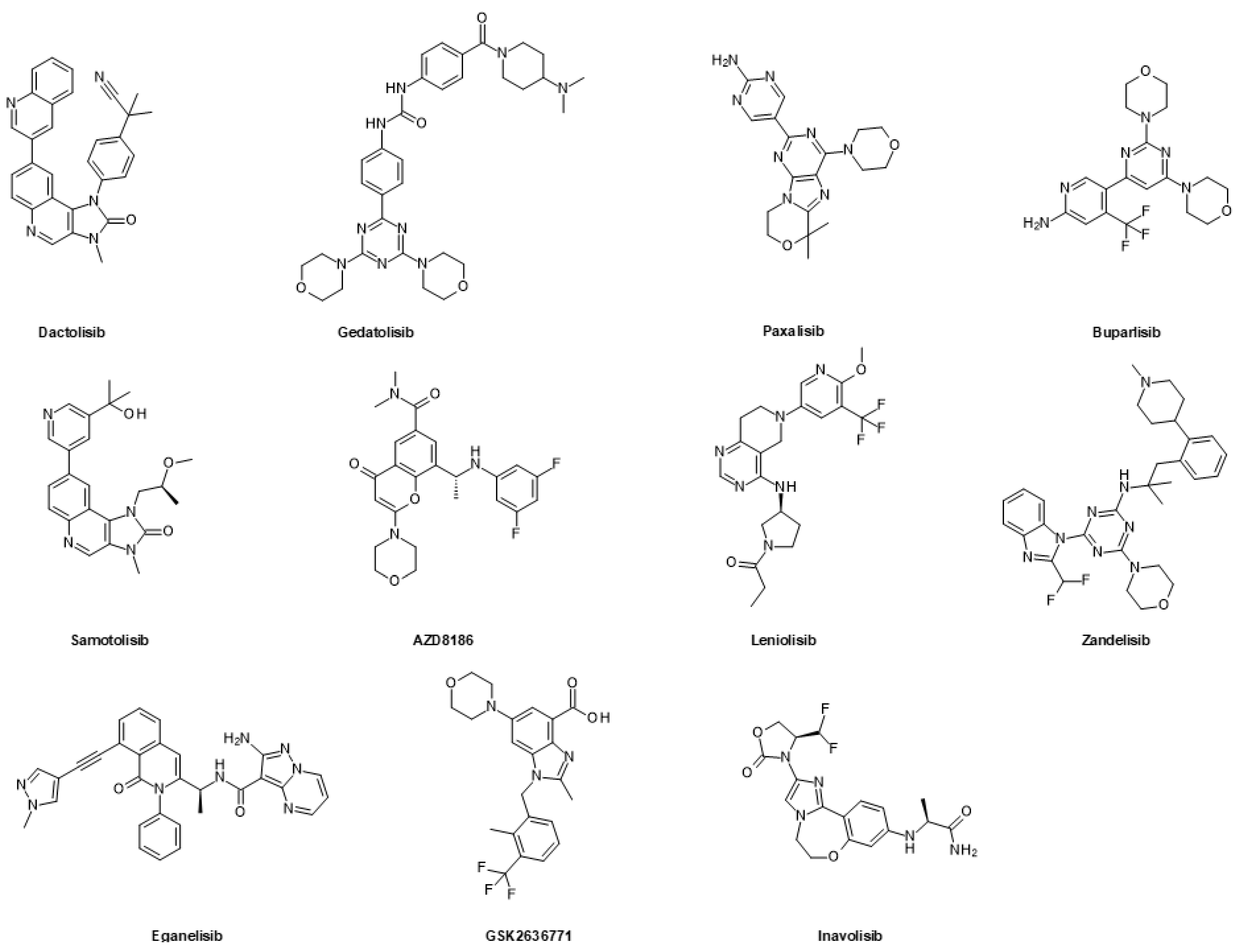
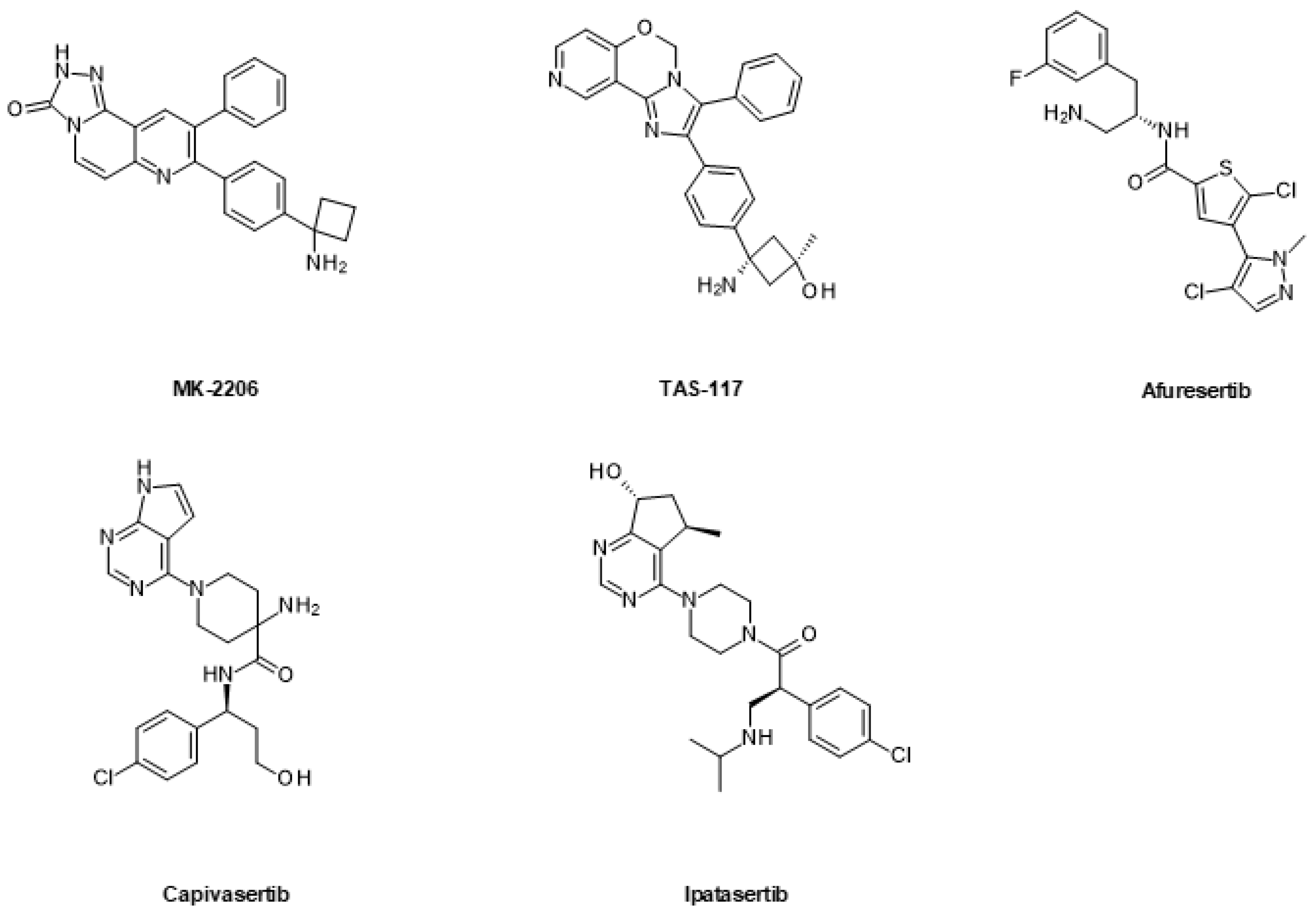
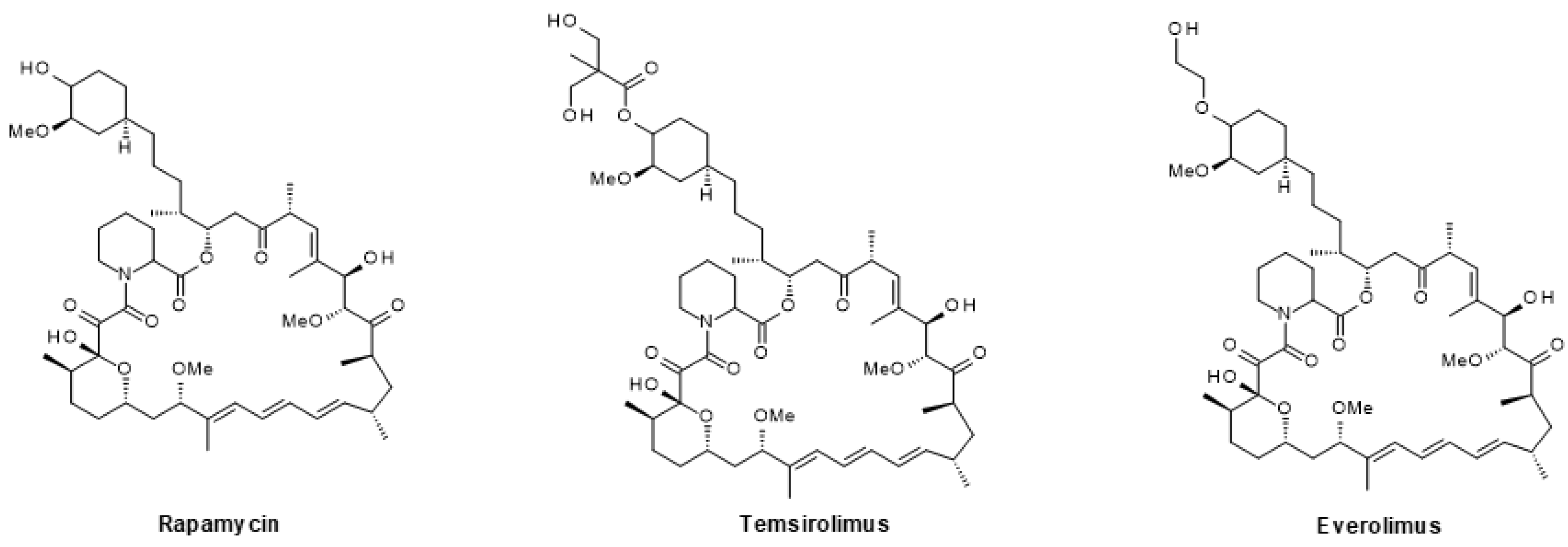


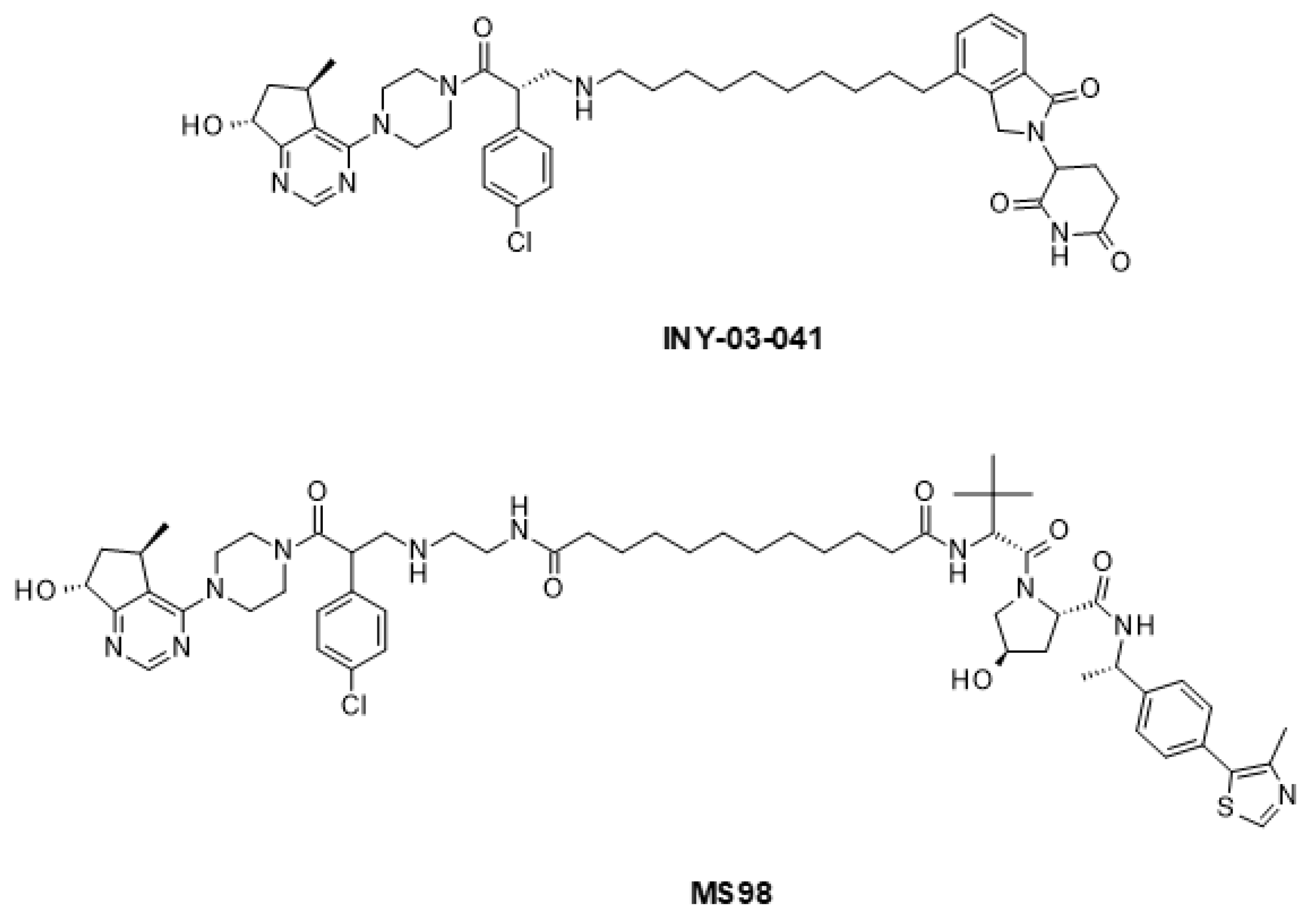
| Drug Combination | Targets | Indication | Status | Phase | Identifier |
|---|---|---|---|---|---|
| Samotolisib + enzalutamide | PI3K-mTOR/AR | mCRPC | Completed | 2 | NCT02407054 |
| Buparlisib + enzalutamide | PI3K/AR | mCRPC | Terminated | 2 | NCT01385293 |
| Apitolisib + abiraterone acetate | PI3K-mTOR/CYP17A | CRPC | Completed | 1b/2 | NCT01485861 |
| GSK2636771 + enzalutamide | PI3K/AR | mCRPC | Completed | 1 | NCT02215096 |
| AZD8186 + abiraterone acetate | PI3K/CYP17A | Advanced CRPC | Completed | 1 | NCT01884285 |
| Ipatasertib + abiraterone acetate | AKT/CYP17A | mCRPC | Active, not recruiting | 3 | NCT03072238 |
| Capivasertib + abiraterone acetate | AKT/CYP17A | mHSPC with PTEN deficiency | Recruiting | 3 | NCT04493853 |
| MK-2206 + bicalutamide | AKT/AR | Recurrent prostate cancer | Active, not recruiting | 2 | NCT01251861 |
| Capivasertib + abiraterone acetate | AKT/CYP17A | High-risk localized prostate cancer with PTEN deficiency | Not yet recruiting | 2 | NCT05593497 |
| Capivasertib + enzalutamide | AKT/AR | mCRPC | Unknown | 2 | NCT02525068 |
| Ipatasertib + abiraterone acetate | AKT/CYP17A | CRPC | Completed | 1b/2 | NCT01485861 |
| Ipatasertib + darolutamide | AKT/AR | Localized CRPC with PTEN deficiency | Terminated | 1/2 | NCT04737109 |
| Everolimus + bicalutamide | mTOR/AR | Recurrent or metastatic CRPC | Completed | 2 | NCT00814788 |
| Everolimus + enzalutamide | mTOR/AR | mCRPC | Completed | 1 | NCT02125084 |
| Everolimus + apalutamide | mTOR/AR | mCRPC | Completed | 1 | NCT02106507 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raith, F.; O’Donovan, D.H.; Lemos, C.; Politz, O.; Haendler, B. Addressing the Reciprocal Crosstalk between the AR and the PI3K/AKT/mTOR Signaling Pathways for Prostate Cancer Treatment. Int. J. Mol. Sci. 2023, 24, 2289. https://doi.org/10.3390/ijms24032289
Raith F, O’Donovan DH, Lemos C, Politz O, Haendler B. Addressing the Reciprocal Crosstalk between the AR and the PI3K/AKT/mTOR Signaling Pathways for Prostate Cancer Treatment. International Journal of Molecular Sciences. 2023; 24(3):2289. https://doi.org/10.3390/ijms24032289
Chicago/Turabian StyleRaith, Fabio, Daniel H. O’Donovan, Clara Lemos, Oliver Politz, and Bernard Haendler. 2023. "Addressing the Reciprocal Crosstalk between the AR and the PI3K/AKT/mTOR Signaling Pathways for Prostate Cancer Treatment" International Journal of Molecular Sciences 24, no. 3: 2289. https://doi.org/10.3390/ijms24032289
APA StyleRaith, F., O’Donovan, D. H., Lemos, C., Politz, O., & Haendler, B. (2023). Addressing the Reciprocal Crosstalk between the AR and the PI3K/AKT/mTOR Signaling Pathways for Prostate Cancer Treatment. International Journal of Molecular Sciences, 24(3), 2289. https://doi.org/10.3390/ijms24032289







