How Do Plants Cope with DNA Damage? A Concise Review on the DDR Pathway in Plants
Abstract
:1. Introduction
2. Wide Range of Genotoxic Agents Activating DDR in Plants
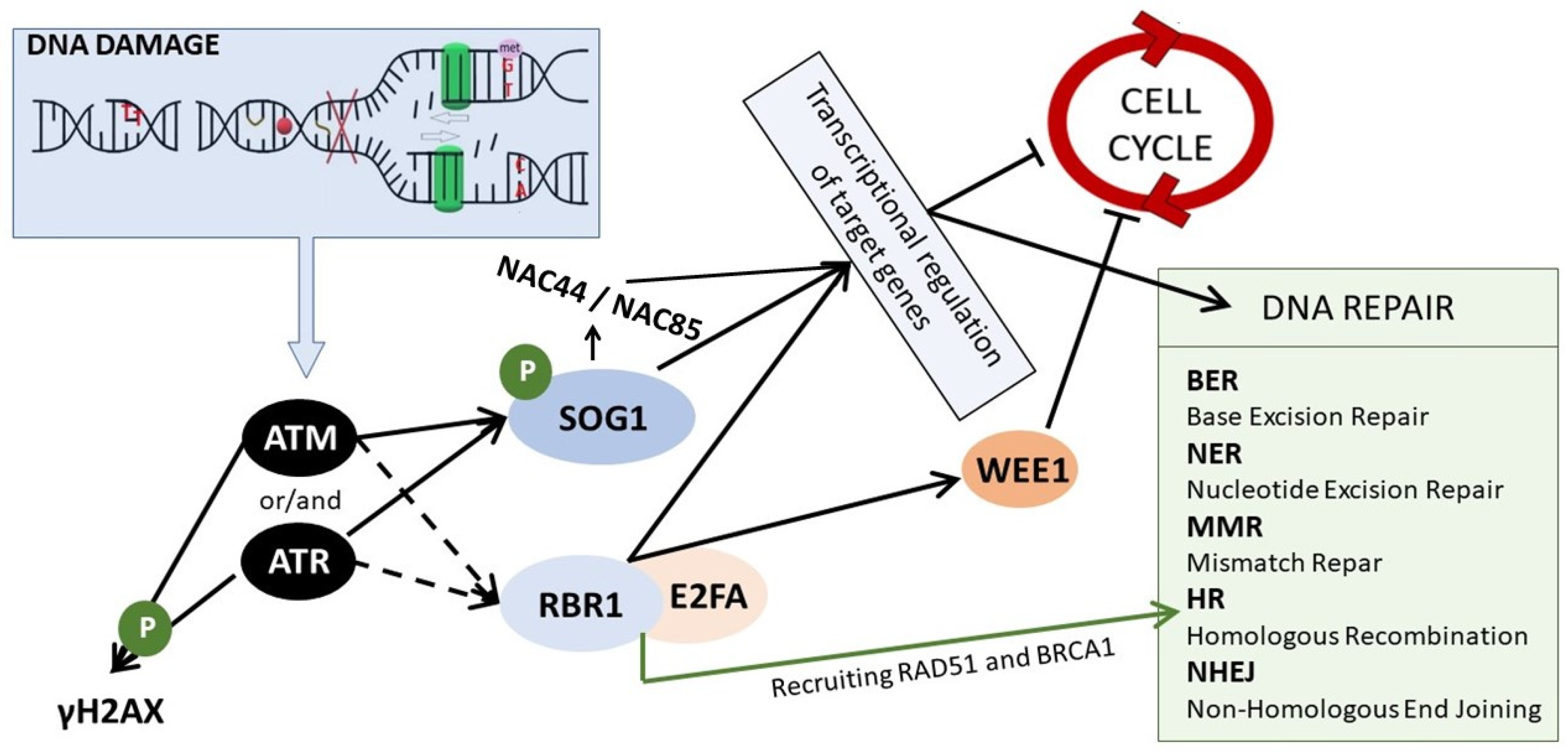
3. DDR–Sensing and Signaling the DNA Damage
4. DNA Repair Mechanisms
4.1. Repair Mechanisms of Damage in Single DNA Strand
4.2. DSB Repair–Homologous Recombination (HR) and Non-Homologous End Joining (NHEJ)
5. Cell Cycle Stoppage—Giving the Cell Time to Repair
6. Endoreduplication and Programmed Cell Death
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Manova, V.; Gruszka, D. DNA damage and repair in plants—From models to crops. Front. Plant Sci. 2015, 6, 885. [Google Scholar] [CrossRef] [Green Version]
- Dudáš, A.; Chovanec, M. DNA double-strand break repair by homologous recombination. Mutat. Res. Rev. Mutat. Res. 2004, 566, 131. [Google Scholar] [CrossRef]
- Sonoda, E.; Hochegger, H. Differential usage of non-homologous end-joining and homologous recombina-tion in double strand break repair. DNA Repair 2006, 5, 1021. [Google Scholar] [CrossRef]
- Kleibl, K. Molecular mechanisms of adaptive response to alkylating agents in Escherichia coli and some remarks on O(6)-methylguanine DNA-methyltransferase in other organisms. Mutat. Res. 2002, 512, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Shrivastav, N.; Li, D.; Essigmann, J.M. Chemical biology of mutagenesis and DNA repair: Cellular responses to DNA alkylation. Carcinogenesis 2010, 31, 59–70. [Google Scholar] [CrossRef] [Green Version]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Hussin, G.; Ramli, A.; Rahim, H.A.; Miah, G.; Usman, M. Principle and application of plant mutagenesis in crop improvement: A review. Biotechnol. Biotechnol. Equip. 2016, 30, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Till, B.J.; Cooper, J.; Tai, T.H.; Colowit, P.; Greene, E.A.; Henikoff, S.; Comai, L. Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 2007, 7, 19. [Google Scholar] [CrossRef] [Green Version]
- Kurowska, M.; Daszkowska-Golec, A.; Gruszka, D.; Marzec, M.; Szurman, M.; Szarejko, I.; Maluszynski, M. TILLING—A shortcut in functional genomics. J. Appl. Genet. 2011, 52, 317. [Google Scholar] [CrossRef] [Green Version]
- Jankowicz-Cieslak, J.; Huynh, O.A.; Brozynska, M.; Nakitandwe, J.; Till, B.J. Induction, rapid fixation and retention of mutations in vegetatively propagated banana. Plant Biotechnol. J. 2012, 10, 1056–1066. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Huang, L.; Min, D.; Phillips, A.; Wang, S.; Madgwick, P.J.; Parry, M.A.; Hu, Y.G. Development and characterization of a new TILLING population of common bread wheat (Triticum aestivum L.). PLoS ONE 2012, 7, e41570. [Google Scholar] [CrossRef] [Green Version]
- Tadele, Z. Mutagenesis and TILLING to dissect gene function in plants. Curr. Genom. 2016, 17, 499–508. [Google Scholar] [CrossRef] [Green Version]
- Espina, M.J.; Ahmed, C.M.S.; Bernardini, A.; Adeleke, E.; Yadegari, Z.; Arelli, P.; Pantalone, V.; Taheri, A. Development and Phenotypic Screening of an Ethyl Methane Sulfonate Mutant Population in Soybean. Front. Plant Sci. 2018, 9, 394. [Google Scholar] [CrossRef] [Green Version]
- Szurman-Zubrzycka, M.; Zbieszczyk, J.; Marzec, M.; Jelonek, J.; Chmielewska, B.; Kurowska, M.; Krok, M.; Daszkowska-Golec, A.; Guzy-Wrobelska, J.; Gruszka, D.; et al. HorTILLUS—A rich and renewable source of induced mutations for forward/reverse genetics and pre-breeding programs in barley (Hordeum vulgare L.). Front. Plant Sci. 2018, 9, 216. [Google Scholar] [CrossRef]
- Siddique, M.I.; Back, S.; Lee, J.H.; Jo, J.; Jang, S.; Han, K.; Venkatesh, J.; Kwon, J.K.; Jo, Y.D.; Kang, B.C. Development and Characterization of an Ethyl Methane Sulfonate (EMS) Induced Mutant Population in Capsicum annuum L. Plants 2020, 9, 396. [Google Scholar] [CrossRef] [Green Version]
- Karaman, K.; Kizil, S.; Başak, M.; Uzun, B.; Yol, E. Development of EMS-induced Mutagenized Groundnut Population and Discovery of Point Mutations in the ahFAD2 and Ara h 1 Genes by TILLING. J. Oleo Sci. 2021, 70, 1631–1640. [Google Scholar] [CrossRef] [PubMed]
- De Silva, I.U.; McHugh, P.J.; Clingen, P.H.; Hartley, J.A. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand crosslinks in mammalian cells. Mol. Cell Biol. 2000, 20, 7980–7990. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, L. DNA crosslinking damage and cancer—A tale of friend and foe. Transl. Cancer Res. 2013, 2, 144–154. [Google Scholar] [CrossRef]
- Weber, G.F. DNA Damaging Drugs. Molecular Therapies of Cancer; Springer: Berlin/Heidelberg, Germany, 2014; pp. 9–112. [Google Scholar] [CrossRef]
- Menke, M.; Chen, I.P.; Angelis, K.J.; Schubert, I. DNA damage and repair in Arabidopsis thaliana as measured by the comet assay after treatment with different classes of genotoxins. Mutat. Res./Genet. Toxicol. Environ. Mutagen. 2001, 493, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.P.; Haehnel, U.; Altschmied, L.; Schubert, I.; Puchta, H. The transcriptional response of Arabidopsis to genotoxic stress—A high-density colony array study (HDCA). Plant J. 2003, 35, 771–786. [Google Scholar] [CrossRef]
- Weimer, A.K.; Biedermann, S.; Harashima, H.; Roodbarkelari, F.; Takahashi, N.; Foreman, J.; Guan, Y.; Pochon, G.; Heese, M.; Van Damme, D.; et al. The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 2016, 35, 2068–2086. [Google Scholar] [CrossRef]
- Papadia, P.; Barozzi, F.; Hoeschele, J.D.; Piro, G.; Margiotta, N.; Di Sansebastiano, G.P. Cisplatin, oxaliplatin, and kiteplatin subcellular effects compared in a plant model. Int. J. Plant Sci. 2017, 18, 306. [Google Scholar] [CrossRef] [Green Version]
- Dorn, A.; Puchta, H. Analyzing Somatic DNA Repair in Arabidopsis Meiotic Mutants. In Plant Meiosis; Pradillo, M., Heckmann, S., Eds.; Humana: New York, NY, USA, 2007; Volume 2061, pp. 359–366. [Google Scholar] [CrossRef]
- Parra-Nunez, P.; Cooper, C.; Sanchez-Moran, E. The Role of DNA Topoisomerase Binding Protein 1 (TopBP1) in Genome Stability in Arabidopsis. Plants 2021, 10, 2568. [Google Scholar] [CrossRef]
- Rosa, M.; Scheid, O.M. DNA Damage Sensitivity Assays with Arabidopsis Seedlings. Bio Protoc. 2014, 4, e1093. [Google Scholar] [CrossRef] [Green Version]
- West, C.E.; Waterworth, W.M.; Sunderland, P.A.; Bray, C.M. Arabidopsis DNA double-strand break repair pathways. Biochem. Soc. Trans. 2004, 32, 964–966. [Google Scholar] [CrossRef] [Green Version]
- Charbonnel, C.; Gallego, M.E.; White, C.I. Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J. 2010, 64, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Stolarek, M.; Gruszka, D.; Braszewska-Zalewska, A.; Maluszynski, M. Functional analysis of the new barley gene HvKu80 indicates that it plays a key role in double-strand DNA break repair and telomere length regulation. Mutagenesis 2015, 30, 785–797. [Google Scholar] [CrossRef] [Green Version]
- Stolarek, M.; Gruszka, D.; Braszewska-Zalewska, A.; Maluszynski, M. Alleles of newly identified barley gene HvPARP3 exhibit changes in efficiency of DNA repair. DNA Repair 2015, 28, 116–130. [Google Scholar] [CrossRef]
- Allawzi, A.; Elajaili, H.; Redente, E.F.; Nozik-Grayck, E. Oxidative toxicology of bleomycin: Role of the extracellular redox environment. Curr. Opin. Toxicol. 2019, 13, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Adachi, S.; Minamisawa, K.; Okushima, Y.; Inagaki, S.; Yoshiyama, K.; Kondou, Y.; Kaminuma, E.; Ka-washima, M.; Toyoda, T.; Matsui, M.; et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 10004–10009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.; Takatsuka, H.; Takahashi, N.; Kurata, R.; Fukao, Y.; Kobayashi, K.; Ito, M.; Umeda, M. Arabidopsis R1R2R3-Myb proteins are essential for inhibiting cell division in response to DNA damage. Nat. Commun. 2017, 8, 635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshiyama, K.O.; Kaminoyama, K.; Sakamoto, T.; Kimura, S. Increased Phosphorylation of Ser-Gln Sites on SUPPRESSOR OF GAMMA RESPONSE1 Strengthens the DNA Damage Response in Arabidopsis thaliana. Plant Cell 2017, 29, 3255–3268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Čížková, M.; Slavková, M.; Vítovám, M.; Zachleder, V.; Bišová, K. Response of the Green Alga Chlamydomonas reinhardtii to the DNA Damaging Agent Zeocin. Cells 2019, 8, 735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saban, N.; Bujak, M. Hydroxyurea and hydroxamic acid derivatives as antitumor drugs. Cancer Chemotheraphy Pharmacol. 2009, 64, 213–221. [Google Scholar] [CrossRef]
- Madaan, K.; Kaushik, D.; Verma, T. Hydroxyurea: A key player in cancer chemotherapy. Expert Rev. Anticancer Ther. 2012, 1, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Musiałek, M.W.; Rybaczek, D. Hydroxyurea—The Good, the Bad and the Ugly. Genes 2021, 12, 1096. [Google Scholar] [CrossRef]
- Gualtieri, C.; Gianella, M.; Pagano, A.; Cadeddu, T.; Araújo, S.; Balestrazzi, A.; Macovei, A. Exploring microRNA Signatures of DNA Damage Response Using an Innovative System of Genotoxic Stress in Medicago truncatula Seedlings. Front. Plant Sci. 2021, 12, 645323. [Google Scholar] [CrossRef]
- Buta, J.G.; Worley, J.F. Camptothecin, a selective plant growth regulator. J. Agric. Food Chem. 1976, 24, 1085–1086. [Google Scholar] [CrossRef]
- Locato, V.; Balestrazzi, A.; De Gara, L.; Carbonera, D. Reduced expression of top1beta gene induces programmed cell death and alters ascorbate metabolism in Daucus carota cultured cells. J. Exp. Bot. 2006, 57, 1667–1676. [Google Scholar] [CrossRef] [Green Version]
- Iakimova, E.T.; Yordanova, Z.P.; Cristescu, S.M.; Harren, F.F.M.; Woltering, E.J. Cell death associated release of volatile organic sulphur compounds with antioxidant properties in chemical-challenged tobacco BY-2 suspension cultured cells. J. Plant Physiol. 2020, 251, 153223. [Google Scholar] [CrossRef]
- Pommier, Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer 2006, 6, 789–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prochazkova, K.; Finke, A.; Tomaštíková, E.D.; Filo, J.; Bente, H.; Dvořák, P.; Ovečka, M.; Šamaj, J.; Pecinka, A. Zebularine induces enzymatic DNA–protein crosslinks in 45S rDNA heterochromatin of Arabidopsis nuclei. Nucleic Acids Res. 2022, 50, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Ryu, T.; Lee, S.; Lee, S.; Chung, B. Ionizing Radiation manifesting DNA damage response in plants: An overview of DNA damage signaling and repair mechanisms in plants. Plant Sci. 2019, 278, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, R.P.; Kumar, A.; Tyagi, M.; Sinha, R. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J. Nucleic Acids 2010, 29, 592980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dany, A.-L.; Douki, T.; Triantaphylides, C.; Cadet, J. Repair of the main UV-induced thymine dimeric lesions within Arabidopsisi thaliana DNA: Evidence for the major involvement of photoreactivation pathways. J. Photochem. Photobiol. B Biol. 2001, 65, 127–135. [Google Scholar] [CrossRef]
- Gill, S.; Anjum, M.; Gill, R.; Jka, M.; Tuteja, N. DNA damage and repair in plants under ultraviolet and ionizing radiations. Sci. World J. 2015, 2015, 250158. [Google Scholar] [CrossRef] [Green Version]
- Limoli, C.L.; Giedzinski, E.; Bonner, W.M.; Cleaver, J.E. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA doublestrand breaks, γ-H2AX formation, and Mre11 relocalization. Proc. Natl. Acad. Sci. USA 2002, 99, 233–238. [Google Scholar] [CrossRef] [Green Version]
- Batista, L.F.Z.; Kaina, B.; Meneghini, R.; Menck, C.F.M. How DNA lesions are turned into powerful killing structures: Insights from UV-induced apoptosis. Mutat. Res. 2009, 681, 197–208. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Chan, Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef] [Green Version]
- Nisa, M.-U.; Huang, Y.; Benhamed, M.; Raynaud, C. The plant DNA Damage Response: Signaling pathways leading to growth inhibition and putative role in response to stress conditions. Front. Plant Sci. 2019, 10, 653. [Google Scholar] [CrossRef] [Green Version]
- Koukalová, B.; Kovarík, A.; Fajkus, J.; Siroký, J. Chromatin fragmentation associated with apoptotic changes in tobacco cells exposed to cold stress. FEBS Lett. 1997, 414, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.H.; Savina, M.; Du, J.; Devendran, A.; Kannivadi Ramakanth, K.; Tian, X.; Sim, W.S.; Mironova, V.V.; Xu, J. A Sacrifice-for-Survival Mechanism Protects Root Stem Cell Niche from Chilling Stress. Cell 2017, 170, 102–113.e14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecinka, A.; Dinh, H.Q.; Baubec, T.; Rosa, M.; Lettner, N.; Mittelsten Scheid, O. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 2010, 22, 3118–3129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, Y.; Kajihara, A.; Kirita, T.; Mori, E. Heat meets DNA: DNA damage and repair. Therm. Med. 2018, 34, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Han, S.H.; Kim, J.Y.; Lee, J.H.; Park, C.M. Safeguarding genome integrity under heat stress in plants. J. Exp. Bot. 2021, 3, erab355. [Google Scholar] [CrossRef]
- Boyko, A.; Golubov, A.; Bilichak, A.; Kovalchuk, I. Chlorine ions but not sodium ions alter genome stability of Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 1066–1078. [Google Scholar] [CrossRef] [Green Version]
- Sihi, S.; Bakshi, S.; Maiti, S.; Nayak, A.; Sengupta, D.N. Analysis of DNA polymerase λ activity and gene expression in response to salt and drought stress in Oryza sativa Indica rice cultivars. J. Plant Growth Regul. 2022, 41, 1499–1515. [Google Scholar] [CrossRef]
- Pedroza-Garcia, J.A.; Xiang, Y.; De Veylder, L. Cell cycle checkpoint control in response to DNA damage by environmental stresses. Plant J. 2022, 109, 490–507. [Google Scholar] [CrossRef]
- Saha, P.; Mukherjee, A.; Biswas, A.K. Modulation of NaCl induced DNA damage and oxidative stress in mungbean by pretreatment with sublethal dose. Biol. Plant. 2014, 59, 139–146. [Google Scholar] [CrossRef]
- Zvanarou, S.; Vágnerová, R.; Mackievic, V.; Usnich, S.; Smolich, I.; Sokolik, A.; Yu, M.; Huang, X.; Angelis, K.J.; Demidchik, V. Salt stress triggers generation of oxygen free radicals and DNA breaks in Physcomitrella patens protonema. Environ. Exp. Bot. 2020, 180, 104236. [Google Scholar] [CrossRef]
- Dutta, S.; Mitra, M.; Agarwal, P.; Mahapatra, K.; De, S.; Sett, U.; Roy, S. Oxidative and genotoxic damages in plants in response to heavy metal stress and maintenance of genome stability. Plant Signal. Behav. 2018, 13, e1460048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nezames, C.; Sjogren, C.; Barajas, J.; Larsen, P. The Arabidopsis cell cycle checkpoint regulators TANMEI/ALT2 and ATR mediate the active process of aluminum-dependent root growth inhibition. Plant Cell 2012, 24, 608–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaskowiak, J.; Tkaczyk, O.; Slota, M.; Kwasniewksa, J.; Szarejko, I. Analysis of aluminium toxicity in Hordeum vulgare roots with an emphasis on DNA integrity and cell cycle. PLoS ONE 2018, 13, e0193156. [Google Scholar] [CrossRef] [PubMed]
- Szurman-Zubrzycka, M.; Nawrot, M.; Jelonek, J.; Dziekanowski, M.; Kwasniewska, J.; Szarejko, I. ATR, a DNA damage signaling kinase, is involved in aluminium response in barley. Front. Plant Sci. 2019, 10, 1299. [Google Scholar] [CrossRef] [PubMed]
- Szurman-Zubrzycka, M.; Chwailkowska, K.; Niemira, M.; Kwasniewski, M.; Nawrot, M.; Gajecka, M.; Larsen, P.; Szarejko, I. Aluminum or low pH—which is the bigger enemy of barley? Transcriptome analysis of barley root meristem under Al and low pH stress. Front. Genet. 2021, 12, 675260. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T.; Barnes, D. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 2000, 65, 127–133. [Google Scholar] [CrossRef]
- Maréchal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef]
- Amiard, S.; Charbonnel, C.; Allain, E.; Depeiges, A.; White, C.I.; Gallego, M.E. Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell 2010, 22, 3020–3033. [Google Scholar] [CrossRef] [Green Version]
- Falck, J.; Coates, J.; Jackson, S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005, 434, 605–611. [Google Scholar] [CrossRef]
- Saldivar, J.C.; Cortez, D.; Cimprich, K.A. The essential kinase ATTR: Ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017, 18, 622–636. [Google Scholar] [CrossRef] [Green Version]
- Garcia, V.; Bruchet, H.; Camescasse, D.; Granier, F.; Bouchez, D.; Tissier, A. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 2003, 15, 119–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Culligan, K.; Tissier, A.; Britt, A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 2004, 16, 1091–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sweeney, P.R.; Britt, A.B.; Culligan, K.M. The Arabidopsis ATRIP ortholog is required for a programmed response to replication inhibitors. Plant J. 2009, 60, 518–526. [Google Scholar] [CrossRef]
- Heitzeberg, F.; Chen, I.-P.; Hartung, F.; Orel, N.; Angelis, K.J.; Puchta, H. The Rad17 homologue of Ara-bidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J. 2004, 38, 954–968. [Google Scholar] [CrossRef] [PubMed]
- Aklilu, B.B.; Soderquist, R.S.; Culligan, K.M. Genetic analysis of the replication protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication. Nucleic Acids Res. 2014, 42, 3104–3118. [Google Scholar] [CrossRef] [Green Version]
- Mah, L.J.; El-Osta, A.; Karagiannis, T. γH2AX: A sensitive molecular marker of DNA damage and repair. Leukemia 2010, 24, 679–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, T.; Kang, H.; Wu, D.; Zhu, X.; Huang, L.; Wu, J.; Zhu, Y. Arabidopsis γ-H2A. X-INTERACTING PROTEIN participates in DNA damage response and safeguards chromatin stability. Nat. Commun. 2022, 13, 1–14. [Google Scholar] [CrossRef]
- Yoshiyama, K.O.; Kobayashi, J.; Ogita, N.; Uead, M.; Kimura, S.; Maki, H.; Umeda, M. ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 2013, 14, 817–822. [Google Scholar] [CrossRef] [Green Version]
- Sjogren, C.A.; Bolaris, S.C.; Larsen, P.B. Aluminum-Dependent Terminal Differentiation of the Arabidopsis Root Tip Is Mediated through an ATR-, ALT2-, and SOG1-Regulated Transcriptional Response. Plant Cell 2015, 27, 2501–2515. [Google Scholar] [CrossRef] [Green Version]
- Preuss, S.B.; Britt, A.B. A DNA-damage-induced cell cycle checkpoint in Arabidopsis. Genetics 2003, 164, 323–334. [Google Scholar] [CrossRef]
- Yoshiyama, K.; Conklin, P.; Huefner, N.D.; Britt, A.B. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. USA 2009, 106, 12843–12848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshiyama, K. SOG1: A master regulator of the DNA damage response in plants. Genes Genet. Syst. 2016, 90, 209–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogita, N.; Okushima, Y.; Tokizawa, M.; Yamamoto, Y.Y.; Tanaka, M.; Seki, M.; Makita, Y.; Matsui, M.; Okamoto-Yoshiyama, K.; Sakamoto, T.; et al. Identifying the target genes of SUPPRESSOR OF GAMMA RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 2018, 94, 439–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourbousse, C.; Vegesna, N.; Law, J.A. SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E12453–E12462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, D.; Kamei, C.L.A.; Cools, T.; Vanderauwera, S.; Takahashi, N.; Okushima, Y.; Eekhout, T.; Yoshiyama, K.O.; Larkin, J.; Van den Daele, H.; et al. The Arabidopsis SIAMESE-RELATED Cyclin-Dependent Kinase Inhibitors SMR5 and SMR7 Regulate the DNA Damage Checkpoint in Response to Reactive Oxygen Species. Plant Cell 2014, 26, 296–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roitinger, E.; Hofer, M.; Köcher, T.; Pichler, P.; Novatchkova, M.; Yang, J.; Schlögelhofer, P.; Mechtler, K. Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol. Cell Proteom. 2015, 14, 556–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berckmans, B.; De Veylder, L. Transcriptional control of the cell cycle. Curr. Opin. Plant Biol. 2009, 12, 599–605. [Google Scholar] [CrossRef]
- Horvath, B.M.; Kourova, H.; Nagy, S.; Nemeth, E.; Magyar, Z.; Papdi, C.; Ahmad, Z.; Sanchez-Perez, G.F.; Perilli, S.; Blilou, I.; et al. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J. 2017, 36, 1261–1278. [Google Scholar] [CrossRef]
- Biedermann, S.; Harashima, H.; Chen, P.; Heese, M.; Bouyer, D.; Sofroni, K.; Schnittger, A. The retinoblas-toma homolog RBR1 mediates localization of the repair protein RAD51 to DNA lesions in Arabidopsis. EMBO J. 2017, 36, 1279–1297. [Google Scholar] [CrossRef]
- Bouyer, D.; Heese, M.; Chen, P.; Harashima, H.; Roudier, F.; Grüttner, C.; Schnittger, A. Genome-wide identification of RETINOBLASTOMA RELATED 1 binding sites in Arabidopsis reveals novel DNA damage regulators. PLoS Genet. 2018, 14, e1007797. [Google Scholar] [CrossRef] [Green Version]
- De Medeiros, N.M.C.; De Medeiros, A.L.M.; Silva, H.C.; Scortecci, K.C. Recent advances in plant DNA repair. In Advances in Plant DNA Repair; IntechOpen: London, UK, 2015. [Google Scholar] [CrossRef] [Green Version]
- Spampinato, C.P. Protecting DNA from errors and damage: An overview of DNA repair mechanisms in plants compared to mammals. Cell Mol. Life Sci. 2017, 74, 1693–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raina, A.; Sahu, P.K.; Laskar, R.A.; Rajora, N.; Sao, R.; Khan, S.; Ganai, R.A. Mechanisms of Genome Maintenance in Plants: Playing It Safe with Breaks and Bumps. Front Genet. 2021, 12, 675686. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Bjoras, M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Drablos, F.; Slupphaug, G. Uracil in DNA—Occurrence, consequences and repair. Oncogene 2002, 21, 8935–8938. [Google Scholar] [CrossRef] [Green Version]
- Córdoba-Cañero, D.; Dubois, E.; Ariza, R.R.; Doutriaux, M.-P.; Roldán-Arjona, T. Arabidopsis Uracil DNA Glycosylase (UNG) Is Required for Base Excision Repair of Uracil and Increases Plant Sensitivity to 5-Fluorouracil. J. Biol. Chem. 2010, 285, 7475–7483. [Google Scholar] [CrossRef] [Green Version]
- Córdoba-Cañero, D.; Morales-Ruiz, T.; Roldán-Arjona, T.; Ariza, R.R. Single-nucleotide and long-patch base excision repair of DNA damage in plants. Plant J. 2009, 60, 716–728. [Google Scholar] [CrossRef] [Green Version]
- Córdoba-Cañero, D.; Roldán-Arjona, T.; Ariza, R.R. Arabidopsis ARP endonuclease functions in a branched base excision DNA repair pathway completed by LIG1. Plant J. 2011, 68, 693–702. [Google Scholar] [CrossRef]
- Santerre, A.; Britt, A.B. Cloning of a 3-methyladenine-DNA glycosylase from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1994, 91, 2240–2244. [Google Scholar] [CrossRef] [Green Version]
- Memisoglu, A.; Samson, L. Base excision repair in yeast and mammals. Mutat. Res 2000, 451, 39–51. [Google Scholar] [CrossRef]
- Srivastava, D.K.; Berg, B.J.; Prasad, R.; Molina, J.T.; Beard, W.A.; Tomkinson, A.E.; Wilson, S.H. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J. Biol. Chem. 1998, 273, 21203–21209. [Google Scholar] [CrossRef] [Green Version]
- Levin, D.S.; Bai, W.; Yao, N.; O’Donnell, M.; Tomkinson, A.E. An interaction between DNA ligase I and proliferating cell nuclear antigen: Implications for Okazaki fragment synthesis and joining. Proc. Natl. Acad. Sci. USA 1997, 94, 12863–12868. [Google Scholar] [CrossRef] [Green Version]
- Nash, R.A.; Caldecott, K.W.; Barnes, D.E.; Lindahl, T. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry 1997, 36, 5207–5211. [Google Scholar] [CrossRef] [PubMed]
- Roldán-Arjona, T.; Ariza, R.R.; Córdoba-Cañero, D. DNA Base Excision Repair in Plants: An Unfolding Story with Familiar and Novel Characters. Front. Plant Sci. 2019, 10, 1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Córdoba-Cañero, D.; Roldán-Arjona, T.; Ariza, R.R. Arabidopsis ZDP DNA 3′-phosphatase and ARP en-donuclease function in 8-oxoG repair initiated by FPG and OGG1 DNA glycosylases. Plant J. 2014, 79, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liang, W.; Li, Y.; Qian, W. Apurinic/apyrimidinic endonuclease2 and zinc finger dna 3′-phosphoesterase play overlapping roles in the maintenance of epigenome and genome stability. Plant Cell 2018, 30, 1954–1970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchiyama, Y.; Suzuki, Y.; Sakaguchi, K. Characterization of plant XRCC1 and its interaction with proliferating cell nuclear antigen. Planta 2008, 227, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, Y.; Takeuchi, R.; Kodera, H.; Sakaguchi, K. Distribution and roles of X-family DNA polymerases in eukaryotes. Biochimie 2009, 91, 165–170. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Masnavi, G.; Bhardwaj, R.M.; Jiang, Q.; Bray, C.M.; West, C.E. A plant DNA ligase is an important determinant of seed longevity. Plant J. 2010, 63, 848–860. [Google Scholar] [CrossRef]
- West, C.E.; Waterworth, W.M.; Jiang, Q.; Bray, C.M. Arabidopsis DNA ligase IV is induced by gamma-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J. 2000, 24, 67–78. [Google Scholar] [CrossRef]
- van Attikum, H.; Bundock, P.; Overmeer, R.M.; Lee, L.Y.; Gelvin, S.B.; Hooykaas, P.J. The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 2003, 31, 4247–4255. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Macías, M.I.; Córdoba-Cañero, D.; Ariza, R.R.; Roldán-Arjona, T. The DNA repair protein XRCC1 functions in the plant DNA demethylation pathway by stimulating cytosine methylation (5-meC) excision, gap tailoring, and DNA ligation. J. Biol. Chem. 2013, 288, 5496–5505. [Google Scholar] [CrossRef] [Green Version]
- Macovei, A.; Balestrazzi, A.; Confalonieri, M.; Faè, M.; Carbonera, D. New insights on the barrel medic MtOGG1 and MtFPG functions in relation to oxidative stress response in planta and during seed imbibition. Plant Physiol. Biochem. 2011, 49, 1040. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Roy, S.; Choudhury, S.R.; Sengupta, D.N. DNA repair and recombination in higher plants: Insights from comparative genomics of Arabidopsis and rice. BMC Genom. 2010, 11, 443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanawalt, P.C.; Spivak, G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008, 9, 958–970. [Google Scholar] [CrossRef] [PubMed]
- Schärer, O.D. Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013, 5, a012609. [Google Scholar] [CrossRef] [Green Version]
- Reed, S.H. Nucleotide excision repair in chromatin: Damage removal at the drop of a HAT. DNA Repair 2011, 10, 734–742. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, H.; Zhang, J.; Guo, G.; Schumaker, K.; Guo, Y. Arabidopsis cockayne syndrome A-like proteins 1A and 1B form a complex with CULLIN4 and damage DNA binding protein 1A and regulate the response to UV irradiation. Plant Cell 2010, 22, 2353–2369. [Google Scholar] [CrossRef] [Green Version]
- Biedermann, S.; Hellmann, H. The DDB1a interacting proteins ATCSA-1 and DDB2 are critical factors for UV-B tolerance and genomic integrity in Arabidopsis thaliana. Plant J. 2010, 62, 404–415. [Google Scholar] [CrossRef]
- Molinier, J.; Lechner, E.; Dumbliauskas, E.; Genschik, P. Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress. PLoS Genet. 2008, 4, e1000093. [Google Scholar] [CrossRef] [Green Version]
- Ganpudi, A.; Schroeder, D. Genetic interactions of Arabidopsis thaliana damaged DNA binding protein 1B (DDB1B) with DDB1A, DET1, and COP1. Genes Genomes Genet. 2013, 3, 493–503. [Google Scholar] [CrossRef] [Green Version]
- Koga, A.; Ishibashi, T.; Kimura, S.; Uchiyama, Y.; Sakaguchi, K. Characterization of T-DNA insertion mutants and RNAi silenced plants of Arabidopsis thaliana UV-damaged DNA binding protein 2 (AtUV-DDB2). Plant Mol. Biol. 2006, 61, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Ly, V.; Hatherell, A.; Kim, E.; Chan, A.; Belmonte, M.; Schroeder, D. Interactions between Arabidopsis DNA repair genes UVH6, DDB1A, and DDB2 during abiotic stress tolerance and floral development. Plant Sci. 2013, 213, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Brettel, K.; Byrdin, M. Reaction mechanisms of DNA photolyase. Curr. Opin. Struct. Biol. 2010, 20, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Hitomi, K.; DiTacchio, L.; Arvai, A.S.; Yamamoto, J.; Kim, S.T.; Todo, T.; Tainer, J.A.; Iwai, S.; Panda, S.; Getzoff, E.D. Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes. Proc. Natl. Acad. Sci. USA 2009, 106, 6962–6967. [Google Scholar] [CrossRef] [Green Version]
- Hitomi, K.; Arvai, A.S.; Yamamoto, J.; Hitomi, C.; Teranishi, M.; Hirouchi, T.; Yamamoto, K.; Iwai, S.; Tainer, J.A.; Hidema, J.; et al. Eukaryotic class II cyclobutane pyrimidine dimer photolyase structure reveals basis for improved ultraviolet tolerance in plants. J. Biol. Chem. 2012, 287, 12060–12069. [Google Scholar] [CrossRef] [Green Version]
- Chirinos-Arias, M.C.; Spampinato, C.P. Growth and development of AtMSH7 mutants in Arabidopsis thaliana. Plant Physiol. Biochem. 2020, 146, 329–336. [Google Scholar] [CrossRef]
- Verma, P.; Tandon, R.; Yadav, G.; Gaur, V. Structural Aspects of DNA Repair and Recombination in Crop Improvement. Front. Genet. 2020, 11, 574549. [Google Scholar] [CrossRef]
- Marti, T.M.; Kunz, C.; Fleck, O. DNA mismatch repair and mutation avoidance pathways. J. Cell. Physiol. 2002, 191, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Culligan, K.; Lamers, M.; Hays, J. Dissimilar mispair-recognition spectra of arabidopsis DNA-mismatch-repair proteins MSH2·MSH6 (MutSα) and MSH2·MSH7 (MutSγ). Nucleic Acids Res. 2003, 31, 6027–6034. [Google Scholar] [CrossRef]
- Tian, L.; Gu, L.; Li, G.M. Distinct nucleotide binding/hydrolysis properties and molar ratio of MutSalpha and MutSbeta determine their differential mismatch binding activities. J. Biol. Chem. 2009, 284, 11557–11562. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Lawrence, C.; Li, G.-M.; Hays, J. Specific Binding of Human MSH2·MSH6 Mismatch-Repair Protein Heterodimers to DNA Incorporating Thymine- or Uracil-containing UV Light Photoproducts Opposite Mismatched Bases. J. Biol. Chem. 1999, 274, 16894–16900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Cao, Q.; Zhao, Q.; Arfan, M.; Liu, W. Mechanisms used by DNA MMR system to cope with Cadmium-induced DNA damage in plants. Chemosphere 2020, 246, 25614. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qin, J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc. Natl. Acad. Sci. USA 2003, 100, 15387–15392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega, J.; Lee, G.S.; Gu, L.; Yang, W.; Li, G.M. Mispair-bound human MutS-MutL complex triggers DNA incisions and activates mismatch repair. Cell Res. 2021, 31, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Goellner, E.M.; Putnam, C.D.; Kolodner, R.D. Exonuclease 1-dependent and independent mismatch repair. DNA Repair. 2015, 32, 24–32. [Google Scholar] [CrossRef] [Green Version]
- Gujar, V.V.; Dahal, B.K.; Kadyrova, L.K.; Kadyrov, F. Exo1-independent MMR at euchromatin is errorprone and involves Pol ζ and Rev1. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Kadyrova, L.Y.; Dahal, B.K.; Gujar, V.; Daley, J.M.; Sung, P.; Kadyrov, F.A. The nuclease activity of DNA2 promotes exonuclease 1-independent mismatch repair. J. Biol. Chem. 2022, 298, 101831. [Google Scholar] [CrossRef]
- Jia, N.; Liu, X.; Gao, H. A DNA2 Homolog Is Required for DNA Damage Repair, Cell Cycle Regulation, and Meristem Maintenance in Plants. Plant Physiol. 2016, 171, 318–333. [Google Scholar] [CrossRef] [Green Version]
- Modrich, P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006, 281, 30305–30309. [Google Scholar] [CrossRef] [Green Version]
- Qiu, S.; Huang, J. MRN complex is an essential effector of DNA damage repair. J. Zhejiang Univ. Sci. B 2021, 22, 31–37. [Google Scholar] [CrossRef]
- Schmidt, C.; Pacher, M.; Puchta, H. DNA break repair in plant and its application for genome engineering. In Transgenic Plants. Methods in Molecular Biology; Kumar, S., Barone, P., Smith, M., Eds.; Humana Press: New York, NY, USA, 2019; Volume 1864, pp. 237–266. [Google Scholar] [CrossRef]
- Mannuss, A.; Trapp, O.; Puchta, H. Gene regulation in response to DNA damage. Biochim. Biophys. Acta 2012, 1819, 154. [Google Scholar] [CrossRef] [PubMed]
- Schröpfer, S.; Knoll, A.; Trapp, O.; Puchta, H. DNA Repair and Recombination in Plants. In Molecular Biology; Plant Sciences book series; Springer: New York, NY, USA, 2014; Volume 2, pp. 51–93. [Google Scholar] [CrossRef]
- Gorbunova, V.; Levy, A. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 1999, 4, 263–269. [Google Scholar] [CrossRef]
- Bray, C.M.; West, C.E. DNA repair mechanisms in plants: Crucial sensors and effectors for the maintenance of genome integrity. New Phytol. 2005, 168, 511–528. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, R.; Onyango, D.; Stark, J. Regulation of Single Strand Annealing and its role in genome maintenance. Trends Genet. 2016, 32, 566–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mladenov, E.; Iliakis, G. Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutat. Res. 2011, 711, 61–72. [Google Scholar] [CrossRef]
- Waterworth, W.; Drury, G.; Bray, C.; West, C. Repairing breaks in the plant genome: The importance of keeping it together. New Phytol. 2011, 192, 805–822. [Google Scholar] [CrossRef]
- Shen, H.; Li, Z. DNA double-strand break repairs and their application in plant DNA integration. Genes 2022, 13, 322. [Google Scholar] [CrossRef] [PubMed]
- Knoll, A.; Fauser, F.; Puchta, H. DNA recombination in somatic plant cells: Mechanisms and evolutionary consequences. Chromosome Res. 2014, 22, 191–201. [Google Scholar] [CrossRef]
- Sanchez-Moran, E.; Armstrong, S.J. Meiotic chromosome synapsis and recombination in Arabidopsis thali-ana: New ways of integrating cytological and molecular approaches. Chromosome Res. 2014, 22, 179–190. [Google Scholar] [CrossRef]
- Chen, H.; He, C.; Wang, C.; Wang, X.; Ruan, F.; Yan, J.; Yin, P.; Wang, Y.; Yan, S. RAD51 supports DMC1 by inhibiting the SMC5/6 complex during meiosis. Plant Cell 2021, 33, 2869–2882. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.; Barbas, C.F., 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacher, M.; Puchta, H. From classical mutagenesis to nuclease-based breeding–directing natural DNA repair for a natural end-product. Plant J. 2017, 90, 819–833. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Lin, Q.; Jin, S.; Gao, C. The CRISPR-Cas toolbox and gene editing technologies. Mol. Cell 2022, 82, 333–347. [Google Scholar] [CrossRef]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.; Danielsen, J.; Yang, Y.; Qi, Y. A role for small RNAs in DNA double-strand break repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Ou, X.; Han, D.; He, Z.; Liu, S.; Mao, N.; Zhang, Z.; Peng, C.L.; Lai, J.; Yang, C. A diRNA–protein scaffold module mediates SMC5/6 recruitment in plant DNA repair. Plant Cell 2022, 34, 3899–3914. [Google Scholar] [CrossRef] [PubMed]
- Serrano, D.; Cordero, G.; Kawamura, R.; Sverzhinsky, A.; Sarker, M.; Roy, S.; Malo, C.; Pascal, J.M.; Marko, J.F.; D’Amours, D. The Smc5/6 Core Complex Is a Structure-Specific DNA Binding and Compacting Machine. Mol. Cell. 2020, 80, 1025–1038.e5. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Mao, N.; Hu, H.; Tang, J.; Han, D.; Liu, S.; Wu, Q.; Liu, Y.; Peng, C.; Lai, J.; et al. A SWI/SNF subunit regulates chromosomal dissociation of structural maintenance complex 5 during DNA repair in plant cells. Proc. Natl. Acad. Sci. USA 2019, 116, 15288–15296. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, S.; Umeda, M. Cell-cycle control and plant development. Int. Rev. Cell Mol. Biol. 2011, 291, 227–261. [Google Scholar] [CrossRef]
- Gentric, N.; Genschik, P.; Noir, S. Connections between the Cell Cycle and the DNA Damage Response in Plants. Int. J. Mol. Sci. 2021, 22, 9558. [Google Scholar] [CrossRef]
- Andersen, S.U.; Buechel, S.; Zhao, Z.; Ljung, K.; Novák, O.; Busch, W.; Schuster, C.; Lohmann, J.U. Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 2008, 20, 88–100. [Google Scholar] [CrossRef] [Green Version]
- Schnittger, A.; De Veylder, L. The Dual Face of Cyclin B1. Trends Plant Sci. 2018, 23, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Ogita, N.; Takahashi, T.; Taniguchi, S.; Tanaka, M.; Seki, M.; Umeda, M. A regulatory module controlling stress-induced cell cycle arrest in Arabidopsis. eLife 2019, 9, e43944. [Google Scholar] [CrossRef] [PubMed]
- Lang, L.; Pettkó-Szandtner, A.; Tunçay Elbaşı, H.; Takatsuka, H.; Nomoto, Y.; Zaki, A.; Dorokhov, S.; De Jaeger, G.; Eeckhout, D.; Ito, M.; et al. The DREAM complex represses growth in response to DNA damage in Arabidopsis. Life Sci. Alliance. 2021, 4, e202101141. [Google Scholar] [CrossRef] [PubMed]
- Cools, T.; Iantcheva, A.; Weimer, A.K.; Boens, S.; Takahashi, N.; Maes, S.; Van den Daele, H.; Van Isterdael, G.; Schnittger, A.; De Veylder, L. The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell 2011, 23, 1435–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Schutter, K.; Joubès, J.; Cools, T.; Verkest, A.; Corellou, F.; Babiychuk, E.; Van Der Schueren, E.; Beeckman, T.; Kushnir, S.; Inzé, D.; et al. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 2007, 19, 211–225. [Google Scholar] [CrossRef] [Green Version]
- Pan, T.; Qin, Q.; Nong, C.; Gao, S.; Wang, L.; Cai, B.; Zhang, M.; Wu, C.; Chen, H.; Li, T.; et al. A novel WEE1 pathway for replication stress responses. Nat. Plants 2021, 7, 209–218. [Google Scholar] [CrossRef]
- Wang, L.; Zhan, L.; Zhao, Y.; Huang, Y.; Wu, C.; Pan, T.; Qin, Q.; Xu, Y.; Deng, Z.; Li, J.; et al. The ATR-WEE1 kinase module inhibits the MAC complex to regulate replication stress response. Nucleic Acids Res. 2021, 49, 1411–1425. [Google Scholar] [CrossRef]
- De Veylder, L.; Larkin, J.C.; Schnittger, A. Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 2011, 16, 624–634. [Google Scholar] [CrossRef]
- Lang, L.; Schnittger, A. Endoreplication—A means to an end in cell growth and stress response. Curr. Opin. Plant Biol. 2020, 54, 85–92. [Google Scholar] [CrossRef]
- Fox, D.T.; Duronio, R.J. Endoreplication and polyploidy: Insights into development and disease. Development 2013, 140, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto-Shirasu, K.; Stacey, N.J.; Corsar, J.; Roberts, K.; McCann, M.C. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 2002, 12, 1782–1786. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto-Shirasu, K.; Roberts, G.R.; Stacey, N.J.; McCann, M.C.; Maxwell, A.; Roberts, K. RHL1 is an essential component of the plant DNA topoisomerase VI complex and is required for ploidy-dependent cell growth. Proc. Natl. Acad. Sci. USA 2005, 102, 18736–18741. [Google Scholar] [CrossRef] [Green Version]
- Kirik, V.; Schrader, A.; Uhrig, J.F.; Hulskamp, M. MIDGET unravels functions of the Arabidopsis topoiso-merase VI complex in DNA endoreduplication, chromatin condensation, and transcriptional silencing. Plant Cell. 2007, 19, 3100–3110. [Google Scholar] [CrossRef] [Green Version]
- Endo, M.; Nakayama, S.; Umeda-Hara, C.; Ohtsuki, N.; Saika, H.; Umeda, M.; Toki, S. CDKB2 is involved in mitosis and DNA damage response in rice. Plant J. 2012, 69, 967–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danon, A.; Delorme, V.; Mailhac, N.; Gallois, P. Plant programmed cell death: A common way to die. Plant Physiol. Biochem. 2000, 38, 647–655. [Google Scholar] [CrossRef]
- Daneva, A.; Gao, Z.; Van Durme, M.; Nowack, M.K. Functions and Regulation of Programmed Cell Death in Plant Development. Annu. Rev. Cell Dev. Biol. 2016, 32, 441–468. [Google Scholar] [CrossRef] [PubMed]
- Burke, R.; Schwarze, J.; Sherwood, O.L.; Jnaid, Y.; McCabe, P.F.; Kacprzyk, J. Stressed to Death: The Role of Transcription Factors in Plant Programmed Cell Death Induced by Abiotic and Biotic Stimuli. Front. Plant Sci. 2020, 11, 1235. [Google Scholar] [CrossRef]
- Chowdhury, I.; Tharakan, B.; Bhat, G.K. Current concepts in apoptosis: The physiological suicide program revisited. Cell Mol. Biol. Lett. 2006, 11, 506–525. [Google Scholar] [CrossRef]

| Protein | Full Name | Acc. No. | Function |
|---|---|---|---|
| FPG | FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE | At1g52500 | Bifunctional glycosylases (glycosylase and AP lyase activity) |
| OGG1 | 8-OXOGUANINE-DNA GLYCOSYLASE1 | At1g21710 | |
| NTH1 | ENDONUCLEASE III 1 | At2g31450 | |
| NTH2 | ENDONUCLEASE III 2 | At1g05900 | |
| DME | DEMETER | At5g04560 | |
| DML2 | DEMETER-LIKE PROTEIN 2 | At3g10010 | |
| DML3 | DEMETER-LIKE PROTEIN 3 | At4g34060 | |
| ROS1 | REPRESSOR OF SILENCING 1 | At2g36490 | |
| UNG (UDG) | URACIL DNA GLYCOSYLASE | At3g18630 | Monofunctional glycosylases |
| AAG | ALKYLADENINE-DNA GLYCOSYLASE | At3g12040 | |
| MBD4L | METHYL-CPG-BINDING DOMAIN PROTEIN 4 LIKE | At3g07930 | |
| ARP | APURINIC ENDONUCLEASE-REDOX PROTEIN | At2g41460 | AP endonucleases |
| APE1L | APURINIC/APYRIMIDINIC ENDONUCLEASE1-LIKE PROTEIN | At3g48425 | |
| APE2 | APURINIC/APYRIMIDINIC ENDONUCLEASE2 | At4g36050 | |
| ZDP | ZINC 4 FINGER DNA 3′-PHOSPHOESTERASE | At3g14890 | 3′ DNA phosphatase |
| XRCC1 | HOMOLOG OF X-RAY REPAIR CROSS COMPLEMENTING 1 | At1g80420 | Stimulation of phosphatase activity of ZDP and enhancement of DNA ligation by interaction with LIG1 |
| FEN1 (SAV6) | FLAP ENDONUCLEASE I | At5g26680 | Flap endonuclease |
| Pol δ | DNA POLYMERASE DELTA | At1g09815 At2g42120 At5g63960 | Polymerases |
| Pol ε | DNA POLYMERASE EPSILON | At1g08260 At2g27120 At5g22110 | |
| Pol λ | DNA POLYMERASE LAMBDA | At1g10520 | |
| LIG1 | LIGASE 1 | At1g08130 | Ligase involved in the final ligation of DNA ends in SP and LP-BER |
| LIG4 | LIGASE 4 | At5g57160 | Ligases critical for seed longevity |
| LIG6 | LIGASE 6 | At1g66730 |
| Protein | Full Name | Acc. No. | Function | |
|---|---|---|---|---|
| XPC-HR23B-CEN2 complex | RAD4 (XPC homolog) | RADIATION SENSITIVE PROTEIN 4 | At5g16630 | Recognition of lesions in GG-NER (Global Genomic Repair–NER) |
| RAD23A | RADIATION SENSITIVE PROTEIN 23A | At1g16190 | ||
| RAD23B | RADIATION SENSITIVE PROTEIN 23B | At1g79650 | ||
| RAD23C | RADIATION SENSITIVE PROTEIN 23C | At3g02540 | ||
| RAD23D | RADIATION SENSITIVE PROTEIN 23D | At5g38470 | ||
| CEN2 | CENTRIN 2 | At4g37010 | ||
| DDB complex | DDB1A | DAMAGED DNA-BINDING PROTEIN 1A | At4g05420 | |
| DDB1B | DAMAGED DNA-BINDING PROTEIN 1B | At4g21100 | ||
| DDB2 | DAMAGED DNA-BINDING PROTEIN 2 | At5g58760 | ||
| CSA1 | COCKAYNE SYNDROME GROUP A 1 | At1g27840 | Recognition of lesions in TC-NER (Transcriptional-Coupled Repair–NER) | |
| CSA2 | COCKAYNE SYNDROME GROUP A 2 | At1g19750 | ||
| CHR8 (CSB homolog) | CHROMATIN REMODELING 8 | At2g18760 | ||
| CHR24 (CSB homolog) | CHROMATIN REMODELING 24 | At5g63950 | ||
| Preincision complex | XPB1 | XERODERMA PIGMENTOSUM GROUP B 1 | At5g41370 | TFIIH core complex with helicase activity |
| XPB2 | XERODERMA PIGMENTOSUM GROUP B 2 | At5g41360 | ||
| XPD (UVH6) | XERODERMA PIGMENTOSUM GROUP D (ULTRAVIOLET HYPERSENSITIVE 6) | At1g03190 | ||
| TTDA | TRICHOTHIODYSTROPHY GROUP A | At1g12400 At1g62886 | ||
| GTF2H2 | GENERAL TRANSCRIPTION FACTOR II H2 | At1g05055 | ||
| At1g18340 | At1g18340 | |||
| At1g55750 | At1g55750 | |||
| At3g61420 | At3g61420 | |||
| At4g17020 | At4g17020 | |||
| TTDA | TRICHOTHIODYSTROPHY GROUP A | At1g12400 | ||
| At1g62886 | ||||
| CDKD1;1 | CYCLIN-DEPENDENT KINASE D1; 1 | At1g73690 | Module of THIIH with kinase activity | |
| CDKD1;2 | CYCLIN-DEPENDENT KINASE D1; 2 | At1g66750 | ||
| CDKD1;3 | CYCLIN-DEPENDENT KINASE D1; 3 | At1g18040 | ||
| CYCH;1 | CYCLIN H;1 | At5g27620 | ||
| MAT1 | MÉNAGE À TROIS | At4g30820 | ||
| UVH1 (XPF homolog) | ULTRAVIOLET HYPERSENSITIVE 1 | At5g41150 | Nucleases performing incisions 5′ or 3′ to the lesion | |
| UVH3 (XPG homolog) | ULTRAVIOLET HYPERSENSITIVE 3 | At3g28030 | ||
| ERCC1 | EXCISION REPAIR CROSS-COMPLEMENTATION GROUP 1 | At3g05210 | ||
| RPA (three subunits: RPA1/2/3) | REPLICATION PROTEIN A 1 | Multiple loci *: At2g06510 At5g08020 At5g45400 At5g61000 At4g19130 At2g24490 At3g02920 At3g52630 At4g18590 | Complex involved in the excision of the damaged nucleotide | |
| REPLICATION PROTEIN A 2 | ||||
| REPLICATION PROTEIN A 3 | ||||
| PCNA1 | PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1 | At1g07370 | Cofactors of polymerases | |
| PCNA2 | PROLIFERATING CELL NUCLEAR ANTIGEN 2 | At2g29570 | ||
| RFC (five subunits: RFC1/2/3/4/5) | REPLICATION FACTOR C | At1g21690 At1g63160 At1g77470 At5g22010 At5g27740 | Complex involved in the loading of PCNAs onto the DNA strand | |
| POL δ | DNA POLYMERASE DELTA | see Table 1 | Polymerases | |
| Pol ε | DNA POLYMERASE EPSILON | see Table 1 | ||
| LIG1 | LIGASE 1 | At1g08130 | Ligase | |
| Protein | Full Name | Acc. No. | Function | |
|---|---|---|---|---|
| MutS complexes | MSH2 | MUTS HOMOLOG 2 | At3g18524 | Recognition of mismatches |
| MSH3 | MUTS HOMOLOG 3 | At4g25540 | ||
| MSH6 | MUTS HOMOLOG 6 | At4g02070 | ||
| MSH7 | MUTS HOMOLOG 7 | At3g24495 | ||
| MutLα complex | MLH1 | MUTL HOMOLOG 1 | At4g09140 | Endonuclease activity |
| PMS1 | POSTMEIOTIC SEGREGATION 1 | At4g02460 | ||
| EXO1 | EXONUCLEASE 1 | At1g29630 | Exonuclease | |
| RFC | REPLICATION FACTOR C | see Table 2 | Complex involved in the loading of PCNAs onto the DNA strand | |
| PCNA1 | PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1 | At1g07370 | Cofactors of polymerase | |
| PCNA2 | PROLIFERATING CELLULAR NUCLEAR ANTIGEN 2 | At2g29570 | ||
| DNA2 (JHS1) | JING HE SHENG 1 | Helicase and nuclease activity | ||
| RPA | REPLICATION PROTEIN A | see Table 2 | Complex involved in the excision of the lesion | |
| POL δ | DNA POLYMERASE DELTA | see Table 1 | Polymerase | |
| LIG1 | LIGASE 1 | At1g08130 | Ligase | |
| Protein | Full Name | Acc. No. | Function | |
|---|---|---|---|---|
| MRN complex | MRE11 | MEIOTIC RECOMBINATION 11 | At5g54260 | Bind to DNA ends and preserve them in close proximity, induce excision of 5′ end |
| RAD50 | RADIATION SENSITIVE PROTEIN 50 | At2g31970 | ||
| NBS1 | NIJMEGEN BREAKAGE SYNDROME 1 | At3g02680 | ||
| RAD51 | RADIATION SENSITIVE PROTEIN 51 | At5g20850 | Recombinase initiating homology search and strand invasion | |
| RPA | REPLICATION PROTEIN A | see Table 2 | Coats 3′ overhangs–protection from exonucleolytic degradation | |
| GR1/COM1 | GAMMA RESPONSE 1 | At3g52115 | DSB end processing | |
| EXO1 | EXONUCLEASE 1 | At1g29630 At1g18090 | ||
| RECQ4A RECQ4B | ATP-DEPENDENT DNA HELICASE Q-LIKE 4A AND 4B | At1g10930 At1g60930 | ||
| RAD52 | RADIATION SENSITIVE PROTEIN 52 | At1g71310 At5g47870 | Recombination mediators | |
| BRCA1 | BREAST CANCER 1 | At4g21070 | ||
| BRCA2 | BREAST CANCER 2 | At5g01630 At4g00020 | ||
| RAD51B | RADIATION SENSITIVE PROTEIN 51B | At2g28560 | ||
| RAD51C | RADIATION SENSITIVE PROTEIN 51C | At2g45280 | ||
| RAD51D | RADIATION SENSITIVE PROTEIN 51D | At1g07745 | ||
| XRCC2 | X-RAY REPAIR CROSS COMPLEMENTING 2 | At5g64520 | ||
| XRCC3 | X-RAY REPAIR CROSS COMPLEMENTING 3 | At5g54750 | ||
| RAD54 | RADIATION SENSITIVE PROTEIN 54 | At3g19210 | dsDNA translocase–stimulates the D-loop formation | |
| PCNA1 | PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1 | At1g07370 | Cofactors of polymerases | |
| PCNA2 | PROLIFERATING CELL NUCLEAR ANTIGEN 2 | At2g29570 | ||
| RFC | REPLICATION FACTOR C | see Table 2 | Complex involved in the loading of PCNAs onto the DNA strand | |
| POL δ | DNA POLYMERASE DELTA | See Table 1 | Polymerase | |
| SRS2 | SUPPRESSOR OF RAD SIX-SCREEN MUTANT 2 | At4g25120 | Helicase activity | |
| FANCM | FANCONI ANEMIA GROUP M PROTEIN | At1g35530 | ||
| EME1 | ESSENTIAL MEIOTIC ENDONUCLEASE 1 | At2g21800 At2g22140 | Endonuclease activity | |
| MUS81 | MMS AND UV SENSITIVE 81 | At4g30870 | ||
| GEN1 | GEN1 | At1g01880 | Holliday junction 5′ flap endonuclease | |
| SEND1 | SINGLE-STRAND DNA ENDONUCLEASE1 | At3g48900 | ||
| TOP3α | TOPOISOMERASE 3 ALPHA | At5g63920 | Together with RECQ4A involved in HJ dissolution | |
| RMI1 | RECQ-MEDIATED GENOME INSTABILITY PROTEIN 1 | At5g63540 | ||
| Protein | Full Name | Acc. No. | Function |
|---|---|---|---|
| KU70 | KU70 HOMOLOG | At1g16970 | KU70-KU80 heterodimer bind and protect DSB ends |
| KU80 | KU80 HOMOLOG | At1g48050 | |
| PARP | POLY ADP-RIBOSE POLYMERASE | At2g31320 At4g02390 At5g22470 | DNA end binding NAD + ADP-ribosyltransferase |
| MRN complex (MRE11-RAD50-NBS1) | MRE11 | At5g54260 | Bind to DNA ends and preserve them in close proximity, induce excision of 5′end |
| RAD50 | At2g31970 | ||
| NBS1 | At3g02680 | ||
| Snm1 | SENSITIVE TO NITROGEN MUSTARD 1 | At3g26680 | DNA end processing |
| ZDP | ZINC 4 FINGER DNA 3′-PHOSPHOESTERASE | At3g14890 | |
| Rad9 | RADIATION SENSITIVE PROTEIN 9 | At3g05480 | |
| Pol λ | POLYMERASE λ | At1g10520 | Polymerase |
| POLθ | POLYMERASE θ | At4g32700 | |
| LIG4 | LIGASE 4 | At5g57160 | Ligase |
| XRCC4 | X-RAY REPAIR CROSS COMPLEMENTING 4 | At3g23100 | Complex with LIG4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szurman-Zubrzycka, M.; Jędrzejek, P.; Szarejko, I. How Do Plants Cope with DNA Damage? A Concise Review on the DDR Pathway in Plants. Int. J. Mol. Sci. 2023, 24, 2404. https://doi.org/10.3390/ijms24032404
Szurman-Zubrzycka M, Jędrzejek P, Szarejko I. How Do Plants Cope with DNA Damage? A Concise Review on the DDR Pathway in Plants. International Journal of Molecular Sciences. 2023; 24(3):2404. https://doi.org/10.3390/ijms24032404
Chicago/Turabian StyleSzurman-Zubrzycka, Miriam, Paulina Jędrzejek, and Iwona Szarejko. 2023. "How Do Plants Cope with DNA Damage? A Concise Review on the DDR Pathway in Plants" International Journal of Molecular Sciences 24, no. 3: 2404. https://doi.org/10.3390/ijms24032404
APA StyleSzurman-Zubrzycka, M., Jędrzejek, P., & Szarejko, I. (2023). How Do Plants Cope with DNA Damage? A Concise Review on the DDR Pathway in Plants. International Journal of Molecular Sciences, 24(3), 2404. https://doi.org/10.3390/ijms24032404






