Abstract
Mitochondria are double membrane-bound organelles that play critical functions in cells including metabolism, energy production, regulation of intrinsic apoptosis, and maintenance of calcium homeostasis. Mitochondria are fascinatingly equipped with their own genome and machinery for transcribing and translating 13 essential proteins of the oxidative phosphorylation system (OXPHOS). The rest of the proteins (99%) that function in mitochondria in the various pathways described above are nuclear-transcribed and synthesized as precursors in the cytosol. These proteins are imported into the mitochondria by the unique mitochondrial protein import system that consists of seven machineries. Proper functioning of the mitochondrial protein import system is crucial for optimal mitochondrial deliverables, as well as mitochondrial and cellular homeostasis. Impaired mitochondrial protein import leads to proteotoxic stress in both mitochondria and cytosol, inducing mitochondrial unfolded protein response (UPRmt). Altered UPRmt is associated with the development of various disease conditions including neurodegenerative and cardiovascular diseases, as well as cancer. This review sheds light on the molecular mechanisms underlying the import of nuclear-encoded mitochondrial proteins, the consequences of defective mitochondrial protein import, and the pathological conditions that arise due to altered UPRmt.
1. Introduction
Mitochondria are organelles present in almost all eukaryotic cells, and their number per cell depends on their energy demand. Organs with high metabolic activity, for example, heart muscles, kidneys, and the brain, contain the largest number of mitochondria [1,2]. Mitochondria are believed to be the descendants of an ancient prokaryote that underwent an endosymbiotic event with early eukaryotes [3]. Apart from their role in energy production, mitochondria are involved in numerous metabolic processes, including the biosynthesis of amino acids, lipids, heme, and Fe-S clusters [4]. In addition, they also play crucial functions in programmed cell death and maintenance of calcium homeostasis [4,5]. A mitochondrion is a double membrane-bound structure consisting of an outer membrane, intermembrane space (IMS), inner membrane, and matrix [6]. The outer membrane is permeable to solutes up to approximately 5 kDa and characterized by the presence of various enzymes and channels, such as carnitine palmitoyltransferase I, acyl-CoA synthetase, voltage-dependent anion channel (VDAC), and mitochondrial apoptosis-induced channel (MAC) [7]. On the other hand, the inner membrane is impermeable except through specific transporters and contains the enzyme complexes responsible for oxidative phosphorylation [6,8]. The IMS consists of enzymes including caspases, adenylyl kinase, and cytochrome c. Similarly, the mitochondrial matrix consists of several enzymes that take part in metabolic processes such as the tricarboxylic acid cycle and β oxidation of fatty acids.
Each mitochondrion has its own genome, which is a 16.5 kb double-stranded, closed circular DNA present in the mitochondrial matrix. The mtDNA is strictly maternally inherited and packaged into nucleoids, which is done to ensure its proper distribution and propagation [9]. The mitochondrial genome consists of 37 genes that encode approximately 1% of the total mitochondrial proteins (13 OXPHOS proteins), 2 ribosomal RNAs (12S and 16S rRNA), and 22 transfer RNAs [6]. The remaining 99% of mitochondrial proteins (~1500) are encoded by the nuclear genome, synthesized in the cytosol, and imported into the mitochondria by the mitochondrial protein import system (Figure 1). It was previously believed that all mitochondrial precursor proteins are imported via one main pathway and mechanism [10]; however, further studies revealed the existence of a mitochondrial protein import system comprising several machineries responsible for importing mitochondrial proteins into diverse mitochondrial compartments through various mechanisms specific to each machinery.

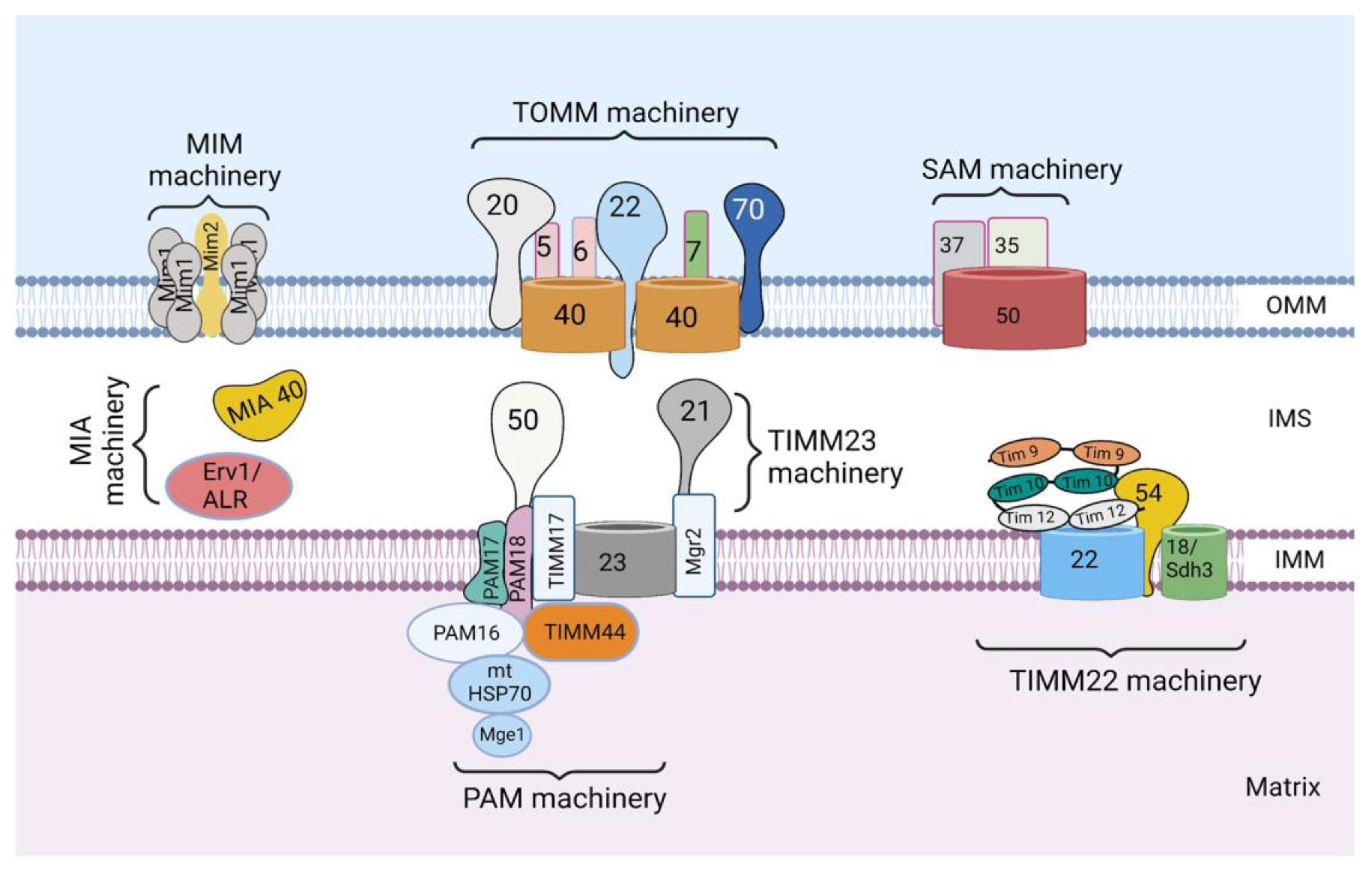
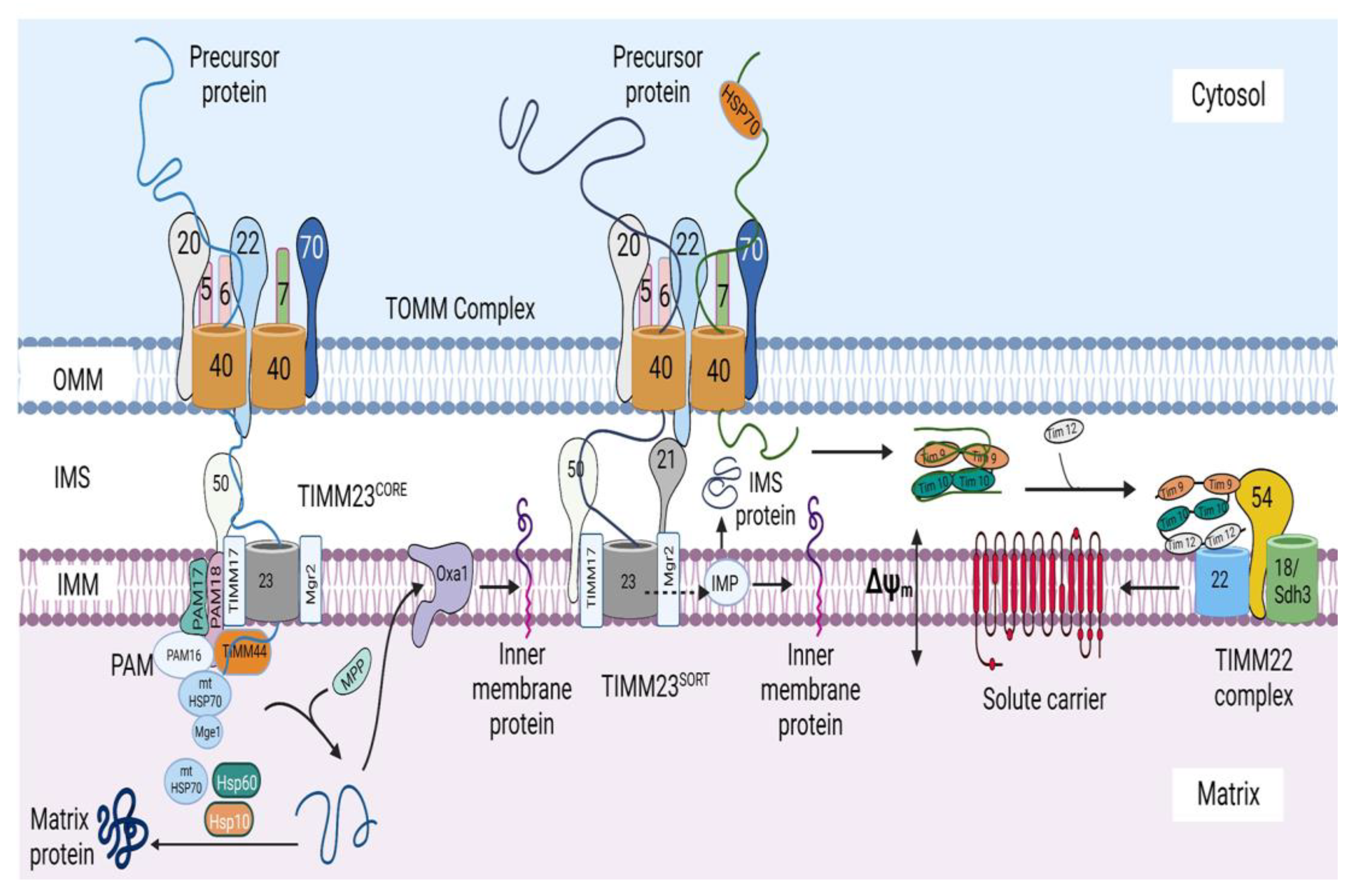
Figure 1.
The mitochondrial protein import system. The mitochondrial protein import system consists of seven machineries, including the translocase of the outer mitochondrial membrane (TOMM) machinery, mitochondrial import machinery (MIM), sorting and assembly machinery (SAM), mitochondrial intermembrane space import and assembly machinery (MIA), translocase of the inner mitochondrial membrane 23 (TIMM 23) machinery, translocase of the inner mitochondrial membrane 22 (TIMM 22) machinery, and a presequence-associated motor (PAM). The figure was created with Biorender.com.
Proper functioning of mitochondria hinges on the efficient operation of the mitochondrial protein import system. Hence, impaired mitochondrial protein import leads to certain consequences including defective mitochondrial operations and proteotoxic stress in both the mitochondrial and cytosolic compartments, which are corrected by certain responses including mitochondrial unfolded protein response (UPRmt). Altered UPRmt is linked to the development of various pathological conditions including neurodegenerative and cardiovascular diseases, as well as cancer. Here, we review the current understanding of mechanisms underlying the import of nuclear-encoded mitochondrial proteins, the consequences of impaired mitochondrial protein import, and different disease conditions that develop due to altered UPRmt.
2. Mitochondrial Protein Import System
The mitochondrial protein import system consists of seven machineries: the translocase of the outer mitochondrial membrane (TOMM) machinery, mitochondrial import machinery (MIM), sorting and assembly machinery (SAM), mitochondrial intermembrane space import and assembly machinery (MIA), translocase of the inner mitochondrial membrane 23 (TIMM 23) machinery, translocase of the inner mitochondrial membrane 22 (TIMM 22) machinery, and a presequence-associated motor (PAM) (Figure 1). These machineries are multiprotein complexes composed of protein subunits, which perform specific roles.
The understanding of how the mitochondrial protein import system works is still evolving; however, the TOMM machinery appears to be the most important, as it is the first to come in contact with majority of the nuclear-encoded mitochondrial proteins to allow their entry into the intermembrane space [4]. Subsequently, these proteins make use of any of the other machineries to get to their final destination. The choice of the next machinery depends on the targeting signal and the destination of the protein. MIM promotes the import of signal-anchored and polytopic outer-membrane proteins, whereas SAM is responsible for the insertion of β-barrel proteins in the OMM [4,11]. MIA machinery promotes the import of many intermembrane space proteins. On the other hand, proteins destined for the inner mitochondrial membrane (IMM) are inserted into the IMM by either TIMM23 or TIMM22 machinery [4,11]. Furthermore, matrix proteins are transported into the matrix through the cooperation of TIMM23 and PAM machineries.
3. Protein Sorting at the Outer Mitochondrial Membrane
The mitochondrial outer membrane possesses two types of integral membrane proteins, including β-barrel proteins that are integrated into the OMM by multiple transmembrane β strands, and α-helical proteins, which are anchored in the OMM by one or more hydrophobic α-helical segments [4,10].
3.1. Sorting of β-Barrel Proteins into the Outer Mitochondrial Membrane
The presence of β-barrel proteins in the OMM is a key feature of the outer membrane of Gram-negative bacteria, reflecting the origin of mitochondria from prokaryotes [12]. Examples of these proteins are voltage-dependent anion channel (VDAC), TOMM40, and SAM50 [4]. The first machinery involved in the import of these β-barrel precursors is the TOMM machinery, which is composed of four receptors—TOMM20, TOMM70, and two molecules of TOMM22, as well as two molecules of the transmembrane channel—TOMM40, and three small subunits—TOMM5, TOMM6, and TOMM7—which are essential for complex stability and assembly [4,13]. However, it is unknown whether both TOMM40 channels take part in the translocation of incoming precursors across the OMM. In addition, how the two TOMM22 receptors cooperate during the recognition and binding of the incoming precursors remains to be elucidated.
After synthesis in the cytosol, the β-barrel precursors are recognized by the TOMM receptors and guided through the TOMM40 channel, through which they enter the IMS [12]. The identification of these β-barrel precursors by the TOMM receptors is directed by a targeting signal that consists of a β-hairpin element containing two adjacent β-strands, which are the two most C-terminal β strands of the precursor, and the connecting loop [4,14]. Although the exact sequence of the recognition of the targeting signal by the TOMM receptors is not completely understood, TOMM20, 70, and 22 have been shown to be crucial for the import of β barrel precursors [15,16]. Upon translocation through the TOMM40 channel, the precursors are bound to the small TIMM chaperones of the IMS, which exist as heterohexameric complexes, including the TIMM9–TIMM10 and TIMM8–TIMM13 complexes [4,12]. Of these, the TIMM9–TIMM10 complexes have been identified to be the main form involved in transfer of many hydrophobic proteins through the IMS [4,17]. These chaperones protect the β-barrel precursors from aggregation in the aqueous IMS and deliver the β-barrel precursors to the SAM complex (Figure 2A) [4,12], which consists of a membrane-integrated protein, SAM50, and two peripheral membrane proteins exposed to the cytosol, SAM35 and SAM37 [4]. The exact mechanisms governing the insertion of β-barrel proteins into the OMM are still not completely understood; however, it is believed that the SAM complex is responsible for the membrane insertion of these proteins (Figure 2A).

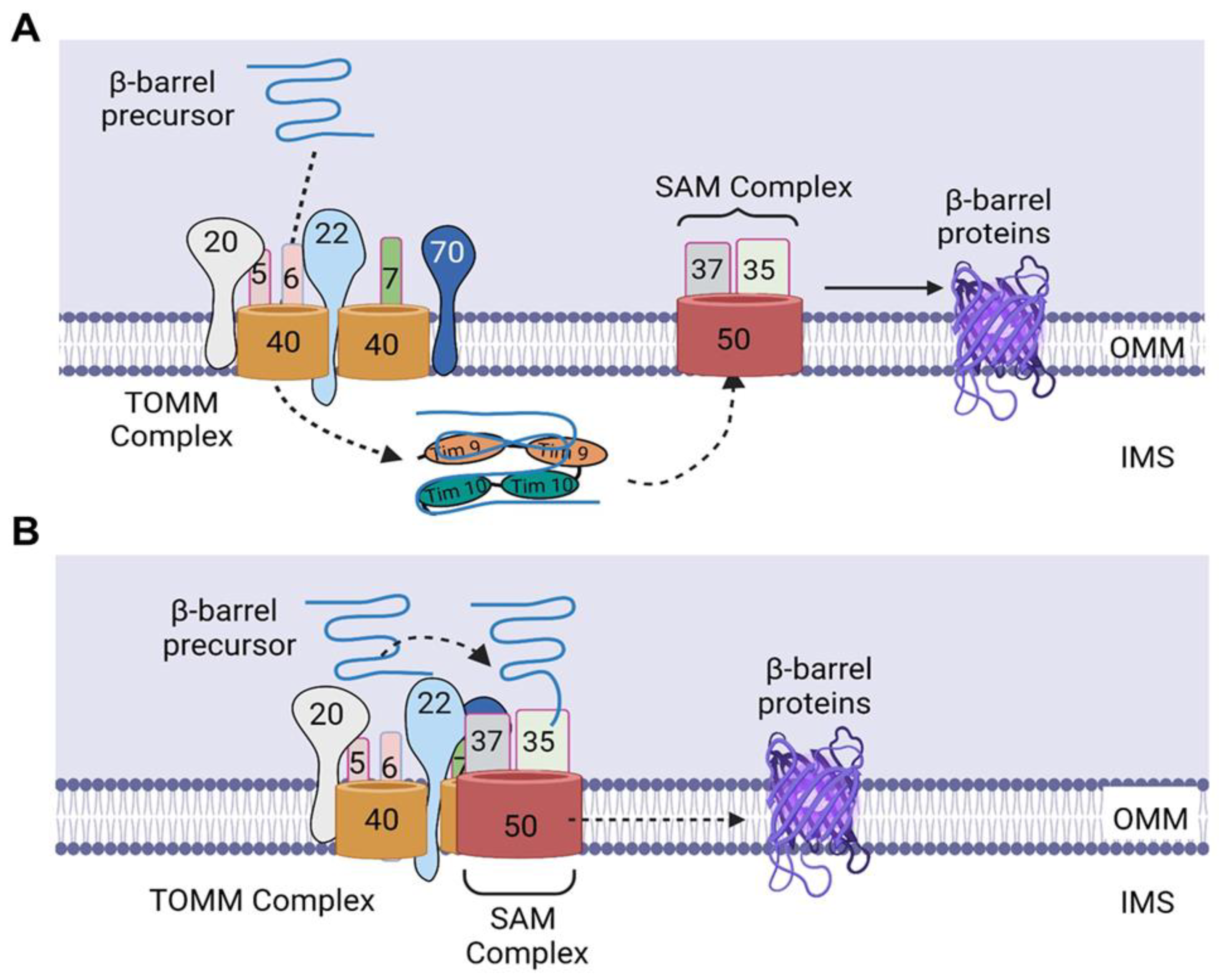
Figure 2.
Import of β-barrel precursors into the outer mitochondrial membrane. (A) Upon translocation through the TOMM40 channel, the precursors bind to TIMM9–TIMM10 complex, which protect the β-barrel precursors from aggregation in the aqueous IMS and deliver the β-barrel precursors to the SAM complex. Subsequently, the precursors are folded in the SAM complex and laterally released into the lipid phase of the outer membrane. (B) SAM37 interacts with the cytosolic receptor domain of TOMM22, thereby linking the two complexes and leading to the formation of a TOMM–SAM supercomplex, which enables the binding of SAM35 to the β signal of the precursor, thereby allowing the direct transfer of the β-barrel precursors from TOMM to the SAM complex. Subsequently, the β-barrel precursors are inserted into the SAM50 channel, after which they are folded in the SAM complex and released laterally into the lipid phase of the outer membrane. IMS, intermembrane space. The figure was created with Biorender.com.
In contrast to the understanding that the β-barrel proteins are imported first into the IMS before being transferred to the SAM complex, Kutik et al. proposed that translocation of β-barrel precursors into the SAM complex is initiated by the binding of the last β-strand (β signal) to SAM35, a SAM subunit located on the cytosolic surface of mitochondria [18,19]. This signal binding then induces a conformational change that leads to opening of the SAM50 channel; thus, several β strands can be inserted into a hydrophilic, proteinaceous membrane environment [18]. Subsequently, the precursors are folded in the SAM complex and laterally released into the lipid phase of the outer membrane [18,19]. The binding of this β signal to a SAM subunit not integrated into the lipid phase of the outer membrane but embedded into a proteinaceous membrane environment by its close association with SAM50 molecules [18] suggests that the β-barrel precursors are not imported into the IMS but transferred directly from the TOMM complex to the SAM complex (Figure 2B).
In addition to the SAM components—SAM50, SAM35, and SAM37—which are required for β-barrel formation, TOMM22 has been shown to be required for β-barrel folding, as the oxidation of TOMM40, a β-barrel protein, was observed to be impaired in mitochondria lacking TOMM22 [15]. The promotion of β-barrel folding by TOMM22 could be due to its interaction with a fraction of SAM subunits, connecting the TOMM and SAM complexes and resulting in the generation of a TOMM–SAM supercomplex [15], suggesting that the formation of the supercomplex is essential for the folding of the β barrel. In another study by Wenz et al., it was shown that SAM37 is the sole SAM subunit responsible for the formation of the TOMM–SAM supercomplex, as deletion of SAM37 blocked the copurification of other SAM subunits with TOMM22His (His-tagged TOMM22) [20]. In addition, overexpression of SAM35 did not restore the interaction of TOMM and SAM complexes in SAM37Δ mitochondria [20]. Furthermore, the authors showed that SAM37 interacts with the cytosolic receptor domain of TOMM22, linking the two complexes and leading to the formation of a TOMM–SAM supercomplex (Figure 2B) [20]. The identification of this supercomplex supports the possibility of β-barrel precursors being transferred directly from TOMM to the SAM complex, as the two complexes are brought close to each other through the formation of the supercomplex, thereby enabling the binding of SAM35 to the β signal and the subsequent insertion of the β barrel precursors in the SAM50 channel. Therefore, further mechanistic studies are needed to determine whether this direct transfer is attainable, as well as to uncover novel insights about the step-by-step mechanisms involved in the import of nuclear-encoded β-barrel mitochondrial proteins.
3.2. Sorting of α-Helical Proteins into the Outer Mitochondrial Membrane
The mechanisms involved in the import of α-helical proteins is only partly understood. Three classes of α-helical proteins have been identified, including signal-anchored proteins, tail-anchored proteins, and polytopic (multispanning) outer-membrane proteins [4]. Signal-anchored and tail-anchored proteins contain an α-helical transmembrane segment at the N terminus and C terminus, respectively, which function as both membrane anchors and targeting signals, in addition to flanking positively charged amino acid residues [4]. In contrast, polytopic proteins possess multiple transmembrane segments that may contain targeting information; however, their exact targeting signals are unknown.
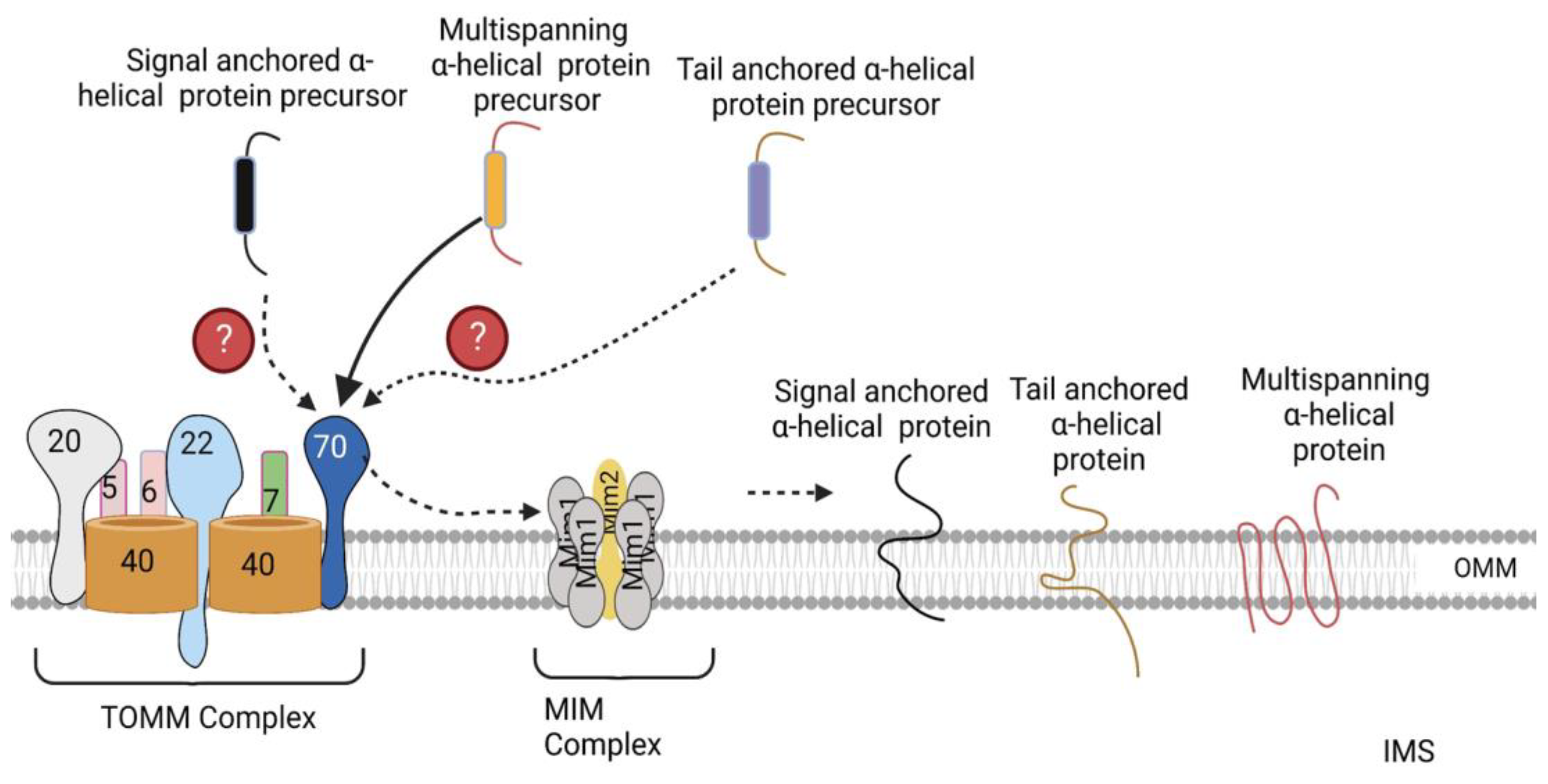
The MIM complex consisting of multiple copies of Mim1 and one or two copies of Mim2 has been identified as the machinery promoting the insertion of signal-anchored and polytopic α-helical proteins into the OMM (Figure 3) [4]. However, the molecular mechanisms through which it inserts these proteins into the OMM has not been elucidated yet [4]. No TOMM receptor has been identified to date that is required for the import of signal-anchored proteins [21,22]. In contrast, TOMM70 has been observed to recognize the precursors of polytopic proteins, after which, it binds them and transfers them to the MIM complex, which inserts them into the OMM (Figure 3) [23,24]. Previously, no proteinaceous machinery had been identified for the import of tail-anchored α-helical proteins; however, a recent study by Doan et al. showed that the import of the radiolabeled precursor of Fis1, a tail-anchored α-helical protein, into isolated mim1Δ and mim1-23 mitochondria was impaired [25]. In addition, re-expression of Mim1 in mim1Δ yeast mitochondria promoted the import of Fis1 [25]. Thus, the MIM complex is essential for the import of all three α-helical proteins. Besides the MIM complex, the low ergosterol content of the outer membrane favors the insertion of tail-anchored proteins into the OMM [26]. Likewise, the precursor of Ugo1, a polytopic protein, has been shown to be inserted into protein-free liposomes that mimic the phospholipid composition of the OMM [27], suggesting that the lipid composition of the mitochondria play a role in the optimal insertion of α-helical proteins.

Figure 3.
Import of α-helical precursors into the outer mitochondrial membrane. Polytopic proteins are recognized by the TOMM70 receptor, after which TOMM70 binds to them and transfers them to the MIM complex, which inserts them into the OMM. Signal- and tail-anchored α-helical precursors are also inserted into the OMM by the MIM complex. The exact TOMM receptors recognizing these precursors have not been identified yet. OMM, outer mitochondrial membrane. The figure was created with Biorender.com.
Some OMM proteins are inserted into the outer membrane via routes distinct from those mentioned above. For example, Mcp3, an OMM protein, contains an N-terminal presequence and a stop-transfer signal, which allows it to be imported by the TOMM and TIMM23 machineries, after which it is released laterally into the inner membrane, where it is processed by the inner-membrane peptidase (IMP). Subsequently, it is released into the IMS and exported into the outer membrane by the MIM complex [28]. This suggests that other import machineries, apart from the import machineries located on the OMM, are involved in the import of OMM proteins. Therefore, more studies are needed to identify additional machineries involved in the import of OMM proteins and the proteins imported by these machineries. Furthermore, because the import of α-helical proteins is only understood in part, more studies with the aim of achieving an improved understanding of the mechanisms underlying the import of these proteins are needed.
4. Protein Sorting into the Intermembrane Space
Many mitochondrial intermembrane space proteins contain characteristic cysteine motifs, such as Cx3C and Cx9C motifs, that form intramolecular disulfide bonds [4,29]. These proteins are kept in a reduced state in the cytosol after synthesis and translocated through the TOMM40 channel across the outer membrane [30,31]; however, none of the TOMM receptors was found to be a requirement for these proteins [31,32]. After emerging on the IMS surface of the TOMM40 channel, the IMS sorting signal of these precursors consisting of a hydrophobic element flanked by a cysteine residue is recognized by mitochondrial intermembrane space import and assembly protein 40 (MIA40), a component of the MIA machinery (Figure 4) [33]. Thereafter, MIA40 binds the precursors and facilitates their entry into the IMS, functioning as an IMS receptor [34]. In addition to aiding in the translocation of precursors into the IMS, binding of MIA40 to the precursors leads to the formation of a disulfide bond between MIA40 and the cysteine of the mitochondrial intermembrane space sorting signal [4], which is later transferred to the imported precursor (Figure 4). The transfer of disulfide bonds to the precursors leads to their oxidation and promotes their conformational stabilization and assembly in the IMS [4].

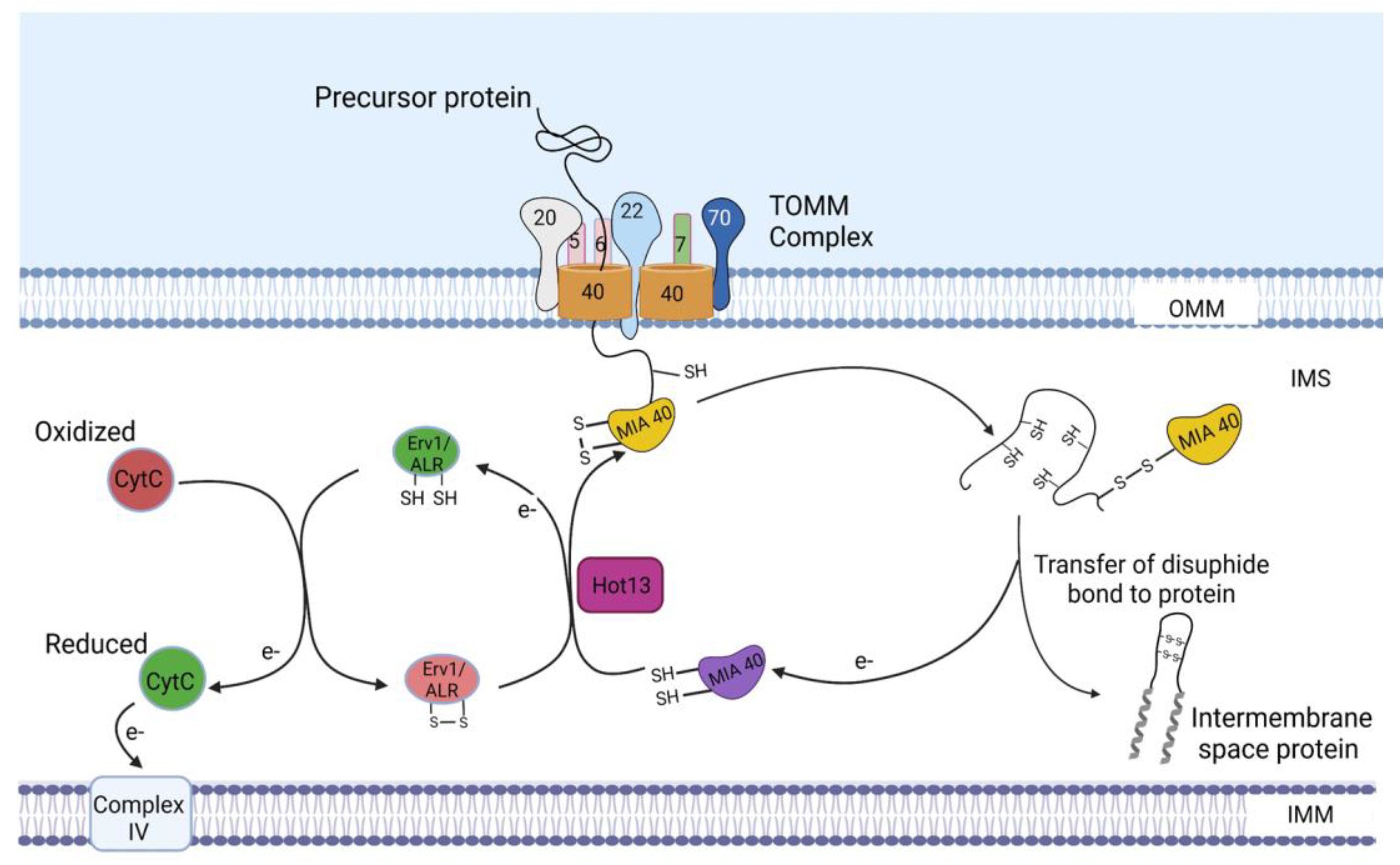
Figure 4.
Import of intermembrane space proteins. As the precursors pass through the TOMM40 channel, the IMS sorting signals of these precursors are recognized by MIA40. Thereafter, MIA40 binds the precursors and facilitates their entry into the IMS. The imported precursors are oxidized by the oxidoreductase activity of MIA40, after which they are assembled in the IMS. In turn, MIA40 becomes reduced and is reoxidized by Erv1/ALR with the assistance of the zinc-binding protein–Helper of Tim protein 13 (Hot13). Electrons derived from the oxidation of the imported precursors by MIA40 are transferred to Erv1/ALR and, subsequently, to cytochrome C and complex IV. IMS, intermembrane space; Erv1, essential for respiration and viability 1 protein; ALR, augmenter of liver regeneration. The figure was created with Biorender.com.
As these precursors become oxidized, electrons are transferred to MIA40, leading to its reduction. MIA40 then becomes reoxidized by essential for respiration and viability 1 protein/Augmenter of liver regeneration (Erv1/ALR), the second component of the MIA machinery. In addition to Erv1/ALR, the zinc-binding protein-Helper of Tim protein 13 (Hot13) promotes the oxidation of MIA40 by keeping it in a zinc-free state [35]. Oxidation of MIA40 by Erv1 leads to the transfer of a disulfide bond to MIA40, which serves as the source of the disulfide bond being transferred from MIA40 to its substrates. During this process of disulfide bond transfer from Erv1 to MIA40, electrons transferred from the oxidized substrates to MIA40 are transferred to Erv1, which then passes the electrons to cytochrome C and, subsequently, to complex IV (Figure 4) [36,37].
Apart from the MIA machinery, some IMS proteins can be imported by the TOMM/TIMM23SORT machineries (see Section 5) (Figure 5). Furthermore, some matrix and inner-membrane proteins are substrates of the MIA machinery. For example, the precursor of TIMM17, a subunit of the TIMM23 machinery, interacts with the hydrophobic binding pocket of MIA40, which facilitates its import [4,38]; however, the exact signal for recognition by MIA40 was not identified. Additionally, TIMM17 was found to be directly oxidized by Erv1, leading to the formation of a disulfide bond, which is critical for efficient protein translocation through the TIMM23 complex and dynamic gating of the TIMM23 channel [38]. The direct oxidation of TIMM17 by Erv1 raises the possibility that Erv1 may directly interact with a number of substrates of the MIA machinery [4,38], leading to the formation of disulfide bonds independent of the oxidoreductase activity of MIA40.

Figure 5.
Import of inner-mitochondrial membrane and matrix proteins. The presequence-carrying precursors are first recognized by TOMM20, which binds to them and transfers them to TOMM22, after which they are translocated through the TOMM40 channel, through which they enter the IMS, where they bind to the IMS domain of TOMM22. TIMM21 then binds the IMS domain of TOMM22, thereby promoting the dissociation of the precursors. Thereafter, TIMM50 binds to the precursor proteins and transfers the precursors into the TIMM23 channel. The Δψm exerts an electrophoretic effect on the positively charged N terminus of precursors and activates the TIMM23 channel, thereby aiding in the movement of the precursors through the TIMM23 channel. The hydrophobic sorting signals of the precursors are then recognized by Mgr2, which then binds to the sorting signals and controls the release of the precursors into the inner membrane. Subsequently, the inner-membrane peptidase (IMP) removes the hydrophobic sorting sequences, and the mature proteins are either released into the IMS or remain anchored in the inner membrane by an additional hydrophobic segment. Precursor proteins containing presequences devoid of hydrophobic sorting signals are destined for the matrix and are imported through the cooperation of the TOMM, TIMM23CORE, and PAM machineries. After translocation through the TOMM40 channel, these precursors bind to the IMS domain of TOMM22, after which TIMM50 binds these precursors and transfers them to the TIMM23 channel. TIMM44 then binds to the precursor as it emerges on the matrix side of the TIMM23 channel and transfers it to mtHsp70, which imports the protein into the matrix. The presequences are removed by the matrix processing peptidase (MMP), and the proteins are folded into their mature forms by the soluble form of mtHSP70 and the HSP60-HSP10 chaperonin complex. Oxa1 aids in the export of some of the transmembrane segments of some inner-membrane precursors from the matrix into the inner mitochondrial membrane. Following their synthesis in the cytosol, carrier precursors are bound to cytosolic chaperones of the Hsp70 and Hsp90 classes to prevent aggregation. Thereafter, these chaperones deliver the precursors to TOMM70, which then transfers this precursor to TOMM22, after which they are transferred to the TOMM40 channel and translocated across the OMM in a loop formation. The small TIM chaperones of the IMS are recruited to the channel exit by an N-terminal segment of the channel protein TOMM40. TIMM54, a subunit of the TIMM22 machinery, recruits the small TIMM chaperones to the TIMM22 complex, after which, the precursors are delivered to the TIMM22 channel. The Δψm activates the TIMM22 channel and exerts an electrophoretic effect on the carrier precursors, which aids in the movement of the precursors through the channel. Finally, the precursors are released laterally from the TIMM22 complex into the IMM. OMM, outer mitochondrial membrane; IMM, inner mitochondrial membrane, IMS, intermembrane space; Δψm, mitochondrial membrane potential. The figure was created with Biorender.com.
5. Protein Sorting at the Inner Mitochondrial Membrane and Matrix
Precursor proteins destined for the IMM contain a cleavable presequence at their N terminus, which serves as their targeting signal. Additionally, they contain a hydrophobic sorting signal, also known as a stop-transfer sequence [4]. However, not all precursors destined for the IMM contain presequences or the stop-transfer sequence. The carrier precursors are the second group of proteins destined for the IMM and contain internal targeting elements, which function as their targeting signals [4]. Both groups of IMM proteins are imported by the TOMM machinery into the IMS; however, their import into the IMM involves two different IMM-localized machineries. The precursors containing presequences are imported into the IMM by the TIMM23 machinery, whereas those containing internal targeting elements are imported into the IMM by the TIMM22 machinery [39]. Precursors destined for the matrix are synthesized with only cleavable presequences and are imported into the matrix through the cooperation of the TOMM, TIMM23, and PAM machineries [4,39].
5.1. Import of Precursors Containing Presequences
An important characteristic of mitochondrial presequences is the formation of an amphipathic α-helix that contains a positively charged face and a hydrophobic face [40]. TOMM20 forms the initial receptor for each presequence. It specifically recognizes and binds the hydrophobic surface of the amphipathic helix [41], after which the presequence is handed over to TOMM22, which binds to the positively charged surface. Subsequently, the presequence is transferred to TOMM40, through which it enters the IMS, where it binds to the IMS domain of TOMM22 [42,43] (Figure 5).
Two different forms of TIMM23 machinery have been described for the import of presequence-carrying precursors [12,44] known as TIMM23SORT and TIMM23CORE (Figure 5). The TIMM23SORT machinery is responsible for the import of presequences with a hydrophobic sorting signal into the IMM, whereas the TIMM23CORE machinery is involved in the import of presequence-carrying proteins devoid of hydrophobic sorting signals into the matrix.
5.1.1. Sorting of Precursors Containing Presequences into the Inner Mitochondrial Membrane
The TIMM23SORT machinery is composed of five subunits: TIMM21, TIMM17, TIMM50, TIMM23, and Mgr2. TIMM21 is known to bind to the IMS domain of TOMM22, thereby promoting the dissociation of the precursor, as well as bringing the TOMM and TIMM23SORT machineries in close contact [44,45]. Thereafter, TIMM50 binds to the precursor protein, after which the precursor is transferred to the TIMM23 channel, a subunit of the TIMM23 machinery, through which precursor proteins enter the IMM and the matrix. TIMM50 is also responsible for regulating the gating of the TIMM23 channel. Although the underlying mechanism is not well understood, Meinecke et al. demonstrated that TIMM50 induces the closure of the TIMM23 channel to prevent ion leakage and dissipation of mitochondrial membrane potential (Δψm) in the absence of precursors, whereas upon binding of a presequence, the channel is activated and opened [46], allowing precursor proteins to pass through. Similarly, TIMM17, a subunit known to be closely associated with TIMM23, has also been reported to regulate the gating of the TIMM23 channel [38].
Once the precursor proteins are in the TIMM23 channel, the hydrophobic sorting signal is recognized by Mgr2, which then binds to the sorting signal and controls the release of the precursor into the inner membrane [4]. Afterwards, the inner-membrane peptidase (IMP) removes the hydrophobic sorting sequence, and the mature protein is either released into the intermembrane space or remains anchored in the inner membrane by an additional hydrophobic segment (Figure 5) [4]. Thus, the TIMM23SORT complex promotes the import of precursors containing both presequences and hydrophobic sorting signals destined for the IMM or IMS. However, it is presently unclear how the presequences of these precursors are removed.
5.1.2. Sorting of Precursors Containing Presequences into the Matrix
In contrast to the TIMM23SORT machinery, the TIMM23CORE machinery lacks a TIMM21 subunit [44]. Upon translocation of precursors containing only cleavable presequences through the TOMM40 channel, these precursors bind to the IMS domain of TOMM22 [42,43]. Subsequently, TIMM50 binds these precursors and transfers them to the TIMM23 channel.
The PAM machinery composed of an ATP-driven molecular chaperone, mitochondrial heat-shock protein 70 (mtHsp70), TIMM44, PAM16, PAM17, PAM18, and nucleotide exchange factor Mge1, is responsible for completing the import of these precursors into the matrix (Figure 5) [4]. It is anchored to the TIMM23CORE machinery via the interaction between the N-terminal domain of PAM18 and TIMM17 (Figure 5). As the precursor emerges on the matrix side of the TIMM23 channel, TIMM44 binds to it and transfers it to mtHsp70, which tightly binds the unfolded polypeptide chain with its peptide-binding domain [4]. Its ATP-hydrolyzing activity is regulated by PAM18 (also known as TIMM14) and PAM16 (TIMM16), whereas Mge1 promotes the release of ADP from it to initiate a new round of the reaction cycle [4]. Although the mechanism through which the PAM machinery imports the precursors into the matrix is not completely understood, two models have been proposed, known as the trapping and pulling models. In the trapping/Brownian ratchet model, binding of mtHsp70 to the precursor in transit prevents backsliding of the polypeptide chain through the import channels, and thus, through Brownian motion, the polypeptide moves inward until another mtHsp70 molecule binds to it. The precursor is thus imported in a stepwise manner into the matrix by ATP-dependent cycles of mtHsp70 binding and release [4]. In contrast to this model, in the pulling model, the import motor plays a more active role. MtHsp70 binds to TIMM44 and interacts with the precursor in transit. Conformational changes of mtHsp70 during its ATPase cycle generate an inwardly directed force on the precursor. Afterwards, another mtHsp70 binds to the polypeptide chain, pulling it further into the matrix [4].
After the import of these proteins into the matrix, their presequences are removed by the matrix processing peptidase (MPP). In addition, other proteases, such as the octapeptidyl aminopeptidase and the intermediate cleaving peptidase (Icp55), remove destabilizing N-terminal amino acid residues from the imported proteins [4,47]. Subsequently, the soluble form of mtHSP70 and other chaperones, for example, the HSP60-HSP10 chaperonin complex, promote folding of the proteins into active forms [39,48].
Although it is understood that TIMM21 dissociates from the TIMM23SORT machinery in order for the TIMM23CORE machinery to associate with the PAM machinery for the import of matrix proteins [44], the exact mechanism underlying the decision is made to switch from a TIMM23SORT to a TIMM23CORE complex and vice versa is unknown. Thus, studies aimed at understanding how this decision is made are needed. It was suggested by van der Lann et al. that the interaction between TIMM21 and the IMS domain of TOMM22 is regulated by the import signals of incoming precursors [49]. Therefore, it is possible that TIMM21 is able to recognize the presence of a hydrophobic sorting signal in presequences, which triggers its binding to the IMS domain of TOMM22, thereby promoting the dissociation of the precursor from TOMM22. On the other hand, in the absence of a hydrophobic sorting signal in the presequence-carrying precursor, TIMM21 dissociates from the TIMM23SORT complex [44], resulting in the formation of the TIMM23CORE machinery.
Apart from the subunits of the TIMM23 machinery, the Δψm also plays critical roles in the translocation of precursors across the inner membrane by exerting an electrophoretic effect on the positively charged N terminus of precursors and activating the TIMM23 channel [50,51]. Furthermore, TIMM21 has been shown to link the TIMM23SORT complex to the respiratory chain III–IV supercomplex (bc1 complex and cytochrome c oxidase (COX)), thereby bringing the TIMM23SORT complex in close proximity to proton-pumping complexes, where the electrochemical proton gradient is higher, which, in turn, promotes precursor translocation [52].
Some inner-membrane precursors are imported into the inner membrane by a combination of stop-transfer and conservative sorting mechanisms [4]. In the conservative sorting route, precursors are imported by TOMM, TIMM23CORE, and PAM machineries into the matrix, followed by export into the inner membrane via Oxa1, the main component of the OXA machinery (Figure 5) [4]. Examples of such precursors imported via the combination of stop-transfer and conservative sorting mechanisms are multispanning inner-membrane precursors. The ABC transporter Mdl1, a multispanning inner-membrane protein, contains six α-helical transmembrane segments, of which two middle transmembrane segments are imported into the matrix in a PAM-dependent manner and inserted into the inner membrane by Oxa1, whereas the other transmembrane segments are sorted by a stop-transfer mechanism [53]. Similarly, TIMM18 and Sdh3, subunits of the TIMM22 machinery are cleavable multispanning inner-membrane proteins, which have also been shown to be imported by a combination of conservative sorting (the first two transmembrane segments) and stop transfer (the third transmembrane segment) [54].
5.2. Import of Carrier Precursors into the Inner Mitochondrial Membrane
Carrier precursors are synthesized without presequences but contain several internal targeting elements distributed over the mature primary structure, which function as targeting signals [4,10]. Examples include ADP/ATP carrier and phosphate carrier. After synthesis in the cytosol, the carrier precursors are bound to cytosolic chaperones of the Hsp70 and Hsp90 classes to prevent aggregation [4]. Thereafter, these chaperones deliver the precursors to TOMM70 [55], which possesses binding sites for both the precursors and chaperones [56]. TOMM70 then transfers this precursor to TOMM22, after which it is inserted into the TOMM40 channel.
In contrast to cleavable precursors, which are translocated across the OMM in a linear manner, carrier precursors are inserted into the TOMM40 channel in a loop formation such that both termini are still on the cytosolic side, whereas the middle portion of the precursor passes through the channel [57]. The N-terminal segment of TOMM40 that passes from the cytosolic side through the interior of the β-barrel channel into the intermembrane space then recruits small TIMM chaperones of the IMS to the channel exit such that the hydrophobic precursors can be directly transferred to the chaperones as they enter the IMS [4]. As stated above (in Section 3), the main form of the small TIMM chaperone complex is the soluble TIMM9–TIMM10 complex. After binding the precursors, another small TIMM protein, TIMM12, associates with the TIMM9–TIMM10 complex, thereby forming the TIMM9–TIMM10–TIMM12 hexameric chaperone complex [12,58]. Subsequently, TIMM54, a subunit of the TIMM22 machinery, recruits the TIMM9–TIMM10–TIMM12 complex to the outer surface of the TIMM22 complex, where it binds the complex [59], thereby delivering the precursors to the TIMM22 channel, another subunit of the TIMM22 machinery, which mediates the insertion of precursor proteins into the inner membrane (Figure 5). Although the mechanism through which carrier precursors are inserted into the IMM is not completely understood, Rehling et al. proposed that Δψm promotes the opening of the TIMM22 channel in the presence of a carrier signal peptide and exerts an electrophoretic force on the incoming precursor [60], thereby aiding the movement of the precursor into the TIMM22 channel, after which it is laterally released from the TIMM22 complex into the IMM, where it assembles into its mature, functional form [12].
Apart from TIMM54, TIMM22, and the TIMM9–TIMM0–TIMM12 complex, other subunits of TIMM22 machinery include TIMM18 and Sdh3, which together form a TIMM18–Sdh3 module, the second pore of the TIMM22 complex [4]. Although, their exact roles in the import of carrier proteins are unknown, it is believed that they play a role in the assembly of the TIMM22 complex [4,61]. TIMM18 and Sdh3 are evolutionarily related to the membrane-integral part of respiratory complex II (succinate dehydrogenase, SDH) of mitochondria and bacteria [4]. While TIMM18 shows similarity to Sdh4, Sdh3 possesses a dual localization as a subunit of both respiratory complex II and of the TIMM22 complex [61]. However, it is unknown whether the dual localization of Sdh3 leads to a functional cross talk between the respiratory chain and carrier translocase [4]. Therefore, further studies are needed to understand whether a functional cross talk occurs between these two complexes. Furthermore, additional investigations are required with the aim of elucidating the exact roles played by Sdh3 and TIMM18 during the import of carrier precursors, as well as the exact mechanisms involved in the insertion of carrier precursors into the IMM.
In addition to carrier precursors that contain six transmembrane segments [62], the TIMM22 machinery also imports other non-cleavable proteins with only four transmembrane segments into the IMM, including TIMM17, TIMM22, and TIMM23 [12].
6. Consequences of Defective Mitochondrial Protein Import
Besides cytosolic proteostasis, mitochondrial proteostasis is also vital for cellular stress resistance and organismal health [63]. Therefore, proteostasis in these compartments needs to be maintained for proper cellular functioning. Defective mitochondrial protein import leads to reduced levels of nuclear-encoded mitochondrial proteins and impaired mitochondrial functioning, as well as proteotoxic stress, which triggers a mitochondrial-to-nuclear signal transduction pathway called mitochondrial unfolded protein response (UPRmt) [64,65]. This pathway reacts to proteotoxic stress in mitochondria by increasing the expression of mitochondrial chaperones, such as mitochondrial presequence translocase-associated motor complex protein (mtDNAj), heat shock 10 kDa protein (Hsp10), and heat shock 60 kDa protein (Hsp60), as well as proteases such as ATP-dependent Clp protease proteolytic subunit (ClpP), Lon protease homolog, mitochondrial (LONP1), YME1-like 1 ATPase, (YME1L), and OMA1 zinc metallopeptidase (OMA1) [66,67], thereby promoting the refolding of misfolded proteins, as well as degradation of damaged mitochondrial proteins. In addition, impaired mitochondrial protein import results in the accumulation of mitochondrial precursor proteins in the cytosol, which leads to proteotoxic stress, also called mitochondrial precursor overaccumulation stress (mPOS) [68,69]. Cells are equipped with several tiers of responses to alleviate such stress and restore homeostasis [68,70,71,72]. These responses include increased expression of cytoplasmic chaperones, attenuation of translation, and increased proteasome activity via the upregulation of proteasome expression and the promotion of proteasome assembly, which supports the clearance of mislocalized precursors from the cytosol [66,68,71].
When these protective mechanisms occurring in the mitochondria and cytosol become defective, the accumulated misfolded proteins in the mitochondria, as well as the unimported mitochondrial precursors in the cytosol, form aggregates [66,73], which lead to the development of diseases such as neurodegenerative and heart diseases, among many others [66]. Although UPRmt activation is a promising therapeutic option for many conditions, its overactivation could lead to undesired side effects, such as cancer development [74,75]. Similarly, the overactivation of cytosolic responses to proteotoxic stress including overexpression of cytosolic chaperones and excessive proteasome activity promotes cancer development and progression [76,77,78,79]. The following section of this review focuses on the pathological conditions that occur due to altered UPRmt.
6.1. Pathobiological Implications of Altered Mitochondrial Unfolded Protein Response
6.1.1. Parkinson’s Disease
Parkinson’s disease (PD) is the second most prevalent neurodegenerative disorder, affecting 1–3% of the population over 65 years of age [80], with most cases being sporadic and approximately 10–15% of patients with a family history of the disease [6]. Clinical features include tremors, akinesia or bradykinesia, progressive rigidity, and postural instability [6]. The neuropathological hallmarks of this pathology include loss of dopaminergic neurons in the midbrain substantia nigra pars compacta (SNpc) and accumulation of Lewy bodies (LBs) containing α-synuclein [80]. Although the molecular mechanisms underlying this neurodegeneration are still not fully understood, various studies have shown that impaired mitochondrial functioning (i.e., mitochondrial dysfunction) and abnormal protein aggregation are two of the major contributors to PD [81]. As mentioned earlier, proper functioning of the mitochondrial protein import system is crucial for optimal mitochondrial functioning, as defective mitochondrial protein import leads to reduced bioenergetic capacity and increased oxidative stress, thereby contributing to the development of PD [82,83]. This reduction in mitochondrial protein import also leads to accumulation of misfolded proteins in the mitochondria, resulting in proteotoxic stress, which activates UPRmt. Although it is not clear how defective UPRmt leads to Parkinson’s disease, studies have shown that the induction of UPRmt protects against neurodegeneration [84]. In addition, activation of UPRmt promotes dopamine neuronal survival and longevity in Parkinson’s disease animal models [85]. Similarly, the inhibition of UPRmt results in decreased lifespan and dopamine neuronal loss [85].
Because UPRmt is essential for the folding of misfolded proteins and the degradation of damaged proteins, one possible mechanism through which its inhibition can promote the development of PD is that inhibition of UPRmt leads to increased levels of misfolded mitochondrial proteins, which are dysfunctional, resulting in defective mitochondrial respiration and enhanced production of ROS that contribute to the induction of neuronal cell death [86]. Furthermore, impaired UPRmt can lead to aggregation of misfolded proteins, thereby causing further impairment in mitochondrial function. For example, the aggregated form of α-synuclein has been shown to inhibit mitochondrial complex I, leading to reduced respiration and ATP production in neurons, in addition to an increase in ROS production, resulting in neuronal death [87]. Interestingly, this aggregated α-synuclein is the main component of the Lewy bodies in PD [88]. Despite various pieces of evidence showing mitochondrial localization of α-synuclein and interaction with components of the electron transport chain [87,89,90], a study by Wang et al. suggested that pathogenic α-synuclein is not present inside mitochondria but is membrane-bound and associated with these organelles. In support of this theory, Di Maio suggested that there is no specific interaction between monomeric α-synuclein and the TOMM machinery; however, the oligomeric forms of α-synuclein bound to TOMM20, which prevented the interaction of TOMM20 with TOMM22, thereby resulting in impaired mitochondrial import of endogenous or exogenous presequence-containing, nuclear-encoded proteins, which, in turn, led to deficient mitochondrial respiration, enhanced production of ROS and loss of MMP [91]. Activation of UPRmt in response to decreased mitochondrial protein import helps to improve mitochondrial function [92]. On the other hand, the impairment of UPRmt in this case would lead to increased mitochondrial stress and dysfunction, resulting in increased neuronal death.
Conversely, overactivation of UPRmt has been suggested to increase the accumulation of defective mitochondria [93]. Furthermore, Martinez et al. showed that overactivation of UPRmt leads to increased neurotoxicity of α-synuclein, which promotes the death of dopaminergic neurons in a nonapoptotic manner [93]. This suggests that the degree of activation of UPRmt determines its effect in PD. Therefore, further studies are needed to confirm the exact roles of UPRmt in the pathogenesis of PD and how optimal UPRmt can be maintained; this would help to achieve an improved understanding of how mitochondrial dysfunction leads to the development of PD and provide novel insights about the development of mitochondria-targeted therapies for better management of PD patients.
6.1.2. Alzheimer’s Disease
Alzheimer’s disease (AD) is the most common form of dementia in the elderly population. Clinically, AD is defined by cognitive impairment that is pervasive enough to interfere with a person’s ability to work or complete daily activities [75]. AD can be divided into different types based on age of onset and genetic predisposition. Sporadic or late-onset AD accounts for more than 95% of cases and begins after the age of 65 years. Early-onset or familial AD is rare and usually manifests by age 60 [94]. The main neuropathology features of AD include extracellular amyloid plaques consisting of polymers of amyloid-β peptides (Aβ) and intracellular neurofibrillary tangles formed mainly by hyperphosphorylated protein tau [95]. Although the exact cause of AD is unknown, genetic evidence predicts that oligomeric species of Aβ are the likely cause [96]. Evidence has also shown that mitochondrial dysfunction plays a key role in AD pathogenesis, as proper mitochondrial functioning is essential for neuronal health [97,98].
Pathologies of AD such as the deposition of amyloid β and tau are known to result in impairments in mitochondrial function [99]. This could occur through the inhibition of mitochondrial protein import. Devi et al. showed that Alzheimer’s amyloid precursor protein 695 (APP), a plasma membrane protein known to be the source of the toxic amyloid β (Aβ) peptide associated with the pathogenesis of Alzheimer’s disease (AD), formed a stable ~480 kDa complex with the TOMM40 import channel in mitochondria of human AD brains but not in age-matched controls, thereby inhibiting mitochondrial protein import, which led to defective mitochondrial functioning [100]. Aβ has been reported to localize to the mitochondrial matrix, where it interacts with the mitochondrial matrix protein, amyloid-beta-binding alcohol dehydrogenase (ABAD), leading to mitochondrial dysfunction and increased ROS generation [101]. Furthermore, pathological tau impairs mitochondrial dynamics by regulating mitochondrial fission/fusion proteins, resulting in mitochondrial dysfunction [102]. In addition, abnormal tau disturbs mitochondrial bioenergetics by inhibiting complex 1 activity, decreasing Δψm and ATP levels, and increasing ROS production [103].
The defect in mitochondrial protein import occurring as a result of APP clogging the TOMM40 channel could lead to an increase in misfolded proteins in the mitochondria, resulting in the activation of UPRmt. Likewise, oxidative stress occurring as a result of excessive ROS generation disrupts the protein folding mechanism, thereby enhancing the production of misfolded proteins [104], which, in turn, results in UPRmt activation. Although the role of UPRmt in AD pathogenesis is not yet completely understood, most studies show that UPRmt activation plays a protective role in AD. Sorrentino et al. showed that inhibition of UPRmt aggravates the disease. Conversely, boosting mitochondrial proteostasis by increasing UPRmt decreased protein aggregation and delayed disease progression [105]. Another study showed that Honokiol, a small-molecule polyphenol isolated from the bark of Magnolia officinalis, activated UPRmt, which improved cognitive impairment in AD mouse models [106]. In addition, UPRmt is strongly activated and exerts a protective role against Aβ protein toxicity in PITRM1-knockout iPSC-derived cortical neurons and cerebral organoids. In line with this, pharmacological inhibition of UPRmt exacerbates Aβ proteotoxicity in cerebral organoids generated from PITRM1-knockout iPSCs [107]. Thus, these studies indicate that defective UPRmt promotes AD progression.
Although no studies have been conducted to date on the effect of UPRmt overactivation on AD pathogenesis, Beck et al. showed that compared to control subjects, the expression of UPRmt-related genes was increased by 40–60% in sporadic AD subjects and 70–90% in familial AD subjects [108]. The activation of these genes in the postmortem frontal cortex samples of these subjects indicate that this physiologically important cellular response may be chronically activated in AD, perhaps as a compensatory neuroprotective response to a sustained accumulation of unfolded, misfolded, and damaged mitochondrial proteins [108]. Thus, further studies are needed to examine the consequences of persistent UPRmt activation. Furthermore, additional studies are needed with the aim of determining the time period during which UPRmt becomes activated in disease development. Such studies would help to provide guidance about the development of UPRmt-based therapeutics for AD management.
6.1.3. Cardiovascular Diseases
Cardiovascular diseases, especially acute myocardial infarction (MI) and chronic heart failure (HF), account for numerous deaths and severely undermine quality of life [109]. Mitochondrial dysfunction has been identified as a crucial etiological factor for these diseases by contributing to energetic dysfunction, oxidative stress, calcium dysregulation, and cardiomyocyte death and is considered a potential therapeutic target [109,110].
A crucial stress response triggered by mitochondrial dysfunction is UPRmt, which has been shown to play cardioprotective roles in cardiovascular diseases [111]. Venkatesh et al. showed that upregulation of LONP1 mitigates cardiac injury by preventing oxidative damage of proteins and lipids, preserving mitochondrial redox balance, and reprogramming bioenergetics by reducing Complex I content and activity [112]. On the other hand, LONP1 activity, which is reduced by oxidative stress, leads to the accumulation of dysfunctional respiratory chain subunits and left ventricle contractile dysfunction [113], indicating that defective UPRmt promotes the development of cardiovascular diseases. In another study by Smyrnias et al., it was demonstrated that UPRmt was induced in the hearts of mice subjected to chronic hemodynamic overload [67]. Additionally, boosting UPRmt with nicotinamide riboside reduced cardiomyocyte death and contractile dysfunction in mice subjected to pressure overload [67]. A study by Wang et al. also confirmed that pharmacological UPRmt activation exerts cardioprotective effects in an activating transcription factor 5 (ATF5)-dependent manner in mouse models of ischemia–reperfusion injury [111]. Moreover, myocardial tissue from patients with aortic stenosis also showed evidence of UPRmt activation, which correlated with reduced tissue cardiomyocyte death and fibrosis, as well as lower plasma levels of biomarkers of cardiac damage (high-sensitivity troponin T) and dysfunction (N-terminal pro-B-type natriuretic peptide) [67], supporting a protective role of UPRmt in pathological heart conditions. Conversely, enhanced sympathoexcitation leads to decreased UPRmt, resulting in increased proteotoxic stress, which results in decreased OXPHOS and Δψm as well as increased ROS generation, which, in turn, induces mitochondrial transition pore opening, activating pro hypertrophy/fibrosis factors that induce pathological cardiac hypertrophy and fibrosis [114]. These findings suggest that although UPRmt is cardioprotective, its activation status varies in different heart conditions. Therefore, further studies are required to determine the status of UPRmt activation in different heart conditions and the period during which this response becomes activated in the development of the disease.
On the other hand, some studies have shown that overactivation of UPRmt promotes the progression of heart diseases. Liu et al. showed that elevated activity of Omi/HtrA2 protease promotes mitochondrial depolarization and apoptosis in aged rat heart [115]. Likewise, in vitro and in vivo overexpression of mitochondrial Omi/HtrA2 induces cardiac apoptosis and dysfunction [116]. A potential explanation for these contradictory findings is that UPRmt is cardioprotective when moderately active, whereas its excessive activity may be cardiotoxic [109]. A moderate activation of UPRmt may be beneficial for removing/repairing damaged mitochondrial proteins, thereby maintaining optimal mitochondrial and cardiac function, whereas excessive UPRmt activation could result in a massive cleavage of mitochondrial proteins, exacerbating mitochondrial dysfunction and promoting heart damage [109]. Thus, further studies are needed to determine the causes of UPRmt overactivation and how a balance can be properly maintained.
6.1.4. Cancer
The initiation and development of cancer is a multistep process that involves the acquisition of diverse functions, such as resistance to apoptotic cell death, prevention of growth inhibition, and activation of proliferation signals [117]. Cancer cells experience mitochondrial stress as they undergo unchecked cellular proliferation and generate ROS, which damage mtDNA and mitochondrial proteins, including components of the OXPHOS family, causing mitochondrial dysfunction [118]. Cancer cells rely on functional mitochondria to generate macromolecules, such as amino acids, nucleotides, and cholesterol, to maintain their high proliferative capacity. Thus, cancer cells activate the mitochondrial stress response to alleviate mitochondrial dysfunction and protein aggregation, which subsequently promotes tumor growth and progression [118].
Numerous studies have validated these notions by showing that activation of the UPRmt is indispensable for cancer development and progression. In recent studies, Chen et al. showed that UPRmt components (Hsp10, Hsp60, and ClpP) are abundantly expressed in breast cancer tissues compared to adjacent noncancerous tissues [119]. In addition, Kenny et al. demonstrated that persistent activation of UPRmt provides survival advantage to cancer cells, leading to tumor progression [120]. Moreover, high expression of UPRmt-related genes is significantly associated with poor clinical outcomes [120]. In the following section, we discuss the roles of key UPRmt components in cancer.
ATF5, a key UPRmt regulator in mammals, has been observed to be upregulated in glioblastoma, breast, pancreas, rectal, and ovarian cancers [121,122,123,124,125]. In addition, high ATF5 expression correlates with reduced survival in glioma and lung cancer patients [126,127]. Besides promoting cancer growth via recovery of mitochondrial functions, ATF5 has also been shown to promote proliferation and survival of glioma and breast cancer cells by regulating Egr-1 expression [128]. Similarly, Dluzen et al. demonstrated that ATF5 promotes survival of glioblastoma and breast cancer cells by transactivating Bcl2 [129].
CIpP, a key protease involved in UPRmt, is known to be upregulated in breast cancer, prostate cancer, and acute myeloid leukemia [130,131,132]. Knockdown of ClpP is associated with reduced proliferation, migration, and invasion of breast cancer cells [130]. Additionally, ClpP silencing was reported to lead to decreased oxidative phosphorylation and mitochondrial metabolism, as well as viability of AML cells [132]. Moreover, xenograft studies demonstrate that ClpP knockdown suppresses the growth of prostate-cancer-derived liver metastases [131]. In contrast, overexpression of ClpP increases migratory and invasive activity in breast cancer [130].
LONP1, another key protease involved in UPRmt, is upregulated in various cancers, including melanoma, colorectal cancer, and pancreatic cancer [133,134]. Its increased expression has also been reported to correlate with reduced overall survival in neuroblastoma, breast cancer, renal cell carcinoma, and colorectal cancer [135]. LONP1 silencing leads to reduced proliferation in melanoma, colorectal cancer, and pancreatic cancer [133,134]. In addition, a deficiency in LONP1 expression in mice was reported to inhibit the formation and growth of azoxymethane and dextran sulfate-induced colorectal tumors [133]. This could be due to the fact that loss of LONP1 inhibits the formation of OXPHOS complexes I and III, thereby impairing normal mitochondrial functioning [133], which is required for the growth and development of tumors. In another study, silencing of LONP1 in prostate cancer cells resulted in the accumulation of misfolded subunits of OXPHOS complexes II and V; reduced assembly of OXPHOS complexes I, III, IV, and V; and inhibition of activities of OXPHOS complexes I, II, and V [135], resulting in impaired oxidative bioenergetics and heightened ROS production. This, in turn, suppressed mitochondrial trafficking to the cortical cytoskeleton, shut off tumor cell migration and invasion, and inhibited primary and metastatic tumor growth in vivo [135], indicating that defective UPRmt inhibits tumor growth and progression.
Although not much is known about the role of HSP10 in cancer, evidence has shown that it is overexpressed in multiple forms of cancer, including astrocytoma, oral squamous cell carcinoma, and nasopharyngeal carcinoma [136,137,138]. In addition, increased HSP10 expression is associated with reduced overall survival in astrocytoma, oral squamous cell carcinoma, and nasopharyngeal carcinoma [136,137,138]. Fan et al. showed that increased expression of HSP10 promoted survival and tumorigenesis via the inhibition of apoptosis [136].
HSP60, another key component of the UPRmt, is overexpressed in many cancer types, and its expression has been correlated with the metastatic potential of cancers and overall survival of cancer patients [139,140,141,142]. HSP60 knockdown leads to severe deficiencies in mitochondrial functions, which hinder cell growth and survival [143]. Specifically, in pancreatic cancer, HSP60 knockdown reduces the expression of subunits in OXPHOS complexes I, III, IV, and V; disrupts the formation of complexes I and III; and blocks mitochondrial respiration and ATP production [144], indicating that increased activation of HSP60 promotes optimal mitochondrial functioning, which, in turn, promotes tumor growth and progression. In addition to direct mitochondrial functions, HSP60 regulates the expression and release of IL-8 in prostate and colon cancers, possibly via transforming growth factor-beta (TGF-β), to enhance cell survival [145]. Besides its established localization in the mitochondria, HSP60 also localizes to other cellular compartments, such as the cytosol, plasma membrane, and extracellular space of cancer cells [118,143]. Cytosolic HSP60 interacts with the IKK complex and enhances activation of IKK [146], resulting in the increased expression of NF- ΚB targets and promotion of cell survival [146]. Interestingly, HSP60 is not overexpressed in all cancers and is not necessarily associated with a poor prognosis in certain patients [143]. For example, HSP60 is downregulated in bronchial cancer, colorectal cancer, and hepatocellular carcinoma [147,148,149]. Furthermore, overexpression of HSP60 suppresses cell proliferation in clear cell renal cell carcinoma and inhibits invasive activity in hepatocellular carcinoma [149,150]. This suggests that HSP60 has multimodal functions in diverse cancers. In addition, because HSP60 is a key component of UPRmt, these findings suggest that increased UPRmt activation might have diverse roles in different cancers. Therefore, more studies are needed to investigate whether UPRmt activation has tumor-inhibitory roles.
7. Conclusions
Highly orchestrated governance and legitimate activity of the mitochondrial protein import system are pivotal for both mitochondrial and cellular homeostasis. At present, knowledge about how precisely the mitochondrial protein import system operates is still maturing. Thus, further in-depth studies are warranted to decipher the structures of these machineries, as well as a complete understanding of the various mechanisms involved in the import, sorting, and assembly of mitochondria-targeted proteins. From the above discussion, it is clear that defective mitochondrial protein import could lead to the generation of proteotoxic stress in both mitochondrial and cytosolic compartments. Cells try to combat these stresses via specific responses, including UPRmt and other compensatory mechanisms. Alterations in these responses result in the development of various disease conditions. On the other hand, the abundant activity of the proteins of these machineries (TOMM, TIMM, SAM, etc.) may lead to sustained functions of the mitochondrial energy metabolism and other relevant pathways that are beneficial for aggressive tumor growth and metastasis.
Whereas most studies report that increased activation of UPRmt plays a protective role in neurodegenerative and heart diseases, few studies show that overactivation of UPRmt promotes the progression of these diseases. In contrast to the findings observed in neurodegenerative and cardiovascular diseases, most studies have demonstrated that increased UPRmt activation is essential for cancer development and progression, whereas few studies suggest that increased expression of UPRmt components inhibits cancer progression. Therefore, further studies are needed with the aim of determining the exact roles of UPRmt in various disease conditions and identifying the relevant mechanisms through which alterations in UPRmt result in the development of pathological conditions. The understanding gained from such studies could aid in directing the development of UPRmt-based therapies for the management of various pathological conditions.
Author Contributions
Conceptualization: S.D.; Supervision: S.D. and A.P.S.; Resources: M.O.H., K.S.V., S.D., A.P.S. and S.S.; Writing—review and editing: M.O.H., K.S.V., S.D., A.P.S. and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors gratefully acknowledge the funding and necessary support and resources from the University of South Alabama and the Mitchell Cancer Institute.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, Z.; Ying, Z.; Bosy-Westphal, A.; Zhang, J.; Schautz, B.; Later, W.; Heymsfield, S.B.; Müller, M.J. Specific metabolic rates of major organs and tissues across adulthood: Evaluation by mechanistic model of resting energy expenditure. Am. J. Clin. Nutr. 2010, 92, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Pagliarini, D.J.; Calvo, S.E.; Chang, B.; Sheth, S.A.; Vafai, S.B.; Ong, S.-E.; Walford, G.A.; Sugiana, C.; Boneh, A.; Chen, W.K.; et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008, 134, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Archibald, J.M. Endosymbiosis and Eukaryotic Cell Evolution. Curr. Biol. 2015, 25, R911–R921. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, N.; Pfanner, N. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef]
- Srinivasan, S.; Guha, M.; Kashina, A.; Avadhani, N.G. Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection. Biochim. Biophys. Acta Bioenerg. 2017, 1858, 602–614. [Google Scholar] [CrossRef]
- Adebayo, M.; Singh, S.; Singh, A.P.; Dasgupta, S. Mitochondrial fusion and fission: The fine-tune balance for cellular homeostasis. FASEB J. 2021, 35, e21620. [Google Scholar] [CrossRef]
- O’Rourke, B. Mitochondrial ion channels. Annu. Rev. Physiol. 2007, 69, 19–49. [Google Scholar] [CrossRef]
- Lemasters, J.J. Modulation of mitochondrial membrane permeability in pathogenesis, autophagy and control of metabolism. J. Gastroenterol. Hepatol. 2007, 22, S31–S37. [Google Scholar] [CrossRef]
- Gilkerson, R.W. Mitochondrial DNA nucleoids determine mitochondrial genetics and dysfunction. Int. J. Biochem. Cell Biol. 2009, 41, 1899–1906. [Google Scholar] [CrossRef]
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing mitochondrial proteins: Machineries and mechanisms. Cell 2009, 138, 628–644. [Google Scholar] [CrossRef]
- Harbauer, A.B.; Zahedi, R.P.; Sickmann, A.; Pfanner, N.; Meisinger, C. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab. 2014, 19, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Dudek, J.; Rehling, P.; van der Laan, M. Mitochondrial protein import: Common principles and physiological networks. Biochim. Biophys. Acta 2013, 1833, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, X.; Zhang, L.; Yi, J.; Ma, Q.; Yin, J.; Zhuo, W.; Gu, J.; Yang, M. Atomic structure of human TOM core complex. Cell Discov. 2020, 6, 67. [Google Scholar] [CrossRef] [PubMed]
- Jores, T.; Klinger, A.; Groß, L.E.; Kawano, S.; Flinner, N.; Duchardt-Ferner, E.; Wöhnert, J.; Kalbacher, H.; Endo, T.; Schleiff, E.; et al. Characterization of the targeting signal in mitochondrial β-barrel proteins. Nat. Commun. 2016, 7, 12036. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Wenz, L.-S.; Zerbes, R.M.; Oeljeklaus, S.; Bohnert, M.; Stroud, D.A.; Wirth, C.; Ellenrieder, L.; Thornton, N.; Kutik, S.; et al. Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell 2013, 154, 596–608. [Google Scholar] [CrossRef]
- Krimmer, T.; Rapaport, D.; Ryan, M.; Meisinger, C.; Kassenbrock, C.K.; Blachly-Dyson, E.; Forte, M.; Douglas, M.G.; Neupert, W.; Nargang, F.E.; et al. Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J. Cell Biol. 2001, 152, 289–300. [Google Scholar] [CrossRef]
- Wiedemann, N.; Truscott, K.N.; Pfannschmidt, S.; Guiard, B.; Meisinger, C.; Pfanner, N. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: Intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem. 2004, 279, 18188–18194. [Google Scholar] [CrossRef]
- Kutik, S.; Stojanovski, D.; Becker, L.; Becker, T.; Meinecke, M.; Krüger, V.; Prinz, C.; Meisinger, C.; Guiard, B.; Wagner, R.; et al. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell 2008, 132, 1011–1024. [Google Scholar] [CrossRef]
- Diederichs, K.A.; Ni, X.; Rollauer, S.E.; Botos, I.; Tan, X.; King, M.S.; Kunji, E.R.S.; Jiang, J.; Buchanan, S.K. Structural insight into mitochondrial β-barrel outer membrane protein biogenesis. Nat. Commun. 2020, 11, 3290. [Google Scholar] [CrossRef]
- Wenz, L.S.; Ellenrieder, L.; Qiu, J.; Bohnert, M.; Zufall, N.; van der Laan, M.; Pfanner, N.; Wiedemann, N.; Becker, T. Sam37 is crucial for formation of the mitochondrial TOM-SAM supercomplex, thereby promoting β-barrel biogenesis. J. Cell Biol. 2015, 210, 1047–1054. [Google Scholar] [CrossRef]
- Ahting, U.; Waizenegger, T.; Neupert, W.; Rapaport, D. Signal-anchored proteins follow a unique insertion pathway into the outer membrane of mitochondria. J. Biol. Chem. 2005, 280, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Meineke, B.; Engl, G.; Kemper, C.; Vasiljev-Neumeyer, A.; Paulitschke, H.; Rapaport, D. The outer membrane form of the mitochondrial protein Mcr1 follows a TOM-independent membrane insertion pathway. FEBS Lett. 2008, 582, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.; Wenz, L.-S.; Krüger, V.; Lehmann, W.; Müller, J.M.; Goroncy, L.; Zufall, N.; Lithgow, T.; Guiard, B.; Chacinska, A.; et al. The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J. Cell Biol. 2011, 194, 387–395. [Google Scholar] [CrossRef]
- Papic, D.; Krumpe, K.; Dukanovic, J.; Dimmer, K.S.; Rapaport, D. Multispan mitochondrial outer membrane protein Ugo1 follows a unique Mim1-dependent import pathway. J. Cell Biol. 2011, 194, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Doan, K.N.; Grevel, A.; Mårtensson, C.U.; Ellenrieder, L.; Thornton, N.; Wenz, L.-S.; Opaliński, Ł.; Guiard, B.; Pfanner, N.; Becker, T. The Mitochondrial Import Complex MIM Functions as Main Translocase for α-Helical Outer Membrane Proteins. Cell Rep. 2020, 31, 107567. [Google Scholar] [CrossRef]
- Kemper, C.; Habib, S.; Engl, G.; Heckmeyer, P.; Dimmer, K.S.; Rapaport, D. Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J. Cell Sci. 2008, 121, 1990–1998. [Google Scholar] [CrossRef]
- Vögtle, F.-N.; Keller, M.; Taskin, A.A.; Horvath, S.E.; Guan, X.L.; Prinz, C.; Opalińska, M.; Zorzin, C.; van der Laan, M.; Wenk, M.R.; et al. The fusogenic lipid phosphatidic acid promotes the biogenesis of mitochondrial outer membrane protein Ugo1. J. Cell Biol. 2015, 210, 951–960. [Google Scholar] [CrossRef]
- Sinzel, M.; Tan, T.; Wendling, P.; Kalbacher, H.; Özbalci, C.; Chelius, X.; Westermann, B.; Brügger, B.; Rapaport, D.; Dimmer, K.S. Mcp3 is a novel mitochondrial outer membrane protein that follows a unique IMP-dependent biogenesis pathway. EMBO Rep. 2016, 17, 965–981. [Google Scholar] [CrossRef]
- Stojanovski, D.; Müller, J.M.; Milenkovic, D.; Guiard, B.; Pfanner, N.; Chacinska, A. The MIA system for protein import into the mitochondrial intermembrane space. Biochim. Biophys. Acta 2008, 1783, 610–617. [Google Scholar] [CrossRef]
- Durigon, R.; Wang, Q.; Pavia, E.C.; Grant, C.M.; Lu, H. Cytosolic thioredoxin system facilitates the import of mitochondrial small Tim proteins. EMBO Rep. 2012, 13, 916–922. [Google Scholar] [CrossRef]
- Gornicka, A.; Bragoszewski, P.; Chroscicki, P.; Wenz, L.-S.; Schulz, C.; Rehling, P.; Chacinska, A. A discrete pathway for the transfer of intermembrane space proteins across the outer membrane of mitochondria. Mol. Biol. Cell 2014, 25, 3999–4009. [Google Scholar] [CrossRef] [PubMed]
- Kurz, M.; Martin, H.; Rassow, J.; Pfanner, N.; Ryan, M.T. Biogenesis of Tim proteins of the mitochondrial carrier import pathway: Differential targeting mechanisms and crossing over with the main import pathway. Mol. Biol. Cell 1999, 10, 2461–2474. [Google Scholar] [CrossRef] [PubMed]
- Milenkovic, D.; Ramming, T.; Müller, J.M.; Wenz, L.-S.; Gebert, N.; Schulze-Specking, A.; Stojanovski, D.; Rospert, S.; Chacinska, A. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol. Biol. Cell 2009, 20, 2530–2539. [Google Scholar] [CrossRef]
- Peleh, V.; Cordat, E.; Herrmann, J.M. Mia40 is a trans-site receptor that drives protein import into the mitochondrial intermembrane space by hydrophobic substrate binding. eLife 2016, 5, e16177. [Google Scholar] [CrossRef]
- Mesecke, N.; Bihlmaier, K.; Grumbt, B.; Longen, S.; Terziyska, N.; Hell, K.; Herrmann, J.M. The zinc-binding protein Hot13 promotes oxidation of the mitochondrial import receptor Mia40. EMBO Rep. 2008, 9, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Dabir, D.V.; Leverich, E.P.; Kim, S.-K.; Tsai, F.D.; Hirasawa, M.; Knaff, D.B.; Koehler, C.M. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 2007, 26, 4801–4811. [Google Scholar] [CrossRef]
- Bihlmaier, K.; Mesecke, N.; Terziyska, N.; Bien, M.; Hell, K.; Herrmann, J.M. The disulfide relay system of mitochondria is connected to the respiratory chain. J. Cell Biol. 2007, 179, 389–395. [Google Scholar] [CrossRef]
- Ramesh, A.; Peleh, V.; Martinez-Caballero, S.; Wollweber, F.; Sommer, F.; van der Laan, M.; Schroda, M.; Alexander, R.T.; Campo, M.L.; Herrmann, J.M. A disulfide bond in the TIM23 complex is crucial for voltage gating and mitochondrial protein import. J. Cell Biol. 2016, 214, 417–431. [Google Scholar] [CrossRef]
- Zhao, F.; Zou, M.H. Role of the Mitochondrial Protein Import Machinery and Protein Processing in Heart Disease. Front. Cardiovasc. Med. 2021, 8, 749756. [Google Scholar] [CrossRef]
- Roise, D.; Horvath, S.J.; Tomich, J.M.; Richards, J.H.; Schatz, G. A chemically synthesized pre-sequence of an imported mitochondrial protein can form an amphiphilic helix and perturb natural and artificial phospholipid bilayers. EMBO J. 1986, 5, 1327–1334. [Google Scholar] [CrossRef]
- Abe, Y.; Shodai, T.; Muto, T.; Mihara, K.; Torii, H.; Nishikawa, S.-I.; Endo, T.; Kohda, D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 2000, 100, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Bolliger, L.; Junne, T.; Schatz, G.; Lithgow, T. Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J. 1995, 14, 6318–6326. [Google Scholar] [CrossRef] [PubMed]
- Moczko, M.; Bömer, U.; Kübrich, M.; Zufall, N.; Hönlinger, A.; Pfanner, N. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell. Biol. 1997, 17, 6574–6584. [Google Scholar] [CrossRef]
- Chacinska, A.; Lind, M.; Frazier, A.E.; Dudek, J.; Meisinger, C.; Geissler, A.; Sickmann, A.; Meyer, H.E.; Truscott, K.N.; Guiard, B.; et al. Mitochondrial presequence translocase: Switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 2005, 120, 817–829. [Google Scholar] [CrossRef]
- Albrecht, R.; Rehling, P.; Chacinska, A.; Brix, J.; Cadamuro, S.A.; Volkmer, R.; Guiard, B.; Pfanner, N.; Zeth, K. The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes. EMBO Rep. 2006, 7, 1233–1238. [Google Scholar] [CrossRef]
- Meinecke, M.; Wagner, R.; Kovermann, P.; Guiard, B.; Mick, D.U.; Hutu, D.P.; Voos, W.; Truscott, K.N.; Chacinska, A.; Pfanner, N.; et al. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 2006, 312, 1523–1526. [Google Scholar] [CrossRef]
- Naamati, A.; Regev-Rudzki, N.; Galperin, S.; Lill, R.; Pines, O. Dual targeting of Nfs1 and discovery of its novel processing enzyme, Icp55. J. Biol. Chem. 2009, 284, 30200–30208. [Google Scholar] [CrossRef]
- Ostermann, J.; Horwich, A.L.; Neupert, W.; Hartl, F.U. Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature 1989, 341, 125–130. [Google Scholar] [CrossRef]
- van der Laan, M.; Rissler, M.; Rehling, P. Mitochondrial preprotein translocases as dynamic molecular machines. FEMS Yeast Res. 2006, 6, 849–861. [Google Scholar] [CrossRef]
- Martin, J.; Mahlke, K.; Pfanner, N. Role of an energized inner membrane in mitochondrial protein import. Delta psi drives the movement of presequences. J. Biol. Chem. 1991, 266, 18051–18057. [Google Scholar] [CrossRef] [PubMed]
- Truscott, K.N.; Kovermann, P.; Geissler, A.; Merlin, A.; Meijer, M.; Driessen, A.J.; Rassow, J.; Pfanner, N.; Wagner, R. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 2001, 8, 1074–1082. [Google Scholar] [CrossRef] [PubMed]
- van der Laan, M.; Wiedemann, N.; Mick, D.U.; Guiard, B.; Rehling, P.; Pfanner, N. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr. Biol. 2006, 16, 2271–2276. [Google Scholar] [CrossRef] [PubMed]
- Bohnert, M.; Rehling, P.; Guiard, B.; Herrmann, J.M.; Pfanner, N.; van der Laan, M. Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr. Biol. 2010, 20, 1227–1232. [Google Scholar] [CrossRef]
- Stiller, S.B.; Höpker, J.; Oeljeklaus, S.; Schütze, C.; Schrempp, S.G.; Vent-Schmidt, J.; Horvath, S.E.; Frazier, A.; Gebert, N.; van der Laan, M.; et al. Mitochondrial OXA Translocase Plays a Major Role in Biogenesis of Inner-Membrane Proteins. Cell Metab. 2016, 23, 901–908. [Google Scholar] [CrossRef]
- Young, J.C.; Hoogenraad, N.J.; Hartl, F.U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 2003, 112, 41–50. [Google Scholar] [CrossRef]
- Wu, Y.; Sha, B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 2006, 13, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Curran, S.P.; Leuenberger, D.; Schmidt, E.; Koehler, C.M. The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J. Cell Biol. 2002, 158, 1017–1027. [Google Scholar] [CrossRef]
- Gebert, N.; Chacinska, A.; Wagner, K.; Guiard, B.; Koehler, C.M.; Rehling, P.; Pfanner, N.; Wiedemann, N. Assembly of the three small Tim proteins precedes docking to the mitochondrial carrier translocase. EMBO Rep. 2008, 9, 548–554. [Google Scholar] [CrossRef]
- Wagner, K.; Gebert, N.; Guiard, B.; Brandner, K.; Truscott, K.N.; Wiedemann, N.; Pfanner, N.; Rehling, P. The assembly pathway of the mitochondrial carrier translocase involves four preprotein translocases. Mol. Cell. Biol. 2008, 28, 4251–4260. [Google Scholar] [CrossRef]
- Rehling, P.; Model, K.; Brandner, K.; Kovermann, P.; Sickmann, A.; Meyer, H.E.; Kühlbrandt, W.; Wagner, R.; Truscott, K.N.; Pfanner, N. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 2003, 299, 1747–1751. [Google Scholar] [CrossRef]
- Gebert, N.; Gebert, M.; Oeljeklaus, S.; von der Malsburg, K.; Stroud, D.A.; Kulawiak, B.; Wirth, C.; Zahedi, R.P.; Dolezal, P.; Wiese, S.; et al. Dual function of Sdh3 in the respiratory chain and TIM22 protein translocase of the mitochondrial inner membrane. Mol. Cell 2011, 44, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Bolender, N.; Sickmann, A.; Wagner, R.; Meisinger, C.; Pfanner, N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008, 9, 42–49. [Google Scholar] [CrossRef]
- Boos, F.; Labbadia, J.; Herrmann, J.M. How the Mitoprotein-Induced Stress Response Safeguards the Cytosol: A Unified View. Trends Cell Biol. 2020, 30, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Nargund, A.M.; Pellegrino, M.W.; Fiorese, C.J.; Baker, B.M.; Haynes, C.M. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 2012, 337, 587–590. [Google Scholar] [CrossRef] [PubMed]
- Horibe, T.; Hoogenraad, N.J. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS ONE 2007, 2, e835. [Google Scholar] [CrossRef] [PubMed]
- Lenkiewicz, A.M.; Krakowczyk, M.; Bragoszewski, P. Cytosolic Quality Control of Mitochondrial Protein Precursors-The Early Stages of the Organelle Biogenesis. Int. J. Mol. Sci. 2021, 23, 7. [Google Scholar] [CrossRef]
- Smyrnias, I.; Gray, S.P.; Okonko, D.O.; Sawyer, G.; Zoccarato, A.; Catibog, N.; López, B.; Gonzalez, A.; Ravassa, S.; Díez, J.; et al. Cardioprotective Effect of the Mitochondrial Unfolded Protein Response During Chronic Pressure Overload. J. Am. Coll. Cardiol. 2019, 73, 1795–1806. [Google Scholar] [CrossRef]
- Wang, X.; Chen, X.J. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 2015, 524, 481–484. [Google Scholar] [CrossRef]
- Coyne, L.P.; Chen, X.J. mPOS is a novel mitochondrial trigger of cell death-implications for neurodegeneration. FEBS Lett. 2018, 592, 759–775. [Google Scholar] [CrossRef]
- Weidberg, H.; Amon, A. MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science 2018, 360, eaan4146. [Google Scholar] [CrossRef]
- Wrobel, L.; Topf, U.; Bragoszewski, P.; Wiese, S.; Sztolsztener, M.E.; Oeljeklaus, S.; Varabyova, A.; Lirski, M.; Chroscicki, P.; Mroczek, S.; et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 2015, 524, 485–488. [Google Scholar] [CrossRef]
- Boos, F.; Krämer, L.; Groh, C.; Jung, F.; Haberkant, P.; Stein, F.; Wollweber, F.; Gackstatter, A.; Zöller, E.; Van Der Laan, M.; et al. Mitochondrial protein-induced stress triggers a global adaptive transcriptional programme. Nat. Cell Biol. 2019, 21, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Schlagowski, A.M.; Knöringer, K.; Morlot, S.; Vicente, A.S.; Flohr, T.; Krämer, L.; Boos, F.; Khalid, N.; Ahmed, S.; Schramm, J.; et al. Increased levels of mitochondrial import factor Mia40 prevent the aggregation of polyQ proteins in the cytosol. EMBO J. 2021, 40, e107913. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhou, Q.; He, L.; Chen, L. Mitochondrial unfolded protein response: An emerging pathway in human diseases. Free Radic. Biol. Med. 2021, 163, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Rivero, J.M.; Pastor-Maldonado, C.J.; Povea-Cabello, S.; Álvarez-Córdoba, M.; Villalón-García, I.; Talaverón-Rey, M.; Suárez-Carrillo, A.; Munuera-Cabeza, M.; Reche-López, D.; Cilleros-Holgado, P.; et al. Activation of the Mitochondrial Unfolded Protein Response: A New Therapeutic Target? Biomedicines 2022, 10, 1611. [Google Scholar] [CrossRef]
- Jolly, C.; Morimoto, R.I. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J. Natl. Cancer Inst. 2000, 92, 1564–1572. [Google Scholar] [CrossRef]
- Wawrzynow, B.; Zylicz, A.; Zylicz, M. Chaperoning the guardian of the genome. The two-faced role of molecular chaperones in p53 tumor suppressor action. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 161–174. [Google Scholar] [CrossRef]
- Arlt, A.; Bauer, I.; Schafmayer, C.; Tepel, J.; Müerköster, S.S.; Brosch, M.; Röder, C.; Kalthoff, H.; Hampe, J.; Moyer, M.P.; et al. Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2). Oncogene 2009, 28, 3983–3996. [Google Scholar] [CrossRef]
- Soave, C.L.; Guerin, T.; Liu, J.; Dou, Q.P. Targeting the ubiquitin-proteasome system for cancer treatment: Discovering novel inhibitors from nature and drug repurposing. Cancer Metastasis Rev. 2017, 36, 717–736. [Google Scholar] [CrossRef]
- Franco-Iborra, S.; Vila, M.; Perier, C. Mitochondrial Quality Control in Neurodegenerative Diseases: Focus on Parkinson’s Disease and Huntington’s Disease. Front. Neurosci. 2018, 12, 342. [Google Scholar] [CrossRef]
- Xie, W.; Wan, O.W.; Chung, K.K. New insights into the role of mitochondrial dysfunction and protein aggregation in Parkinson’s disease. Biochim. Biophys. Acta 2010, 1802, 935–941. [Google Scholar] [CrossRef]
- Perier, C.; Vila, M. Mitochondrial biology and Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012, 2, a009332. [Google Scholar] [CrossRef]
- Celardo, I.; Martins, L.M.; Gandhi, S. Unravelling mitochondrial pathways to Parkinson’s disease. Br. J. Pharmacol. 2014, 171, 1943–1957. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yu, S.; Wang, J.; Qiao, J.; Liu, Y.; Wang, S.; Zhao, Y. Ginseng protein protects against mitochondrial dysfunction and neurodegeneration by inducing mitochondrial unfolded protein response in Drosophila melanogaster PINK1 model of Parkinson’s disease. J. Ethnopharmacol. 2020, 247, 112213. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.F.; Machiela, E.; Dues, D.J.; Spielbauer, K.K.; Senchuk, M.M.; Van Raamsdonk, J.M. Activation of the mitochondrial unfolded protein response promotes longevity and dopamine neuron survival in Parkinson’s disease models. Sci. Rep. 2017, 7, 16441. [Google Scholar] [CrossRef]
- Moisoi, N.; Klupsch, K.; Fedele, V.; East, P.; Sharma, S.; Renton, A.; Plun-Favreau, H.; Edwards, R.E.; Teismann, P.; Esposti, M.D.; et al. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 2009, 16, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Reeve, A.K.; Ludtmann, M.H.R.; Angelova, P.R.; Simcox, E.M.; Horrocks, M.H.; Klenerman, D.; Gandhi, S.C.; Turnbull, D.M.; Abramov, A.Y. Aggregated α-synuclein and complex I deficiency: Exploration of their relationship in differentiated neurons. Cell Death Dis. 2015, 6, e1820. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Crowther, R.A.; Jakes, R.; Hasegawa, M.; Goedert, M. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proc. Natl. Acad. Sci. USA 1998, 95, 6469–6473. [Google Scholar] [CrossRef]
- Ludtmann, M.H.; Angelova, P.R.; Ninkina, N.N.; Gandhi, S.; Buchman, V.L.; Abramov, A.Y. Monomeric Alpha-Synuclein Exerts a Physiological Role on Brain ATP Synthase. J. Neurosci. 2016, 36, 10510–10521. [Google Scholar] [CrossRef]
- Devi, L.; Raghavendran, V.; Prabhu, B.M.; Avadhani, N.G.; Anandatheerthavarada, H.K. Mitochondrial import and accumulation of alpha-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. J. Biol. Chem. 2008, 283, 9089–9100. [Google Scholar] [CrossRef]
- Di Maio, R.; Barrett, P.J.; Hoffman, E.K.; Barrett, C.W.; Zharikov, A.; Borah, A.; Hu, X.; McCoy, J.; Chu, C.T.; Burton, E.A.; et al. α-Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson’s disease. Sci. Transl. Med. 2016, 8, 342ra378. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.F.; Haynes, C.M. Metabolism and the UPR(mt). Mol. Cell 2016, 61, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Martinez, B.A.; Petersen, D.A.; Gaeta, A.L.; Stanley, S.P.; Caldwell, G.A.; Caldwell, K.A. Dysregulation of the Mitochondrial Unfolded Protein Response Induces Non-Apoptotic Dopaminergic Neurodegeneration in. J. Neurosci. 2017, 37, 11085–11100. [Google Scholar] [CrossRef] [PubMed]
- Bali, J.; Gheinani, A.H.; Zurbriggen, S.; Rajendran, L. Role of genes linked to sporadic Alzheimer’s disease risk in the production of β-amyloid peptides. Proc. Natl. Acad. Sci. USA 2012, 109, 15307–15311. [Google Scholar] [CrossRef]
- Swerdlow, R.H. Pathogenesis of Alzheimer’s disease. Clin. Interv. Aging 2007, 2, 347–359. [Google Scholar] [PubMed]
- Shen, Y.; Ding, M.; Xie, Z.; Liu, X.; Yang, H.; Jin, S.; Xu, S.; Zhu, Z.; Wang, Y.; Wang, D.; et al. Activation of Mitochondrial Unfolded Protein Response in SHSY5Y Expressing APP Cells and APP/PS1 Mice. Front. Cell. Neurosci. 2019, 13, 568. [Google Scholar] [CrossRef]
- Swerdlow, R.H. Mitochondria and Mitochondrial Cascades in Alzheimer’s Disease. J. Alzheimers Dis. 2018, 62, 1403–1416. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Li, L.; Perry, G.; Lee, H.G.; Zhu, X. Oxidative stress and mitochondrial dysfunction in Alzheimer’s disease. Biochim. Biophys. Acta 2014, 1842, 1240–1247. [Google Scholar] [CrossRef]
- Kim, D.K.; Mook-Jung, I. The role of cell type-specific mitochondrial dysfunction in the pathogenesis of Alzheimer’s disease. BMB Rep. 2019, 52, 679–688. [Google Scholar] [CrossRef]
- Devi, L.; Prabhu, B.M.; Galati, D.F.; Avadhani, N.G.; Anandatheerthavarada, H.K. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer’s disease brain is associated with mitochondrial dysfunction. J. Neurosci. 2006, 26, 9057–9068. [Google Scholar] [CrossRef]
- Lustbader, J.W.; Cirilli, M.; Lin, C.; Xu, H.W.; Takuma, K.; Wang, N.; Caspersen, C.; Chen, X.; Pollak, S.; Chaney, M.; et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer’s disease. Science 2004, 304, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Bai, F. The Association of Tau With Mitochondrial Dysfunction in Alzheimer’s Disease. Front. Neurosci. 2018, 12, 163. [Google Scholar] [CrossRef] [PubMed]
- Szabo, L.; Eckert, A.; Grimm, A. Insights into Disease-Associated Tau Impact on Mitochondria. Int. J. Mol. Sci. 2020, 21, 6344. [Google Scholar] [CrossRef] [PubMed]
- Chong, W.C.; Shastri, M.D.; Eri, R. Endoplasmic Reticulum Stress and Oxidative Stress: A Vicious Nexus Implicated in Bowel Disease Pathophysiology. Int. J. Mol. Sci. 2017, 18, 771. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Bao, W.; Gao, Y.; Chen, J.; Song, G. Honokiol improves cognitive impairment in APP/PS1 mice through activating mitophagy and mitochondrial unfolded protein response. Chem. Biol. Interact. 2022, 351, 109741. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, V.; Romani, M.; Mouchiroud, L.; Beck, J.S.; Zhang, H.; D’Amico, D.; Moullan, N.; Potenza, F.; Schmid, A.W.; Rietsch, S.; et al. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature 2017, 552, 187–193. [Google Scholar] [CrossRef]
- Pérez, M.J.; Ivanyuk, D.; Panagiotakopoulou, V.; Di Napoli, G.; Kalb, S.; Brunetti, D.; Al-Shaana, R.; Kaeser, S.A.; Fraschka, S.A.-K.; Jucker, M.; et al. Loss of function of the mitochondrial peptidase PITRM1 induces proteotoxic stress and Alzheimer’s disease-like pathology in human cerebral organoids. Mol. Psychiatry 2021, 26, 5733–5750. [Google Scholar] [CrossRef]
- Beck, J.S.; Mufson, E.J.; Counts, S.E. Evidence for Mitochondrial UPR Gene Activation in Familial and Sporadic Alzheimer’s Disease. Curr. Alzheimer. Res. 2016, 13, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Svaguša, T.; Martinić, M.; Martinić, M.; Kovačević, L.; Šepac, A.; Miličić, D.; Bulum, J.; Starčević, B.; Sirotković-Skerlev, M.; Seiwerth, F.; et al. Mitochondrial unfolded protein response, mitophagy and other mitochondrial quality control mechanisms in heart disease and aged heart. Croat. Med. J. 2020, 61, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Dietl, A.; Maack, C. Targeting Mitochondrial Calcium Handling and Reactive Oxygen Species in Heart Failure. Curr. Heart Fail Rep. 2017, 14, 338–349. [Google Scholar] [CrossRef]
- Wang, Y.T.; Im, Y.; McCall, M.N.; Huang, K.-T.; Haynes, C.M.; Nehrke, K.; Brookes, P.S. Cardioprotection by the mitochondrial unfolded protein response requires ATF5. Am. J. Physiol. Heart Circ. Physiol. 2019, 317, H472–H478. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, S.; Li, M.; Saito, T.; Tong, M.; Rashed, E.; Mareedu, S.; Zhai, P.; Bárcena, C.; López-Otín, C.; Yehia, G.; et al. Mitochondrial LonP1 protects cardiomyocytes from ischemia/reperfusion injury in vivo. J. Mol. Cell. Cardiol. 2019, 128, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Okawa, Y.; Ariyoshi, M.; Kaimoto, S.; Uchihashi, M.; Fukai, K.; Iwai-Kanai, E.; Matoba, S. Oxidative post-translational modifications develop LONP1 dysfunction in pressure overload heart failure. Circ. Heart Fail 2014, 7, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Nandi, S.S.; Katsurada, K.; Mahata, S.K.; Patel, K.P. Neurogenic Hypertension Mediated Mitochondrial Abnormality Leads to Cardiomyopathy: Contribution of UPR. Front. Physiol. 2021, 12, 718982. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lei, J.; Wang, K.; Ma, L.; Liu, D.; Du, Y.; Wu, Y.; Zhang, S.; Wang, W.; Ma, X.; et al. Mitochondrial Omi/HtrA2 Promotes Caspase Activation Through Cleavage of HAX-1 in Aging Heart. Rejuvenation Res. 2017, 20, 183–192. [Google Scholar] [CrossRef]
- Wang, K.; Yuan, Y.; Liu, X.; Lau, W.B.; Zuo, L.; Wang, X.; Ma, L.; Jiao, K.; Shang, J.; Wang, W.; et al. Cardiac Specific Overexpression of Mitochondrial Omi/HtrA2 Induces Myocardial Apoptosis and Cardiac Dysfunction. Sci. Rep. 2016, 6, 37927. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, J.; Kumar, R.; Inigo, J.; Yadava, N.; Chandra, D. Mitochondrial Stress Response and Cancer. Trends Cancer 2020, 6, 688–701. [Google Scholar] [CrossRef]
- Chen, F.-M.; Huang, L.-J.; Ou-Yang, F.; Kan, J.-Y.; Kao, L.-C.; Hou, M.-F. Activation of mitochondrial unfolded protein response is associated with Her2-overexpression breast cancer. Breast Cancer Res. Treat. 2020, 183, 61–70. [Google Scholar] [CrossRef]
- Kenny, T.C.; Craig, A.J.; Villanueva, A.; Germain, D. Mitohormesis Primes Tumor Invasion and Metastasis. Cell Rep. 2019, 27, 2292–2303.e2296. [Google Scholar] [CrossRef]
- Feldheim, J.; Kessler, A.F.; Schmitt, D.; Wilczek, L.; Linsenmann, T.; Dahlmann, M.; Monoranu, C.M.; Ernestus, R.-I.; Hagemann, C.; Löhr, M. Expression of activating transcription factor 5 (ATF5) is increased in astrocytomas of different WHO grades and correlates with survival of glioblastoma patients. Onco Targets Ther. 2018, 11, 8673–8684. [Google Scholar] [CrossRef] [PubMed]
- Monaco, S.E.; Angelastro, J.M.; Szabolcs, M.; Greene, L.A. The transcription factor ATF5 is widely expressed in carcinomas, and interference with its function selectively kills neoplastic, but not nontransformed, breast cell lines. Int. J. Cancer 2007, 120, 1883–1890. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Wang, B.; Qian, D.; Li, L.; Zhang, L.; Song, X.; Liu, D.X. Interference with ATF5 function enhances the sensitivity of human pancreatic cancer cells to paclitaxel-induced apoptosis. Anticancer Res. 2012, 32, 4385–4394. [Google Scholar] [PubMed]
- Kong, X.; Meng, W.; Zhou, Z.; Li, Y.; Zhou, B.; Wang, R.; Zhan, L. Overexpression of activating transcription factor 5 in human rectal cancer. Exp. Ther. Med. 2011, 2, 827–831. [Google Scholar] [CrossRef]
- Chen, A.; Qian, D.; Wang, B.; Hu, M.; Lu, J.; Qi, Y.; Liu, D.X. ATF5 is overexpressed in epithelial ovarian carcinomas and interference with its function increases apoptosis through the downregulation of Bcl-2 in SKOV-3 cells. Int. J. Gynecol. Pathol. 2012, 31, 532–537. [Google Scholar] [CrossRef]
- Sheng, Z.; Li, L.; Zhu, L.J.; Smith, T.W.; Demers, A.; Ross, A.H.; Moser, R.P.; Green, M.R. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat. Med. 2010, 16, 671–677. [Google Scholar] [CrossRef]
- Ishihara, S.; Yasuda, M.; Ishizu, A.; Ishikawa, M.; Shirato, H.; Haga, H. Activating transcription factor 5 enhances radioresistance and malignancy in cancer cells. Oncotarget 2015, 6, 4602–4614. [Google Scholar] [CrossRef]
- Liu, D.X.; Qian, D.; Wang, B.; Yang, J.M.; Lu, Z. p300-Dependent ATF5 acetylation is essential for Egr-1 gene activation and cell proliferation and survival. Mol. Cell. Biol. 2011, 31, 3906–3916. [Google Scholar] [CrossRef]
- Dluzen, D.; Li, G.; Tacelosky, D.; Moreau, M.; Liu, D.X. BCL-2 is a downstream target of ATF5 that mediates the prosurvival function of ATF5 in a cell type-dependent manner. J. Biol. Chem. 2011, 286, 7705–7713. [Google Scholar] [CrossRef]
- Luo, J.; Zeng, B.; Tao, C.; Lu, M.; Ren, G. ClpP regulates breast cancer cell proliferation, invasion and apoptosis by modulating the Src/PI3K/Akt signaling pathway. PeerJ 2020, 8, e8754. [Google Scholar] [CrossRef]
- Seo, J.H.; Rivadeneira, D.B.; Caino, M.C.; Chae, Y.C.; Speicher, D.W.; Tang, H.Y.; Vaira, V.; Bosari, S.; Palleschi, A.; Rampini, P.; et al. The Mitochondrial Unfoldase-Peptidase Complex ClpXP Controls Bioenergetics Stress and Metastasis. PLoS Biol. 2016, 14, e1002507. [Google Scholar] [CrossRef] [PubMed]
- Cole, A.; Wang, Z.; Coyaud, E.; Voisin, V.; Gronda, M.; Jitkova, Y.; Mattson, R.; Hurren, R.; Babovic, S.; Maclean, N.; et al. Inhibition of the Mitochondrial Protease ClpP as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell 2015, 27, 864–876. [Google Scholar] [CrossRef] [PubMed]
- Quirós, P.M.; Español, Y.; Acín-Pérez, R.; Rodríguez, F.; Bárcena, C.; Watanabe, K.; Calvo, E.; Loureiro, M.; Fernández-García, M.S.; Fueyo, A.; et al. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014, 8, 542–556. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, H.; Li, H.; Chen, X.; Wu, X.; Lu, B.; Zhang, W.; Zhou, Y.; Xiao, G.G.; Gao, G. Inhibition of LONP1 Suppresses Pancreatic Cancer Progression Via c-Jun N-Terminal Kinase Pathway-Meditated Epithelial-Mesenchymal Transition. Pancreas 2019, 48, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, J.C.; Seo, J.H.; Agarwal, E.; Wang, Y.; Kossenkov, A.V.; Tang, H.Y.; Speicher, D.W.; Altieri, D.C. Akt phosphorylation of mitochondrial Lonp1 protease enables oxidative metabolism and advanced tumor traits. Oncogene 2019, 38, 6926–6939. [Google Scholar] [CrossRef]
- Fan, W.; Fan, S.-S.; Feng, J.; Xiao, D.; Fan, S.; Luo, J. Elevated expression of HSP10 protein inhibits apoptosis and associates with poor prognosis of astrocytoma. PLoS ONE 2017, 12, e0185563. [Google Scholar] [CrossRef]
- Feng, J.; Luo, J.; Wang, H.; Lu, J.; Zhan, Y.; Zang, H.; Wen, Q.; Wang, W.; Chen, L.; Xu, L.; et al. High expression of heat shock protein 10 (Hsp10) is associated with poor prognosis in oral squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2017, 10, 7784–7791. [Google Scholar]
- Feng, J.; Zhan, Y.; Zhang, Y.; Zheng, H.; Wang, W.; Fan, S. Increased expression of heat shock protein (HSP) 10 and HSP70 correlates with poor prognosis of nasopharyngeal carcinoma. Cancer Manag. Res. 2019, 11, 8219–8227. [Google Scholar] [CrossRef]
- Hjerpe, E.; Egyhazi, S.; Carlson, J.; Stolt, M.F.; Schedvins, K.; Johansson, H.; Shoshan, M.; Avall-Lundqvist, E. HSP60 predicts survival in advanced serous ovarian cancer. Int. J. Gynecol. Cancer 2013, 23, 448–455. [Google Scholar] [CrossRef]
- Ghosh, J.C.; Dohi, T.; Kang, B.H.; Altieri, D.C. Hsp60 regulation of tumor cell apoptosis. J. Biol. Chem. 2008, 283, 5188–5194. [Google Scholar] [CrossRef]
- Cappello, F.; de Macario, E.C.; Marasà, L.; Zummo, G.; Macario, A.J. Hsp60 expression, new locations, functions and perspectives for cancer diagnosis and therapy. Cancer Biol. Ther. 2008, 7, 801–809. [Google Scholar] [CrossRef]
- Li, X.S.; Xu, Q.; Fu, X.Y.; Luo, W.S. Heat shock protein 60 overexpression is associated with the progression and prognosis in gastric cancer. PLoS ONE 2014, 9, e107507. [Google Scholar] [CrossRef]
- Inigo, J.R.; Chandra, D. The mitochondrial unfolded protein response (UPRmt): Shielding against toxicity to mitochondria in cancer. J. Hematol. Oncol. 2022, 15, 98. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Sun, H.; Zheng, C.; Gao, J.; Fu, Q.; Hu, N.; Shao, X.; Zhou, Y.; Xiong, J.; Nie, K.; et al. Oncogenic HSP60 regulates mitochondrial oxidative phosphorylation to support Erk1/2 activation during pancreatic cancer cell growth. Cell Death Dis. 2018, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; O’Malley, J.; Chaudhary, A.K.; Inigo, J.R.; Yadav, N.; Kumar, R.; Chandra, D. Hsp60 and IL-8 axis promotes apoptosis resistance in cancer. Br. J. Cancer 2019, 121, 934–943. [Google Scholar] [CrossRef]
- Chun, J.N.; Choi, B.; Lee, K.W.; Lee, D.J.; Kang, D.H.; Lee, J.Y.; Song, I.S.; Kim, H.I.; Lee, S.H.; Kim, H.S.; et al. Cytosolic Hsp60 is involved in the NF-kappaB-dependent survival of cancer cells via IKK regulation. PLoS ONE 2010, 5, e9422. [Google Scholar] [CrossRef]
- Cappello, F.; Di Stefano, A.; D’Anna, S.E.; Donner, C.; Zummo, G. Immunopositivity of heat shock protein 60 as a biomarker of bronchial carcinogenesis. Lancet Oncol. 2005, 6, 816. [Google Scholar] [CrossRef]
- Ruan, W.; Wang, Y.; Ma, Y.; Xing, X.; Lin, J.; Cui, J.; Lai, M. HSP60, a protein downregulated by IGFBP7 in colorectal carcinoma. J. Exp. Clin. Cancer Res. 2010, 29, 41. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, X.; Chang, H.; Huang, X.; Guo, X.; Du, X.; Tian, S.; Wang, L.; Lyv, Y.; Yuan, P.; et al. Hsp60 exerts a tumor suppressor function by inducing cell differentiation and inhibiting invasion in hepatocellular carcinoma. Oncotarget 2016, 7, 68976–68989. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Chen, Y.; Liu, X.; Wang, S.; Lv, Y.; Wu, D.; Wang, Q.; Luo, M.; Deng, H. Downregulation of HSP60 disrupts mitochondrial proteostasis to promote tumorigenesis and progression in clear cell renal cell carcinoma. Oncotarget 2016, 7, 38822–38834. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).