Secrets behind Protein Sequences: Unveiling the Potential Reasons for Varying Allergenicity Caused by Caseins from Cows, Goats, Camels, and Mares Based on Bioinformatics Analyses
Abstract
:1. Introduction
2. Results and Discussion
2.1. Analysis of the Composition and Content of Casein in Human Milk (HM), CM, GM, CAM, and MM
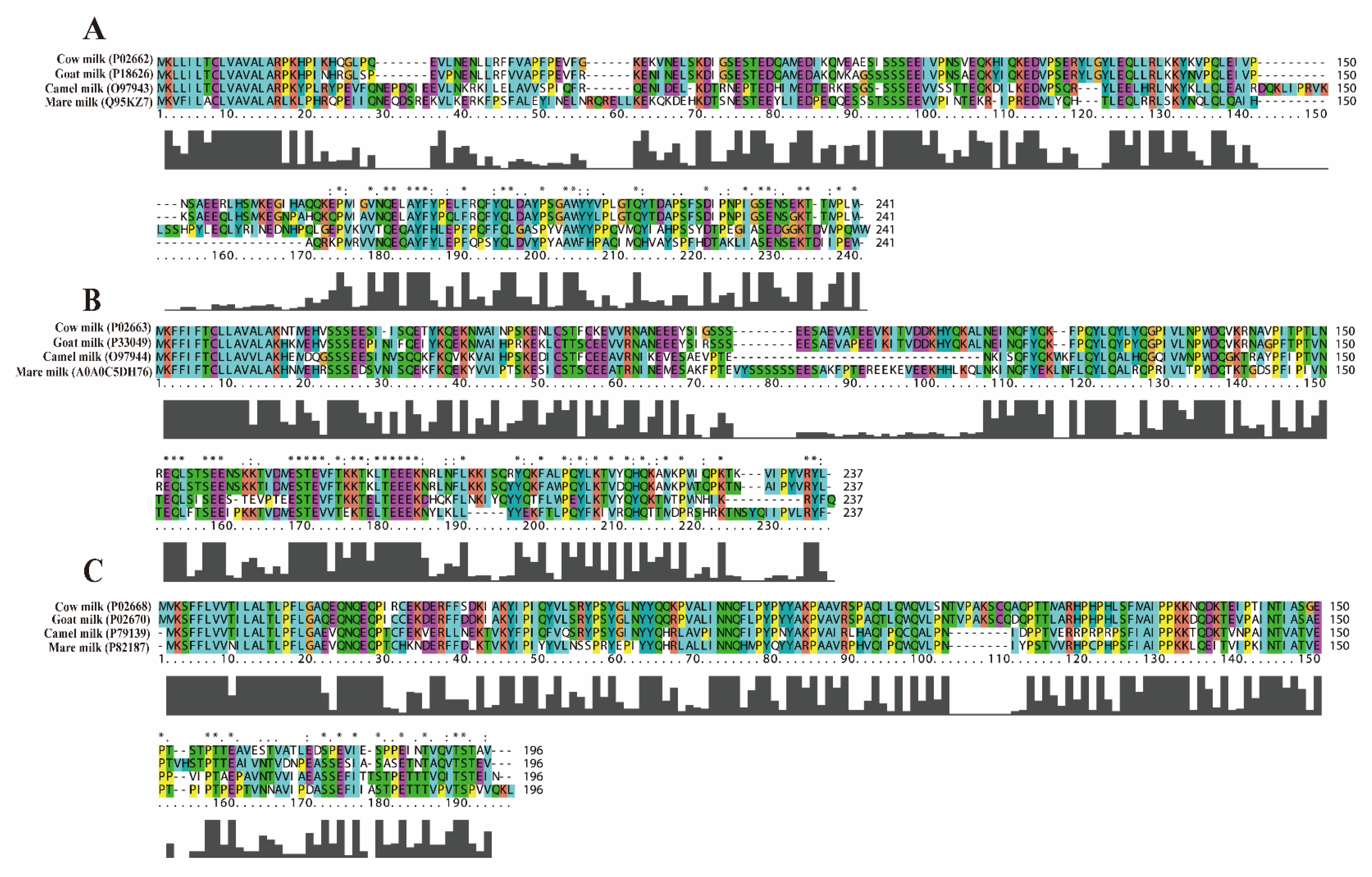
2.2. Sequence Similarity Analysis of αS1-CN, αS2-CN and κ-CN in CM, GM, CAM, and MM
2.3. Analyses of Physicochemical Property
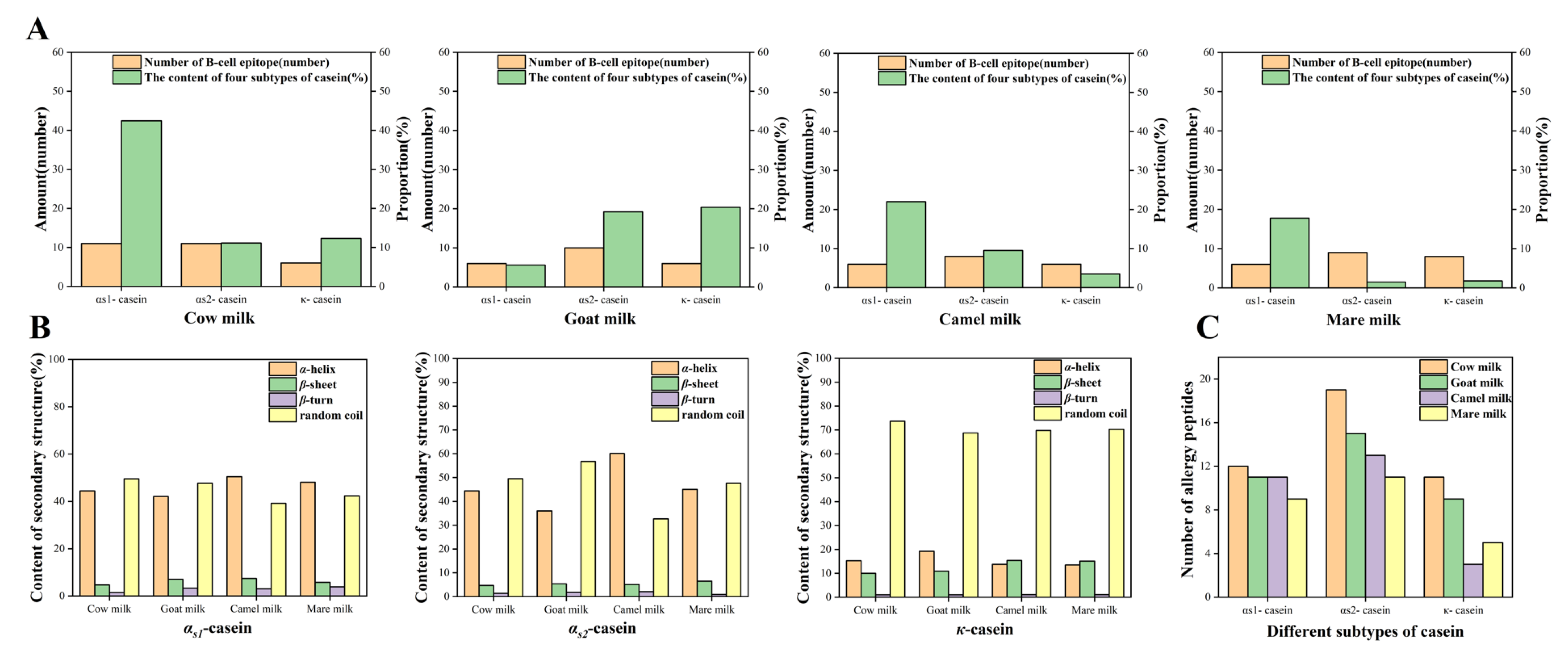
2.4. The Prediction of the Secondary Structure, the Linear B-Cell Epitope of Proteins, and the Screening of Allergenic Peptides
2.5. Prediction of T-Cell Epitope of Proteins
2.6. Analysis of the Method Limitations
3. Material and Methods
3.1. Database and Computational Software
3.2. Compositions and Contents of Caseins in CM, GM, CAM, and MM
3.3. Prediction of Property and Structure of αS1-CN, αS2-CN, and κ-CN
3.4. Allergenicity Definition
3.5. Prediction of Linear B-Cell Epitopes Regarding αS1-CN, αS2-CN, and κ-CN
3.6. Prediction of T-Cell Epitope of αS1-CN, αS2-CN, and κ-CN
3.7. Hydrolysis of αS1-CN, αS2-CN and κ-CN by In Silico
3.8. Prediction of Solubility and the Allergies of Peptides
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CM | cow milk |
| GM | goat milk |
| CAM | camel milk |
| MM | mare milk |
| HM | human milk |
| αS1-CN | αS1-casein |
| αS2-CN | αS2-casein |
| β-CN | β-casein |
| κ-CN | κ-casein |
| α-CN | α-casein |
| β-lg | β-lactoglobulin |
| GI | gastrointestinal |
| GRAVY | grand average of hydropathicity |
| AI | aliphatic index |
| NRPFLB-cellE | number of recognized polypeptides fragments of linear B-cell epitope |
References
- Pu, P.; Zheng, X.; Jiao, L.N.; Chen, L.; Yang, J.; Zhang, Y.H.; Liang, G.Z. Six flavonoids inhibit the antigenicity of β-lactoglobulin by noncovalent interactions: A spectroscopic and molecular docking study. Food Chem. 2021, 339, 128106. [Google Scholar] [CrossRef] [PubMed]
- Villa, C.; Costa, J.; Oliveira, M.B.P.P.; Mafra, I. Bovine Milk Allergens: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 137–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katarzyna, K.; Biela, A.; Loch, J.I.; Lipowska, J.; Siuda, M.; Lewiński, K. Towards understanding the effect of high pressure on food protein allergenicity: β-lactoglobulin structural studies. Food Chem. 2019, 270, 315–321. [Google Scholar]
- Monaci, L.; Tregoat, V.; Hengel, A.J.V.; Anklam, E. Milk allergens, their characteristics and their detection in food: A review. Eur. Food Res. Technol. 2006, 223, 149–179. [Google Scholar] [CrossRef]
- Ehlayel, M.; Bener, A.; Hazeima, K.A.; Mesaifri, F.A. Camel Milk Is a Safer Choice than Goat Milk for Feeding Children with Cow Milk Allergy. Int. Sch. Res. Netw. 2011, 2011, 391641. [Google Scholar] [CrossRef] [Green Version]
- Uniacke-Lowe, T.; Huppertz, T.; Fox, P.F. Equine milk proteins: Chemistry, structure and nutritional significance. Int. Dairy J. 2010, 20, 609–629. [Google Scholar] [CrossRef]
- Ng, S.W.; Lu, P.; Rulikowska, A.; Boehm, D.; O’Neill, G.; Bourke, P. The effect of atmospheric cold plasma treatment on the antigenic properties of bovine milk casein and whey proteins. Food Chem. 2021, 342, 128283. [Google Scholar] [CrossRef]
- Selvaggi, M.; Laudadio, V.; Dario, C.; Tufarelli, V. Major proteins in goat milk: An updated overview on genetic variability. Mol. Biol. Rep. 2014, 41, 1035–1048. [Google Scholar] [CrossRef]
- Brezovečki, A.; Čagalj, M.; Filipović Dermit, Z.; Mikulec, N.; Bendelja Ljoljić, D.; Antunac, N. Camel milk and milk products. Mljekarstvo 2015, 65, 81–90. [Google Scholar]
- Natale, M.; Bisson, C.; Monti, G.; Peltran, A.; Garoffo, L.P.; Valentini, S.; Fabris, C.; Bertino, E.; Coscia, A.; Conti, A. Cow’s milk allergens identification by two-dimensional immunoblotting and mass spectrometry. Mol. Nutr. Food Res. 2004, 48, 363. [Google Scholar] [CrossRef]
- Mackie, A. The Digestive Tract: A Complex System. Interdiscip. Approaches Food Dig. 2019, 11–27. [Google Scholar]
- Han, R.; Maycock, J.; Murray, B.S.; Boesch, C. Identification of angiotensin converting enzyme and dipeptidyl peptidase-IV inhibitory peptides derived from oilseed proteins using two integrated bioinformatic approaches. Food Res. Int. 2019, 115, 283–291. [Google Scholar] [CrossRef]
- Bondili, C.S.; Veeramachaneni, G.K.; Thunuguntla, V.B.S.C.; Manda, N.K.; Bondili, J.S. Specific panallergen peptide of Sorghum Polcalcin showing IgE response identified based on In Silico and In Vivo peptide mapping. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Vieira, D.S.; Polveiro, R.C.; Butler, T.J.; Hackett, T.A.; Braga, C.P.; Puniya, B.L.; Teixeira, W.F.P.; Padilha, P.M.; Adamec, J.; Feitosa, F.L.F. An In Silico, structural, and biological analysis of lactoferrin of different mammals. Int. J. Biol. Macromol. 2021, 187, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Huang, L. Cross-Linking Mass Spectrometry: An Emerging Technology for Interactomics and Structural Biology. Anal. Chem. 2017, 90, 144–165. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Freddolino, P.L.; Zhang, Y. Cofactor: Improved protein function prediction by combining structure, sequence and protein-protein interaction information. Nucleic Acids Res. 2017, 45, 291–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nongonierma, A.B.; Fitzgerald, R.J. Strategies for the discovery and identification of food protein-derived biologically active peptides. Trends Food Sci. Technol. 2017, 69, 289–305. [Google Scholar] [CrossRef] [Green Version]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2—A server for in silico prediction of allergens. J. Mol. Model 2014, 20, 2278. [Google Scholar] [CrossRef] [PubMed]
- Sudheer, G.; Pallavi, K.; Kumardeep, C.; Ankur, G.; Rahul, K.; Open, S.D.D.C.; Gajendra, P.S.R. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar]
- Miranda, G.; Mahé, M.F.; Leroux, C.; Martin, P. Proteomic tools to characterize the protein fraction of Equidae milk. Proteomics 2004, 4, 2496–2509. [Google Scholar] [CrossRef] [PubMed]
- Lönnerdal, B. Nutritional and physiologic significance of human milk proteins. Am. J. Clin. Nutr. 2003, 77, 1537S–1543S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaiswal, L.; Worku, M. Recent perspective on cow’s milk allergy and dairy nutrition. Crit. Rev. Food Sci. Nutr. 2021, 62, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Current protocols in bioinformatics. Curr. Protoc. Bioinform. 2003, 2.3.1–2.3.22. [Google Scholar] [CrossRef]
- Restani, P.; Ballabio, C.; Lorenzo, C.D.; Tripodi, S.; Fiocchi, A. Molecular aspects of milk allergens and their role in clinical events. Anal. Bioanal. Chem. 2009, 395, 47–56. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. The proteomics protocols handbook, Protein identification and analysis tools on the ExPASy server. Proteom. Protoc. Handbook 2005, 571–607. [Google Scholar]
- Kyte, J.; Doolittle, R.F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atsushi, I. Thermostability and Aliphatic Index of Globular Proteins. J. Biochem. 1980, 88, 1895–1898. [Google Scholar]
- Rehman, S.U.; Nadeem, A.; Javed, M.; Hassan, F.; Luo, X.; Khalid, R.B.; Liu, Q.Y. Genomic Identification, Evolution and Sequence Analysis of the Heat-Shock Protein Gene Family in Buffalo. Genes 2020, 11, 1388. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, A.; Boye, J.I. Impact of thermal processing time and cookie size on the detection of casein, egg, gluten and soy allergens in food. Food Res. Int. 2013, 52, 483–489. [Google Scholar] [CrossRef]
- Zhou, F.L.; He, S.D.; Sun, H.J.; Wang, Y.F.; Zhang, Y. Advances in epitope mapping technologies for food protein allergens: A review. Trends Food Sci. Technol. 2020, 107, 226–239. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhanda, S.K.; Mahajan, S.; Paul, S.; Yan, Z.; Kim, H.; Jespersen, M.C.; Jurtz, V.; Andreatta, M.; Greenbaum, J.A.; Marcatili, P.; et al. IEDB-AR: Immune epitope database—Analysis resource in 2019. Nucleic Acids Res. 2019, 47, W502–W506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatchatee, P.; Järvinen, K.M.; Bardina, L.; Beyer, K.; Sampson, H.A. Identification of IgE-and IgG-binding epitopes on αs1-casein: Differences in patients with persistent and transient cow’s milk allergy. J. Allergy Clin. Immunol. 2001, 107, 379–383. [Google Scholar] [CrossRef] [Green Version]
- Busse, P.J.; Järvinen, K.M.; Vila, L.; Beyer, K.; Sampson, H.A. Identification of sequential IgE-binding epitopes on bovine αs2-casein in cow’s milk allergic patients. Int. Arch. Allergy Imm. 2002, 129, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Chatchatee, P.; Järvinen, K.M.; Bardina, L.; Vila, L.; Beyer, K.; Sampson, H.A. Identification of IgE and IgG binding epitopes on β-and κ-casein in cow’s milk allergic patients. Clin. Exp. Allergy 2001, 31, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Kapila, R.; Kavadi, P.K.; Kapila, S. Comparative evaluation of allergic sensitization to milk proteins of cow, buffalo and goat. Small Rumin. Res. 2013, 112, 191–198. [Google Scholar] [CrossRef]
- El-Agamy, E.I. The challenge of cow milk protein allergy. Small Rumin. Res. 2007, 68, 64–72. [Google Scholar] [CrossRef]
- Lafarga, T.; O’Connor, P.; Hayes, M. Identification of novel dipeptidyl peptidase-IV and angiotensin-I-converting enzyme inhibitory peptides from meat proteins using in silico analysis. Peptides 2014, 59, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Bush, R.K.; Hefle, S.L. Food allergens. Crit. Rev. Food Sci. Nutr. 1996, 36, 119–163. [Google Scholar] [CrossRef]
- Gallego, M.; Toldrá, F.; Mora, L. Quantification and In Silico analysis of taste dipeptides generated during dry-cured ham processing. Food Chem. 2022, 370, 130977. [Google Scholar] [CrossRef] [PubMed]
- Bohle, B. T-cell epitopes of food allergens. Clin. Rev. Allerg. Immu. 2006, 30, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.K.; Andreatta, M.; Marcatili, P.; Buus, S.; Greenbaum, J.A.; Yan, Z.; Sette, A.; Peters, B.; Nielsen, M. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 2018, 154, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Fu, Y.; Ma, L.; Dai, H.J.; Wang, H.X.; Chen, H.; Zhu, H.K.; Yu, Y.; Hou, Y.; Zhang, Y.H. Exploration of Dipeptidyl Peptidase-IV (DPP-IV) Inhibitory Peptides from Silkworm Pupae (Bombyx mori) Proteins Based on In Silico and In Vitro Assessments. J. Agric. Food Chem. 2022, 70, 3862–3871. [Google Scholar] [CrossRef] [PubMed]
- Buchan, D.W.A.; Minneci, F.; Nugent, T.C.O.; Bryson, K.; Jones, D.T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013, 41, W349–W357. [Google Scholar] [CrossRef] [Green Version]
- Combet, C.; Blanchet, C.; Geourjon, C.; Deléage, G. NPS@: Network protein sequence analysis. Trends Biochem. 2000, 25, 147–150. [Google Scholar] [CrossRef] [PubMed]
| Protein | Human a,b | Cow c | Goat d | Camel e | Mare c |
|---|---|---|---|---|---|
| Casein (%) * | 40.00 | 80.00 | 64.52 | 52.00 | 55.00 |
| αs1-casein (%) # | 13.79 | 42.46 | 5.60 | 22.00 | 17.78 |
| αs2-casein (%) # | - | 11.11 | 19.20 | 9.50 | 1.48 |
| β-casein (%) # | 68.97 | 34.13 | 54.80 | 65.00 | 78.96 |
| κ-casein (%) # | 17.24 | 12.30 | 20.40 | 3.50 | 1.78 |
| Species Name | Protein | UniProtKB Database Accession Number | Molecular Weight/Da (Mw) | Grand Average of Hydropathicity (GRAVY) | Aliphatic Index(AI) |
|---|---|---|---|---|---|
| Cow | αs1-casein | P02662 | 24,528.94 | −0.481 | 85.19 |
| Goat | P18626 | 24,289.59 | −0.534 | 80.23 | |
| Camel | O97943 | 26,861.40 | −0.661 | 84.30 | |
| Mare | Q95KZ7 | 24,688.89 | −0.801 | 80.67 | |
| Cow | αs2-casein | P02663 | 26,018.69 | −0.704 | 73.74 |
| Goat | P33049 | 26,389.03 | −0.844 | 66.46 | |
| Camel | O97944 | 22,964.10 | −0.661 | 67.62 | |
| Mare | A0A0C5DH76 | 27,262.89 | −0.729 | 70.00 | |
| Cow | κ-casein | P02668 | 21,269.35 | −0.287 | 81.63 |
| Goat | P02670 | 21,441.32 | −0.328 | 79.27 | |
| Camel | P79139 | 20,417.56 | −0.150 | 90.49 | |
| Mare | P82187 | 21,021.43 | −0.191 | 97.41 |
| Species Name | Protein | Hydrolyzed Peptide Number | Allergenic Peptide Number | Allergenic Peptide Number/Hydrolyzed Peptide Number (%) | |
|---|---|---|---|---|---|
| Total | Liner B-Cell Epitope | ||||
| Cow | αs1-casein | 30 | 12 | 8 | 40.00 |
| αs2-casein | 42 | 19 | 6 | 45.24 | |
| κ-casein | 24 | 11 | 6 | 45.83 | |
| Goat | αs1-casein | 31 | 11 | 6 | 35.48 |
| αs2-casein | 42 | 15 | 6 | 35.71 | |
| κ-casein | 23 | 9 | 3 | 39.13 | |
| Camel | αs1-casein | 36 | 11 | 4 | 30.56 |
| αs2-casein | 32 | 13 | 7 | 40.62 | |
| κ-casein | 22 | 3 | 1 | 13.63 | |
| Mare | αs1-casein | 35 | 9 | 2 | 25.71 |
| αs2-casein | 38 | 11 | 5 | 28.95 | |
| κ-casein | 21 | 5 | 3 | 23.81 | |
| Protein Type | Species | Consensus Core Epitope | Binding Type | T-Cell Epitope Number |
|---|---|---|---|---|
| αs1-casein | Cow | IGSESTEDQ, SESTEDQAM | Strong binder, Weak binder | 12 |
| Goat | IGSESTEDQ, SESTEDQAM | 5 | ||
| Camel | - | 4 | ||
| Mare | - | 6 | ||
| αs2-casein | Cow | MEHVSSSEE, VRNANEEEY, EYSIGSSSE, IGSSSEESA | Weak binder | 12 |
| Goat | MEHVSSSEE, VRNANEEEY, EYSIGSSSE, IGSSSEESA | Weak binder | 10 | |
| Camel | - | - | 9 | |
| Mare | - | - | 6 | |
| κ-casein | Cow | FLGAEVQNQ, PYYAKPAAV | Weak binder | 13 |
| Goat | FLGAEVQNQ, PYYAKPAAV | Weak binder | 13 | |
| Camel | FLGAEVQNQ, INTVATVEP | Weak binder | 11 | |
| Mare | FLGAEVQNQ, INTVATVEP | Weak binder | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Pan, F.; Cai, S.; Yi, J.; Zhou, L.; Liu, Z. Secrets behind Protein Sequences: Unveiling the Potential Reasons for Varying Allergenicity Caused by Caseins from Cows, Goats, Camels, and Mares Based on Bioinformatics Analyses. Int. J. Mol. Sci. 2023, 24, 2481. https://doi.org/10.3390/ijms24032481
Zhao S, Pan F, Cai S, Yi J, Zhou L, Liu Z. Secrets behind Protein Sequences: Unveiling the Potential Reasons for Varying Allergenicity Caused by Caseins from Cows, Goats, Camels, and Mares Based on Bioinformatics Analyses. International Journal of Molecular Sciences. 2023; 24(3):2481. https://doi.org/10.3390/ijms24032481
Chicago/Turabian StyleZhao, Shuai, Fei Pan, Shengbao Cai, Junjie Yi, Linyan Zhou, and Zhijia Liu. 2023. "Secrets behind Protein Sequences: Unveiling the Potential Reasons for Varying Allergenicity Caused by Caseins from Cows, Goats, Camels, and Mares Based on Bioinformatics Analyses" International Journal of Molecular Sciences 24, no. 3: 2481. https://doi.org/10.3390/ijms24032481
APA StyleZhao, S., Pan, F., Cai, S., Yi, J., Zhou, L., & Liu, Z. (2023). Secrets behind Protein Sequences: Unveiling the Potential Reasons for Varying Allergenicity Caused by Caseins from Cows, Goats, Camels, and Mares Based on Bioinformatics Analyses. International Journal of Molecular Sciences, 24(3), 2481. https://doi.org/10.3390/ijms24032481








