LHC-like Proteins: The Guardians of Photosynthesis
Abstract
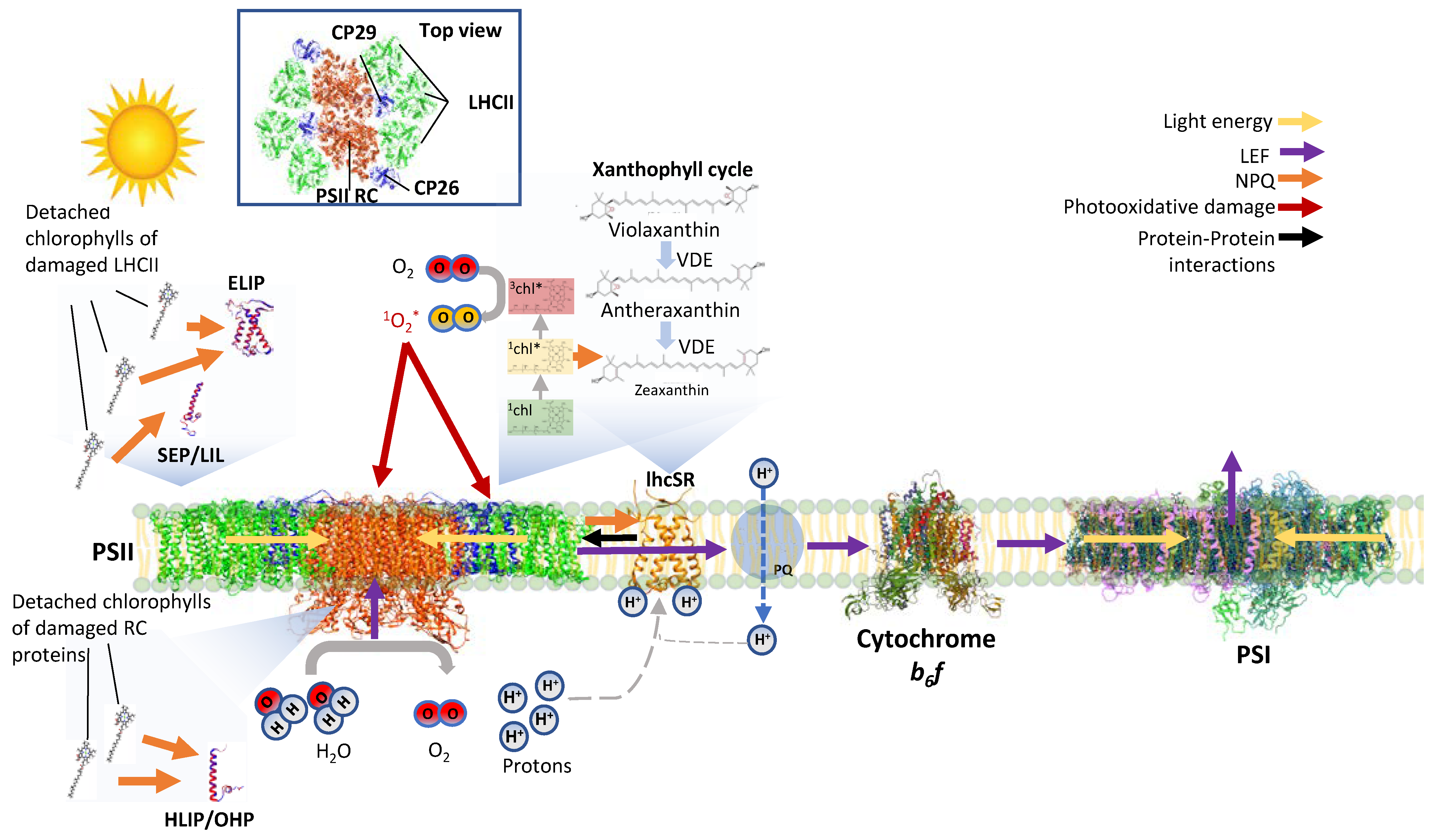
:1. In Addition to Functioning in Light Harvesting, the Light-Harvesting Proteins Are Important for Protection from Photoinhibition
2. LHC-like Proteins Do Not Serve as Antenna but Function in Protection against Photoinhibition
3. lhcSR and psbS Are Stress-Induced Proteins That Protect Photosynthesis against Photoinhibition
4. One-Helix LHC-like Proteins (HLIPs/OHPs/Small-CABs) Protect PSII during Biogenesis and the Photoinhibition Repair Cycle
5. Two-Helix LHC-like Proteins (SEPs/Lils) Function in Chlorophyll Biosynthesis and Protection against Photoinhibition
6. Three-Helix LHC-like Proteins (ELIPs) Play Important Roles in Protection against Photoinhibition
7. Soluble Pigment Binding Proteins also Contribute to Photoprotection
8. LHC-like Proteins Function as Enhancers of Photosynthesis and Survivability during Abiotic Stress and Their Overexpression May Increase Crop Yield
9. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Schuster, G.; Timberg, R.; Ohad, I. Turnover of Thylakoid Photosystem II Proteins during Photoinhibition of Chlamydomonas reinhardtii. Eur. J. Biochem. 1988, 177, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Allakhverdiev, S.I.; Murata, N. A New Paradigm for the Action of Reactive Oxygen Species in the Photoinhibition of Photosystem II. Biochim. Biophys. Acta Bioenerg. 2006, 1757, 742–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pospíšil, P. Production of Reactive Oxygen Species by Photosystem II. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 1151–1160. [Google Scholar] [CrossRef] [Green Version]
- Erickson, E.; Wakao, S.; Niyogi, K.K. Light Stress and Photoprotection in Chlamydomonas reinhardtii. Plant J. 2015, 82, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Levin, G.; Kulikovsky, S.; Liveanu, V.; Eichenbaum, B.; Meir, A.; Isaacson, T.; Tadmor, Y.; Adir, N.; Schuster, G. The Desert Green Algae Chlorella Ohadii Thrives at Excessively High Light Intensities by Exceptionally Enhancing the Mechanisms That Protect Photosynthesis from Photoinhibition. Plant J. 2021, 106, 1260–1277. [Google Scholar] [CrossRef] [PubMed]
- Niyogi, K.K.; Truong, T.B. Evolution of Flexible Non-Photochemical Quenching Mechanisms That Regulate Light Harvesting in Oxygenic Photosynthesis. Curr. Opin. Plant Biol. 2013, 16, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Croce, R.; van Amerongen, H. Natural Strategies for Photosynthetic Light Harvesting. Nat. Chem. Biol. 2014, 10, 492–501. [Google Scholar] [CrossRef]
- Melis, A. Photosystem-II Damage and Repair Cycle in Chloroplasts: What Modulates the Rate of Photodamage in Vivo? Trends Plant Sci. 1999, 4, 130–135. [Google Scholar] [CrossRef]
- Spetea, C.; Nishiyama, Y.; Gollan, P.J.; Liu, J.; Cn, L.; Hua, W.; Lu, Y.; Last, R.L. A New Light on Photosystem II Maintenance in Oxygenic Photosynthesis. Front. Plant Sci. 2019, 10, 975. [Google Scholar] [CrossRef] [Green Version]
- Nixon, P.J.; Michoux, F.; Yu, J.; Boehm, M.; Komenda, J. Recent Advances in Understanding the Assembly and Repair of Photosystem II. Ann. Bot. 2010, 106, 1–16. [Google Scholar] [CrossRef]
- Drop, B.; Webber-Birungi, M.; Yadav, S.K.N.; Filipowicz-Szymanska, A.; Fusetti, F.; Boekema, E.J.; Croce, R. Light-Harvesting Complex II (LHCII) and Its Supramolecular Organization in Chlamydomonas reinhardtii. Biochim. Biophys. Acta Bioenerg. 2014, 1837, 63–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caspy, I.; Neumann, E.; Fadeeva, M.; Liveanu, V.; Savitsky, A.; Frank, A.; Levi Kalisman, Y.; Shkolnisky, Y.; Murik, O.; Treves, H.; et al. Cryo-EM Photosystem I Structure Reveals Adaptation Mechanisms to Extreme High Light in Chlorella ohadii. Nat. Plants 2021, 7, 1314–1322. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Huang, Z.; Chang, S.; Wang, W.; Wang, J.; Kuang, T.; Han, G.; Shen, J.R.; Zhang, X. Structure of a C2S2M2N2-Type PSII–LHCII Supercomplex from the Green Alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2019, 116, 21246–21255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Yan, H.; Wang, K.; Kuang, T.; Zhang, J.; Gui, L.; An, X.; Chang, W. Crystal Structure of Spinach Major Light-Harvesting Complex at 2.72 A˚ Resolution. Nature 2004, 428, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Jansson, S. A Guide to the Lhc Genes and Their Relatives in Arabidopsis. Trends Plant Sci. 1999, 4, 236–240. [Google Scholar] [CrossRef]
- Montané, M.-H.; Kloppstech, K. The Family of Light-Harvesting-Related Proteins (LHCs, ELIPs, HLIPs): Was the Harvesting of Light Their Primary Function? Gene 2000, 258, 1–8. [Google Scholar] [CrossRef]
- Engelken, J.; Brinkmann, H.; Adamska, I. Taxonomic Distribution and Origins of the Extended LHC (Light-Harvesting Complex) Antenna Protein Superfamily. BMC Evol. Biol. 2010, 10, 233. [Google Scholar] [CrossRef] [Green Version]
- Rochaix, J.D.; Bassi, R. LHC-like Proteins Involved in Stress Responses and Biogenesis/Repair of the Photosynthetic Apparatus. Biochem. J. 2019, 476, 581–593. [Google Scholar] [CrossRef]
- Dolganov, N.A.M.; Bhayat, D.; Grossman, A.R. Cyanobacterial Protein with Similarity to the Chlorophyll a/b Binding Proteins of Higher Plants: Evolution and Regulation. Proc. Natl. Acad. Sci. USA 1995, 92, 636–640. [Google Scholar] [CrossRef] [Green Version]
- Heddad, M.; Adamska, I. The Evolution of Light Stress Proteins in Photosynthetic Organisms. Comp. Funct. Genom. 2002, 3, 504–510. [Google Scholar] [CrossRef]
- Bassi, R.; Dall’osto, L. Dissipation of Light Energy Absorbed in Excess: The Molecular Mechanisms. Annu. Rev. Plant Biol. 2021, 72, 47–76. [Google Scholar] [CrossRef] [PubMed]
- Li, X.P.; Gilmore, A.M.; Caffarri, S.; Bassi, R.; Golan, T.; Kramer, D.; Niyogi, K.K. Regulation of Photosynthetic Light Harvesting Involves Intrathylakoid Lumen PH Sensing by the PsbS Protein. J. Biol. Chem. 2004, 279, 22866–22874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinnola, A.; Griffiths, H. The Rise and Fall of Light-Harvesting Complex Stress-Related Proteins as Photoprotection Agents during Evolution. J. Exp. Bot. 2019, 70, 5527–5535. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Chen, X.; Wood, A.J. Two Early Light-Inducible Protein (ELIP) CDNAs from the Resurrection Plant Tortula Ruralis Are Differentially Expressed in Response to Desiccation, Rehydration, Salinity, and High Light. J. Exp. Bot. 2002, 53, 1197–1205. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Wilken, S.; Jimenez, V.; Choi, C.J.; Ansong, C.; Dannebaum, R.; Sudek, L.; Milner, D.S.; Bachy, C.; Nahas Reistetter, E.; et al. Specialized Proteomic Responses and an Ancient Photoprotection Mechanism Sustain Marine Green Algal Growth during Phosphate Limitation. Nat. Microbiol. 2018, 3, 781–790. [Google Scholar] [CrossRef]
- Lee, J.W.; Lee, S.H.; Han, J.W.; Kim, G.H. Early Light-Inducible Protein (ELIP) Can Enhance Resistance to Cold-Induced Photooxidative Stress in Chlamydomonas reinhardtii. Front. Physiol. 2020, 11, 1083. [Google Scholar] [CrossRef]
- Levin, G.; Yasmin, M.; Simanowitz, M.C.; Meir, A.; Tadmor, Y.; Hirschberg, J.; Adir, N.; Schuster, G. A Desert Green Alga That Thrives at Extreme High-Light Intensities Using a Unique Photoinhibition Protection Mechanism. bioRxiv 2022. [Google Scholar] [CrossRef]
- Zhao, L.; Cheng, D.; Huang, X.; Chen, M.; Dall’osto, L.; Xing, J.; Gao, L.; Li, L.; Wang, Y.; Bassi, R.; et al. A Light Harvesting Complex-Like Protein in Maintenance of Photosynthetic Components in Chlamydomonas. Plant Physiol. 2017, 174, 2419–2433. [Google Scholar] [CrossRef] [Green Version]
- Rossini, S.; Casazza, A.P.; Engelmann, E.C.M.; Havaux, M.; Jennings, R.C.; Soave, C. Suppression of Both ELIP1 and ELIP2 in Arabidopsis Does Not Affect Tolerance to Photoinhibition and Photooxidative Stress. Plant Physiol. 2006, 141, 1264–1273. [Google Scholar] [CrossRef] [Green Version]
- Myouga, F.; Takahashi, K.; Tanaka, R.; Nagata, N.; Kiss, A.Z.; Funk, C.; Nomura, Y.; Nakagami, H.; Jansson, S.; Shinozaki, K. Stable Accumulation of Photosystem II Requires ONE-HELIX PROTEIN1 (OHP1) of the Light Harvesting-like Family. Plant Physiol. 2018, 176, 2277–2291. [Google Scholar] [CrossRef]
- Tzvetkova-Chevolleau, T.; Franck, F.; Alawady, A.E.; Dall’Osto, L.; Carrière, F.; Bassi, R.; Grimm, B.; Nussaume, L.; Havaux, M. The Light Stress-Induced Protein ELIP2 Is a Regulator of Chlorophyll Synthesis in Arabidopsis thaliana. Plant J. 2007, 50, 795–809. [Google Scholar] [CrossRef] [PubMed]
- Kromdijk, J.; Głowacka, K.; Leonelli, L.; Gabilly, S.T.; Iwai, M.; Niyogi, K.K.; Long, S.P. Improving Photosynthesis and crop Productivity by Accelerating from Photoprotection. Science (1979) 2016, 354, 857–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.G.; Long, S.P.; Ort, D.R. Improving Photosynthetic Efficiency for Greater Yield. Annu. Rev. Plant Biol. 2010, 61, 235–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Souza, A.P.; Burgess, S.J.; Doran, L.; Hansen, J.; Manukyan, L.; Maryn, N.; Gotarkar, D.; Leonelli, L.; Niyogi, K.K.; Long, S.P. Soybean Photosynthesis and Crop Yield Are Improved by Accelerating Recovery from Photoprotection. Science (1979) 2022, 377, 851–854. [Google Scholar] [CrossRef] [PubMed]
- Treves, H.; Raanan, H.; Kedem, I.; Murik, O.; Keren, N.; Zer, H.; Berkowicz, S.M.; Giordano, M.; Norici, A.; Shotland, Y.; et al. The Mechanisms Whereby the Green Alga Chlorella Ohadii, Isolated from Desert Soil Crust, Exhibits Unparalleled Photodamage Resistance. New Phytol. 2016, 210, 1229–1243. [Google Scholar] [CrossRef] [Green Version]
- Peers, G.; Truong, T.B.; Ostendorf, E.; Busch, A.; Elrad, D.; Grossman, A.R.; Hippler, M.; Niyogi, K.K. An Ancient Light-Harvesting Protein Is Critical for the Regulation of Algal Photosynthesis. Nature 2009, 462, 518–521. [Google Scholar] [CrossRef]
- Prandi, I.G.; Sláma, V.; Pecorilla, C.; Cupellini, L.; Mennucci, B. Structure of the Stress-Related LHCSR1 Complex Determined by an Integrated Computational Strategy. Commun. Biol. 2022, 5, 145. [Google Scholar] [CrossRef]
- Mou, S.; Zhang, X.; Ye, N.; Dong, M.; Liang, C.; Liang, Q.; Miao, J.; Xu, D.; Zheng, Z. Cloning and Expression Analysis of Two Different LhcSR Genes Involved in Stress Adaptation in an Antarctic Microalga, Chlamydomonas sp. ICE-L. Extremophiles 2012, 16, 193–203. [Google Scholar] [CrossRef]
- Ballottari, M.; Truong, T.B.; Re De, E.; Erickson, E.; Stella, G.R.; Fleming, G.R.; Bassi, R.; Niyogi, K.K. Identification of Ph-Sensing Sites in the Light Harvesting Complex Stress-Related 3 Protein Essential for Triggering Non-Photochemical Quenching in Chlamydomonas reinhardtii. J. Biol. Chem. 2016, 291, 7334–7346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liguori, N.; Roy, L.M.; Opacic, M.; Durand, G.; Croce, R. Regulation of Light Harvesting in the Green Alga Chlamydomonas reinhardtii: The C-Terminus of LHCSR Is the Knob of a Dimmer Switch. J. Am. Chem. Soc. 2013, 135, 18339–18342. [Google Scholar] [CrossRef]
- Girolomoni, L.; Cazzaniga, S.; Pinnola, A.; Perozeni, F.; Ballottari, M.; Bassi, R. LHCSR3 Is a Nonphotochemical Quencher of Both Photosystems in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2019, 116, 4212–4217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, M.; Li, M.; Liu, Z.; Cao, P.; Pan, X.; Zhang, H.; Zhao, X.; Zhang, J.; Chang, W. Crystal Structures of the PsbS Protein Essential for Photoprotection in Plants. Nat. Struct. Mol. Biol. 2015, 22, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Guardini, Z.; Bressan, M.; Caferri, R.; Bassi, R.; Dall’Osto, L. Identification of a Pigment Cluster Catalysing Fast Photoprotective Quenching Response in CP29. Nat. Plants 2020, 6, 303–313. [Google Scholar] [CrossRef]
- Alboresia, A.; Gerottob, C.; Giacomettib, G.M.; Bassia, R.; Morosinotto, T. Physcomitrella Patens Mutants Affected on Heat Dissipation Clarify the Evolution of Photoprotection Mechanisms upon Land Colonization. Proc. Natl. Acad. Sci USA 2010, 107, 11128–11133. [Google Scholar] [CrossRef] [Green Version]
- Bonente, G.; Passarini, F.; Cazzaniga, S.; Mancone, C.; Buia, M.C.; Tripodi, M.; Bassi, R.; Caffarri, S. The Occurrence of the PsbS Gene Product in Chlamydomonas reinhardtii and in Other Photosynthetic Organisms and Its Correlation with Energy Quenching. Photochem. Photobiol. 2008, 84, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Correa-Galvis, V.; Redekop, P.; Guan, K.; Griess, A.; Truong, T.B.; Wakao, S.; Niyogi, K.K.; Jahns, P. Photosystem II Subunit PsbS Is Involved in the Induction of LHCSR Protein-Dependent Energy Dissipation in Chlamydomonas Reinhardtii. J. Biol. Chem. 2016, 291, 17478–17487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tibiletti, T.; Auroy, P.; Peltier, G.; Caffarri, S. Chlamydomonas reinhardtii PsbS Protein Is Functional and Accumulates Rapidly and Transiently under High Light. Plant Physiol. 2016, 171, 2717–2730. [Google Scholar] [CrossRef] [Green Version]
- Redekop, P.; Sanz-Luque, E.; Yuan, Y.; Villain, G.; Petroutsos, D.; Grossman, A.R. Transcriptional Regulation of Photoprotection in Dark-to-Light Transition-More than Just a Matter of Excess Light Energy. Sci. Adv. 2022, 8, 1832. [Google Scholar] [CrossRef]
- Nawrocki, W.J.; Liu, X.; Croce, R. Chlamydomonas reinhardtii Exhibits De Facto Constitutive NPQ Capacity in Physiologically Relevant Conditions. Plant Physiol. 2020, 182, 472–479. [Google Scholar] [CrossRef] [Green Version]
- Redekop, P.; Rothhausen, N.; Rothhausen, N.; Melzer, M.; Mosebach, L.; Dülger, E.; Bovdilova, A.; Caffarri, S.; Hippler, M.; Jahns, P. PsbS Contributes to Photoprotection in Chlamydomonas reinhardtii Independently of Energy Dissipation. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148183. [Google Scholar] [CrossRef]
- Allorent, G.; Lefebvre-Legendre, L.; Chappuis, R.; Kuntz, M.; Truong, T.B.; Niyogi, K.K.; Ulm, R.; Goldschmidt-Clermont, M. UV-B Photoreceptor-Mediated Protection of the Photosynthetic Machinery in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2016, 113, 14864–14869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosuge, K.; Tokutsu, R.; Kim, E.; Akimoto, S.; Yokono, M.; Ueno, Y.; Minagawa, J. LHCSR1-Dependent Fluorescence Quenching Is Mediated by Excitation Energy Transfer from LHCII to Photosystem I in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2018, 115, 3722–3727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, U.; Heddad, M.; Adamska, I. Light Stress-Induced One-Helix Protein of the Chlorophyll a/b-Binding Family Associated with Photosystem I. Plant Physiol. 2003, 132, 811–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staleva, H.; Komenda, J.; Shukla, M.K.; Ka, R.; Sobotka, R. Mechanism of Photoprotection in the Cyanobacterial Ancestor of Plant Antenna Proteins. Nat. Chem. Biol. 2015, 11, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Funk, C.; Vermaas, W. A Cyanobacterial Gene Family Coding for Single-Helix Proteins Resembling Part of the Light-Harvesting Proteins from Higher Plants. Biochemistry 1999, 38, 9397–9404. [Google Scholar] [CrossRef]
- Yao, D.; Kieselbach, T.; Komenda, J.; Promnares, K.; Hernández Prieto, M.A.; Tichy, M.; Vermaas, W.; Funk, C. Localization of the Small CAB-like Proteins in Photosystem II. J. Biol. Chem. 2007, 282, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Komenda, J.; Sobotka, R. Cyanobacterial High-Light-Inducible Proteins - Protectors of Chlorophyll-Protein Synthesis and Assembly. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 288–295. [Google Scholar] [CrossRef]
- Wang, Q.; Jantaro, S.; Lu, B.; Majeed, W.; Bailey, M.; He, Q. The High Light-Inducible Polypeptides Stabilize Trimeric Photosystem I Complex under High Light Conditions in Synechocystis PCC 6803. Plant Physiol. 2008, 147, 1239–1250. [Google Scholar] [CrossRef] [Green Version]
- Promnares, K.; Komenda, J.; Bumba, L.; Nebesarova, J.; Vacha, F.; Tichy, M. Cyanobacterial Small Chlorophyll-Binding Protein ScpD (HliB) Is Located on the Periphery of Photosystem II in the Vicinity of PsbH and CP47 Subunits. J. Biol. Chem. 2006, 281, 32705–32713. [Google Scholar] [CrossRef] [Green Version]
- Vavilin, D.; Yao, D.; Vermaas, W. Small Cab-like Proteins Retard Degradation of Photosystem II-Associated Chlorophyll in Synechocystis Sp. PCC 6803: Kinetic Analysis of Pigment Labeling with 15N and 13C. J. Biol. Chem. 2007, 282, 37660–37668. [Google Scholar] [CrossRef]
- Knoppová, J.; Sobotka, R.; Tichý, M.; Yu, J.; Konik, P.; Halada, P.; Nixon, P.J.; Komenda, J. Discovery of a Chlorophyll Binding Protein Complex Involved in the Early Steps of Photosystem II Assembly in Synechocystis. Plant Cell 2014, 26, 1200–1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chidgey, J.W.; Linhartová, M.; Komenda, J.; Jackson, P.J.; Dickman, M.J.; Canniffe, D.P.; Koník, P.; Pilný, J.; Hunter, C.N.; Sobotka, R. A Cyanobacterial Chlorophyll Synthase-HliD Complex Associates with the Ycf39 Protein and the YidC/Alb3 Insertase. Plant Cell 2014, 26, 1267–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansson, S.; Andersson, J.; Kim, S.J.; Jackowski, G. An Arabidopsis Thaliana Protein Homologous to Cyanobacterial High-Light-Inducible Proteins. Plant Mol. Biol. 2000, 42, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, B.; Zhang, J.; Kong, F.; Zhang, L.; Meng, H.; Li, W.; Rochaix, J.D.; Li, D.; Peng, L. OHP1, OHP2, and HCF244 Form a Transient Functional Complex with the Photosystem II Reaction Center. Plant Physiol. 2019, 179, 195–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hey, D.; Grimm, B. ONE-HELIX PROTEIN2 (OHP2) Is Required for the Stability of OHP1 and Assembly Factor HCF244 and Is Functionally Linked to PSII Biogenesis. Plant Physiol. 2018, 177, 1453–1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahns, P.; Sakamoto, W.; Picorel, R.; Funck, D.; Beck, J.; Lohscheider, J.N.; Albert, S.; Andersson, U.; Mendgen, K.W.; Rojas-Stütz, M.C.; et al. Small One-Helix Proteins Are Essential for Photosynthesis in Arabidopsis. Front. Plant Sci. 2017, 8, 7. [Google Scholar] [CrossRef] [Green Version]
- Hey, D.; Grimm, B. One-Helix Protein 1 and 2 Form Heterodimers to Bind Chlorophyll in Photosystem II Biogenesis. Plant Physiol. 2020, 183, 179–193. [Google Scholar] [CrossRef] [Green Version]
- Heddad, M.; Adamska, I. Light Stress-Regulated Two-Helix Proteins in Arabidopsis Thaliana Related to the Chlorophyll a/b-Binding Gene Family. Proc. Natl. Acad. Sci. USA 2000, 97, 3741–3746. [Google Scholar] [CrossRef]
- Reisinger, V.; Plöscher, M.; Eichacker, L.A. Lil3 Assembles as Chlorophyll-Binding Protein Complex during Deetiolation. FEBS Lett. 2008, 582, 1547–1551. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, R.; Rothbart, M.; Oka, S.; Takabayashi, A.; Takahashi, K.; Shibata, M.; Myouga, F.; Motohashi, R.; Shinozaki, K.; Grimm, B.; et al. LIL3, a Light-Harvesting-like Protein, Plays an Essential Role in Chlorophyll and Tocopherol Biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 16721–16725. [Google Scholar] [CrossRef]
- Hey, D.; Rothbart, M.; Herbst, J.; Wang, P.; Müller, J.; Wittmann, D.; Gruhl, K.; Grimm, B. LIL3, a Light-Harvesting Complex Protein, Links Terpenoid and Tetrapyrrole Biosynthesis in Arabidopsis Thaliana. Plant Physiol. 2017, 174, 1037–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohscheider, J.N.; Rojas-Stütz, M.C.; Rothbart, M.; Andersson, U.; Funck, D.; Mendgen, K.; Grimm, B.; Adamska, I. Altered Levels of LIL3 Isoforms in Arabidopsis Lead to Disturbed Pigment-Protein Assembly and Chlorophyll Synthesis, Chlorotic Phenotype and Impaired Photosynthetic Performance. Plant Cell Environ. 2015, 38, 2115–2127. [Google Scholar] [CrossRef] [PubMed]
- Mork-Jansson, A.; Bue, A.K.; Gargano, D.; Furnes, C.; Reisinger, V.; Arnold, J.; Kmiec, K.; Eichacker, L.A. Lil3 Assembles with Proteins Regulating Chlorophyll Synthesis in Barley. PLoS ONE 2015, 10, e0133145. [Google Scholar] [CrossRef] [Green Version]
- Skotnicová, P.; Staleva, M.H.; Kuznetsova, V.; Bína, D.; Konert, M.M.; Lu, S.; Polívka, T.; Sobotka, R. Plant LHC-like Proteins Show Robust Folding and Static Non-Photochemical Quenching. Nat. Commun. 2021, 12, 6890. [Google Scholar] [CrossRef] [PubMed]
- Green, B.R.; Ktihlbrandt, W. Sequence Conservation of Light-Harvesting and Stress-Response Proteins in Relation to the Three-Dimensional Molecular Structure of LHCII. Photosynth. Res. 1995, 44, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Adamska, I.; Ohad, I.; Kloppstech, K. Synthesis of the Early Light-Inducible Protein Is Controlled by Blue Light and Related to Light Stress. Proc. Natl. Acad. Sci. USA 1992, 89, 2610–2613. [Google Scholar] [CrossRef] [Green Version]
- Heddad, M.; Norén, H.; Reiser, V.; Dunaeva, M.; Andersson, B.; Adamska, I. Differential Expression and Localization of Early Light-Induced Proteins in Arabidopsis. Plant Physiol. 2006, 142, 75–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamska, I.; Roobol-Bóza, M.; Lindahl, M.; Andersson, B. Isolation of Pigment-Binding Early Light-Inducible Proteins from Pea. Eur. J. Biochem. 1999, 260, 453–460. [Google Scholar] [CrossRef]
- Adamska, I. ELIPs–Light-Induced Stress Proteins. Physiol. Plant. 1997, 100, 794–805. [Google Scholar] [CrossRef]
- Hutin, C.; Nussaume, L.; Moise, N.; Moya, I.; Kloppstech, K.; Havaux, M. Early Light-Induced Proteins Protect Arabidopsis from Photooxidative Stress. Proc. Natl. Acad. Sci. USA 2003, 100, 4921–4926. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Yang, H.; Liang, Y.; Li, X.; Oliver, M.J.; Zhang, D. Functional Aspects of Early Light-induced Protein (Elip) Genes from the Desiccation-tolerant Moss Syntrichia Caninervis. Int. J. Mol. Sci. 2020, 21, 1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elrad, D.; Grossman, A.R. A Genome’s Eye View of the Light-Harvesting Polypeptides of Chlamydomonas reinhardtii. Curr. Genet. 2004, 45, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Zones, J.M.; Blaby, I.K.; Merchant, S.S.; Umen, J.G. High-Resolution Profiling of a Synchronized Diurnal Transcriptome from Chlamydomonas reinhardtii Reveals Continuous Cell and Metabolic Differentiation. Plant Cell 2015, 27, 2743–2769. [Google Scholar] [CrossRef]
- Lerss, A.; Levy, H.; Zamirq, A. Co-Regulation of a Gene Homologous to Early Light-Induced Genes in Higher Plants and Carotene Biosynthesis in the Alga Dunaiella bardawi. J. Biol. Chem. 1991, 266, 13698–13705. [Google Scholar] [CrossRef]
- Levy, H.; Gokhman, I.; Zamir, A. Regulation and Light-Harvesting Complex Association of a Dunaliella Protein Homologous to Early Light-Induced Proteins in Higher Plants. J. Biol. Chem. 1992, 267, 18831–18836. [Google Scholar] [CrossRef] [PubMed]
- Levy, H.; Tal, T.; Shaish, A.; Zamir, A. Cbr, an Algal Homolog of Plant Early Light-Induced Proteins, Is a Putative Zeaxanthin Binding Protein. J. Biol. Chem. 1993, 268, 20892–20896. [Google Scholar] [CrossRef] [PubMed]
- Banet, G.; Pick, U.; Zamir, A. Light-Harvesting Complex II Pigments and Proteins in association with Cbr, a Homolog of Higher-Plant Early Light-Inducible proteins in the Unicellular Green Alga Dunaliella. Planta 2000, 210, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Holt, T.K.; Krogmann, D.W. A Carotenoid-Protein From Cyanobacteria. Biochim. Biophys. Acta Bioenerg. 1981, 637, 408–414. [Google Scholar] [CrossRef]
- Kerfeld, C.A.; Sawaya, M.R.; Brahmandam, V.; Cascio, D.; Ho, K.K.; Trevithick-Sutton, C.C.; Krogmann, D.W.; Yeates, T.O. The Crystal Structure of a Cyanobacterial Water-Soluble Carotenoid Binding Protein. Structure 2003, 11, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Wilson, A.; Ajlani, G.; Verbavatz, J.M.; Vass, I.; Kerfeld, C.A.; Kirilovsky, D. A Soluble Carotenoid Protein Involved in Phycobilisome-Related Energy Dissipation in Cyanobacteria. Plant Cell 2006, 18, 992–1007. [Google Scholar] [CrossRef]
- Sedoud, A.; López-Igual, R.; Rehman, A.U.; Wilson, A.; Perreau, F.; Boulay, C.; Vass, I.; Krieger-Liszkay, A.; Kirilovsky, D. The Cyanobacterial Photoactive Orange Carotenoid Protein Is an Excellent Singlet Oxygen Quencher. Plant Cell 2014, 26, 1781–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirilovsky, D.; Kerfeld, C.A. Cyanobacterial Photoprotection by the Orange Carotenoid Protein. Nat. Plants 2016, 2, 16180. [Google Scholar] [CrossRef] [PubMed]
- Lé Wilson, A.; Punginelli, C.; Gall, A.; Bonetti, C.; Alexandre, M.; Routaboul, J.-M.; Kerfeld, C.A.; van Grondelle, R.; Robert, B.; Kennis, J.T.M.; et al. A Photoactive Carotenoid Protein Acting as Light Intensity Sensor. Proc. Natl. Acad. Sci. USA 2008, 105, 12075–12080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yakushiji, E.; Uchino, K.; Sugimura, Y.; Shiratori, I.; Takamiya, F. Isolation of Water-Soluble Chlorophyll Protein from the Leaves of Chenopodium album. Biochim. Biophys. Acta 1963, 75, 293–298. [Google Scholar] [CrossRef]
- Horigome, D.; Satoh, H.; Itoh, N.; Mitsunaga, K.; Oonishi, I.; Nakagawa, A.; Uchida, A. Structural Mechanism and Photoprotective Function of Water-Soluble Chlorophyll-Binding Protein. J. Biol. Chem. 2007, 282, 6525–6531. [Google Scholar] [CrossRef] [Green Version]
- Agostini, A.; Palm, D.M.; Schmitt, F.J.; Albertini, M.; di Valentin, M.; Paulsen, H.; Carbonera, D. An Unusual Role for the Phytyl Chains in the Photoprotection of the Chlorophylls Bound to Water-Soluble Chlorophyll-Binding Proteins. Sci. Rep. 2017, 7, 7504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palm, D.M.; Agostini, A.; Pohland, A.C.; Werwie, M.; Jaenicke, E.; Paulsen, H. Stability of Water-Soluble Chlorophyll Protein (WSCP) Depends on Phytyl Conformation. ACS Omega 2019, 4, 7971–7979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damaraju, S.; Schlede, S.; Eckhardt, U.; Lokstein, H.; Grimm, B. Functions of the Water Soluble Chlorophyll-Binding Protein in Plants. J. Plant Physiol. 2011, 168, 1444–1451. [Google Scholar] [CrossRef]
- Evans, J.R. Improving Photosynthesis. Plant Physiol. 2013, 162, 1780–1793. [Google Scholar] [CrossRef] [Green Version]
- Nowicka, B.; Ciura, J.; Szymańska, R.; Kruk, J. Improving Photosynthesis, Plant Productivity and Abiotic Stress Tolerance – Current Trends and Future Perspectives. J. Plant Physiol. 2018, 231, 415–433. [Google Scholar] [CrossRef]
- Vecchi, V.; Barera, S.; Bassi, R.; Dall’osto, L. Potential and Challenges of Improving Photosynthesis in Algae. Plants 2020, 9, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Głowacka, K.; Kromdijk, J.; Kucera, K.; Xie, J.; Cavanagh, A.P.; Leonelli, L.; Leakey, A.D.B.; Ort, D.R.; Niyogi, K.K.; Long, S.P. Photosystem II Subunit S Overexpression Increases the Efficiency of Water Use in a Field-Grown Crop. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubbart, S.; Smillie, I.R.A.; Heatley, M.; Swarup, R.; Foo, C.C.; Zhao, L.; Murchie, E.H. Enhanced Thylakoid Photoprotection Can Increase Yield and Canopy Radiation Use Efficiency in Rice. Commun. Biol. 2018, 1, 22. [Google Scholar] [CrossRef] [PubMed]
- Perozeni, F.; Stella, G.R.; Ballottari, M. LHCSR Expression under HSP70/RBCS2 Promoter as a Strategy to Increase Productivity in Microalgae. Int. J. Mol. Sci. 2018, 19, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuo, C.; Cai, J.; Guo, Z. Overexpression of Early Light-Induced Protein (ELIP) Gene from Medicago Sativa Ssp. Falcata Increases Tolerance to Abiotic Stresses. J. Agron. 2013, 105, 1433–1440. [Google Scholar] [CrossRef]
- Liu, B.; Zhao, F.; Zhou, H.; Xia, Y.; Wang, X. Photoprotection Conferring Plant Tolerance to Freezing Stress through Rescuing Photosystem in Evergreen Rhododendron. Plant Cell Environ. 2022, 45, 2093–2108. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; He, B.; Guo, M.L.; Xie, X.; Huan, L.; Zhang, B.; Shao, Z.; Wang, G. Overexpression of OHPs in Neopyropia Yezoensis (Rhodophyta) Reveals Their Possible Physiological Roles. Algal Res. 2022, 64, 102714. [Google Scholar] [CrossRef]
- Hey, D.; Grimm, B. Requirement of ONE-HELIX PROTEIN 1 (OHP1) in Early Arabidopsis Seedling Development and under High Light Intensity. Plant Signal. Behav. 2018, 13, e1550317. [Google Scholar] [CrossRef]




| Organism | Protein | LHC-Motif Sequence | Location |
|---|---|---|---|
| C. reinhardtii | psbS2 | ELFVGRLAMVGFSAS | 71–85 |
| ELFVGRAAQLGFAFS | 159–173 | ||
| C. reinhardtii | lhcSR1 | EITHGRVAMLAALGF | 81–95 |
| ELNNGRLAMIAIAAF | 191–205 | ||
| C. reinhardtii | lhcSR3 | EITHGRVAMLAALGF | 87–101 |
| ELNNGRLAMIAIAAF | 197–211 | ||
| C. reinhardtii | ELIP (1) | EINNGRIAMVSVVTA | 67–81 |
| EKINGRAAMMGLTSL | 346–360 | ||
| C. reinhardtii | ELIP (2) | EIVNGRLAMLGFVSA | 103–117 |
| ELLNGRAAMIGFAAM | 171–185 | ||
| C. reinhardtii | Lil3 (SEP) | EKLNGRAAMMGYVLA | 162–176 |
| A. thaliana | Lil3 (SEP) | ELLNGRAAMIGFFMA | 174–188 |
| A. thaliana | OHP1 (HLIP) | EIWNSRACMIGLIGT | 69–83 |
| A. thaliana | OHP2 (HLIP) | EISNGRWAMFGFAVG | 130–144 |
| Synechocystis | HliA (HLIP) | EKLNGRLAMIGFVAL | 36–40 |
| Consensus | E**NGR*AM*G |
| TM Helices | Protein | Organism | Pigments | Suggested Function |
|---|---|---|---|---|
| 1 | HLIP (Prokaryota)OHP (Eukaryota) | Cyanobacteria, Algae, Plants | Chlorophyll a, β-carotene | Temporarily binds pigments during PSII repair cycle. |
| 2 | SEP/Lil | Algae, Plants | Chlorophyll a, Lutein/Zeaxanthin | Temporarily binds pigments during LHC turnover, chlorophyll biosynthesis. |
| 3 | ELIP | Algae, Plants | Chlorophyll a, Lutein, Zeaxanthin | Temporarily binds pigments during LHC turnover, suppression of chlorophyll biosynthesis |
| 3 | lhcSR | Algae and Mosses | Chlorophyll a, Lutein | Thylakoid lumen acidification sensing, destination for excess energy transfer from LHC. |
| 4 | psbS | Mostly Plants and Mosses | - | Thylakoid lumen acidification sensing, activation of NPQ in LHCII subunit CP29. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Levin, G.; Schuster, G. LHC-like Proteins: The Guardians of Photosynthesis. Int. J. Mol. Sci. 2023, 24, 2503. https://doi.org/10.3390/ijms24032503
Levin G, Schuster G. LHC-like Proteins: The Guardians of Photosynthesis. International Journal of Molecular Sciences. 2023; 24(3):2503. https://doi.org/10.3390/ijms24032503
Chicago/Turabian StyleLevin, Guy, and Gadi Schuster. 2023. "LHC-like Proteins: The Guardians of Photosynthesis" International Journal of Molecular Sciences 24, no. 3: 2503. https://doi.org/10.3390/ijms24032503
APA StyleLevin, G., & Schuster, G. (2023). LHC-like Proteins: The Guardians of Photosynthesis. International Journal of Molecular Sciences, 24(3), 2503. https://doi.org/10.3390/ijms24032503






