Abstract
Epilepsy is one of the most frequent chronic neurologic disorders that affects nearly 1% of the population worldwide, especially in developing countries. Currently, several antiepileptic drugs (AEDs) are available for its therapy, and although the prognosis is good for most patients, 20%–30% amongst them do not reach seizure freedom. Numerous factors may explain AED-resistance such as sex, age, ethnicity, type of seizure, early epilepsy onset, suboptimal dosing, poor drug compliance, alcohol abuse, and in particular, genetic factors. Specifically, the interindividual differences in drug response can be caused by single nucleotide polymorphisms (SNPs) in genes encoding for drug efflux transporters, for the brain targets of AEDs, and for enzymes involved in drug metabolism. In this review, we used the PubMed database to retrieve studies that assessed the influence of SNPs on the pharmacokinetic (PK), pharmacodynamic (PD), and efficacy of new antiepileptic drugs. Our results showed that polymorphisms in the ABCB1, ABCC2, UGT1A4, UGT2B7, UGT2B15, CYP2C9, and CYP2C19 genes have an influence on the PK and efficacy of AEDs, suggesting that a genetic pre-evaluation of epileptic patients could help clinicians in prescribing a personalized treatment to improve the efficacy and the safety of the therapy.
1. Introduction
Epilepsy is one of the most frequent chronic neurologic disorders, and it affects nearly 1% of the population worldwide, especially in developing countries [1]. It is characterized by recurrent epileptic seizures caused by super-synchronous discharges in the brain neurons [2]. Currently, around 25 antiepileptic drugs (AEDs) are available for epilepsy therapy [3], and they are classified as old or first generation and the new generation of antiepileptics. The older-generation AEDs have been used in clinical practice for more than four decades, and they include valproate, carbamazepine phenobarbital, ethosuximide, phenytoin, primidone, clonazepam, and clobazam [4] (Table 1), while the new generation AEDs have been approved for the therapy of epilepsy since 1989. In particular, they are divided into second generation and third generation, which have been launched in the market since 2004 [5] (Table 1). Older AEDs such as phenytoin, carbamazepine, valproic acid (sodium valproate), and phenobarbital are characterized by a narrow therapeutic range and a pronounced inter-individual variability in their pharmacokinetics. It is often claimed that the new generation of AEDs such as gabapentin and levetiracetam have a major advantage over the older AEDs in that they have more predictable pharmacokinetics [6].

Table 1.
Classification of AEDs.
Moreover, depending on their mechanism of action, the AEDs can be classified in four main categories: (i) voltage-gated ion channels modulators; (ii) neurotransmitter-release modulators; (iii) GABAergic transmission enhancers; and (iv) glutamatergic transmission inhibitors [4].
Amongst the AEDs that block ion channels, the most important class are the voltage-gated sodium channel blockers. These drugs (i.e., lamotrigine and lacosamide) act to prevent the opening of membrane depolarization-activated sodium channels, thus arresting the initiation of action potentials [7]. Regarding the neurotransmitter release modulators, the most widely used drugs are levetiracetam and brivaracetam, and both these compounds facilitate the action of synaptic vesicle protein 2A (SV2A) [8,9], a protein that reduces the fusion between the vesicle and presynaptic membrane by altering the cell calcium responsivity [10]. Moreover, barbiturates, benzodiazepines, vigabatrin, and tiagabine are the main actors in the modulation of GABAergic transmission. The first two drugs act directly on the GABAA channel, which, when open, leads to a membrane hyperpolarization by allowing a chloride current within the cell. On the other hand, tiagabine and vigabatrin act by increasing the quantity of GABA available in the synaptic cleft. Finally, the only molecule that specifically acts on glutamatergic receptors is perampanel [11], a specific non-competitive AMPA receptor antagonist, even if an antagonistic action on the metabotropic mGluR5 receptor has also been suggested as an associated mechanism of action of rufinamide.
Although these drugs ameliorate the prognosis for most patients affected by epilepsy, 20%–30% of subjects do not reach seizure freedom, even when treated with multiple AEDs [12]. Several factors may be addressed to partly explain AED-resistance such as early epilepsy onset, type of seizure, suboptimal dosing, poor drug compliance, alcohol abuse, and a high frequency of seizures in the diagnostic assessment period [13]. In addition, it has been suggested that genetic factors may contribute to this variability in clinical outcomes [14]. In fact, single nucleotide polymorphisms (SNPs), which are variations of a single DNA base, may affect the AEDs’ efficacy, tolerability, safety, and duration of action [15]. Specifically, the interindividual variability in drug response can be affected by SNPs in genes encoding for drug efflux transporters (ABCB1, ABCC2) localized in the gastrointestinal tract and blood–brain barrier (BBB), for brain targets of AEDs (voltage-dependent Na+ channels, synapse vesicle protein SV2A), and for enzymes involved in drug metabolism (CYP2C19, UGT1A4).
Therefore, the aim of the present review was to evaluate the effects of SNPs in genes involved in the pharmacokinetic (PK) and pharmacodynamic (PD) of AEDs and assess if these polymorphisms could affect the efficacy, blood levels, and clinical outcomes of AEDs in epileptic patients. In particular, we chose to focus our attention on the second and third generation of antiepileptic drugs in light of their rapid increase in clinical practice.
2. Polymorphisms Affecting AEDs Transporters
SNPs in genes encoding for drug efflux transporters may affect the absorption and distribution of AEDs, and the most studied polymorphisms are those concerning the genes encoding for the transporters ABCB1 and ABCC2, also known as P-glycoprotein (P-gp), which transport several AEDs [16]. One of the most common variants in the ABCB1 gene is the C3435T polymorphism in exon 26 of the MDR1(Multi-Drug Resistance 1) gene, which was correlated with drug resistance in epilepsy among Caucasians. In fact, it has been observed that individuals with 3435 TT carriers have reduced P-gp activity and higher plasma drug concentrations after oral administration [17,18]. P-gp is present in duodenal cells, and this explains why ABCB1 SNPs can influence the absorption and plasma concentrations of AEDs [19]. Moreover, P-gp is also present in the membrane of cerebral capillary endothelial cells, in order to protect the brain from intoxication caused by lipophilic xenobiotics. Therefore, an increased expression of this gene causes higher amounts of P-gp in the endothelial cells and in astrocytes, leading to a decreased drug parenchymal concentration, regardless of the fact that they reach blood therapeutic levels [20]. In this context, Shen et al. found that carriers of the ABCB1 3435C > T CC genotype had a higher oxcarbazepine concentration than TT. They also observed more oxcarbazepine-responsive patients amongst the CC genotype than CT and TT. However, when ABCC2 SNPs were taken into account, they did not find an effect on the oxcarbazepine plasma concentrations and therapeutic efficacy [21]. On the other hand, an influence of ABCC2 1249 G > A on the oxcarbazepine dose was observed in a study conducted by Ma et al., which showed that ABCC2 1249 G > A carriers needed a significantly higher oxcarbazepine dose than non-carriers [22].
Moreover, Yao et al. found that child carriers of the ABCB1 C3435 > T CC genotype had a higher concentration–dose ratio (CDR) of oxcarbazepine when compared with CT carriers [23]. Two studies by Zhao and colleagues assessed lacosamide PK and drug-resistance in a population of Asiatic pediatric patients. The first one showed that patients with ABCC2 1249G > A GA and AA genotypes and with ABCC2 -24C > T CT and TT genotypes had lower lacosamide CDR than the ABCC2 1249G > A GG and ABCC2 -24C > T CC genotypes [24]. In the second study, they found that the drug-resistant group had a higher frequency of the ABCB1 G2677T/A GT genotype, while a higher CDR value was found in the GG genotype and in the ABCB1 C3435T CC genotype when compared to the G2677T/A AT and ABCB1 C3435T CT genotypes. Moreover, they observed higher lacosamide plasma levels in ABCB1 C3435T CC carriers rather than the CT and TT carriers [25]. Furthermore, Zhou et al. reported that rs3114020 TT carriers had lower lamotrigine CDR values than C allele carriers and patients carrying the rs2231142 CA or AA genotype showed higher lamotrigine concentrations compared with the CC carriers [26]. Shen et al. studied the association between SNPs in ABCG2 or ABCC2 genes and lamotrigine pharmacokinetics and pharmaco-resistance. They also investigated the influence on these parameters of SNPs in another subfamily of influx transporters, the organic cation transporters (OCTs), and of the hepatocyte nuclear factor 4 alpha (HNF4a), a liver-enriched regulator of liver development that binds several gene promoters in human and mouse liver including CYPs, UGTs, and transporters [27,28]. They reported higher lamotrigine levels in the patients that were carriers of the rs628031 AA and AG genotype and in the rs2231142 AA carriers when compared with the OCT GG genotype and ABCG2 CC and CA genotype, respectively. On the other hand, they found no significant influence of the other studied SNPs (OCT1 rs2282143, ABCG2 rs2231137, ABCC2 rs2273697, and HNF4 rs2071197, rs3212183) on lamotrigine concentrations and therapeutic response [29]. Lovric et al. studied the influence on lamotrigine levels of ABCB1 C1236T, G2677T/A, and C3435T SNPs. They reported that 1236CT and TT genotype carriers had lower lamotrigine levels and dose-corrected concentrations than CC carriers. They also found no significant influence on the lamotrigine parameters of ABCB1 G2677T/A and the C3435T genotypes, although the concentrations in the monotherapy group were higher in the 2677GG and C3435T CC genotypes [30]. Two studies analyzed the correlation between SNPs in transporters and the pharmaco-resistance of AEDs. In the first, Stasiołek et al. found that ABCB1 C3435T CC carriers had a higher incidence of drug resistant epilepsy in a population of Polish children [31]. In the second, Ufer et al. observed, in a Caucasian population, a higher frequency of the ABCC2 1249G > A variant in the responder’s patients. Instead, they reported that ABCC2 -24C > T and 3972C > T did not influence therapy response [32]. On the other hand, some studies have reported a lack of association between polymorphisms in transporter genes and the effects of AEDs. For example, Wang et al. did not find a significant correlation between SNPs in the ABCB1 genes and oxcarbazepine PK [33]. Lin et al. studied the ABCB1 3435C > T and ABCB2 1249G > A genotypes and found that they had no influence on oxcarbazepine PK in a pediatric population [34]. Petrenaite et al. found no association between the ABCB1 1236 C > T and ABC1B1 3435 C > T SNPs and lamotrigine [35]. Finally, Zhao et al. did not find a correlation between the presence of the ABCB1 C1236T genotype and the levetiracetam levels [36]. In summary, several studies found a correlation between the presence of SNPs in the ABCB1, ABCC2, and ABCG2 genes and PK, PD, or efficacy of AEDs, which suggests that a pharmacogenetic pre-evaluation could be useful for prescribing the most appropriate dose-drug, according to the patient’s genetic profile (Figure 1 and Table 2).
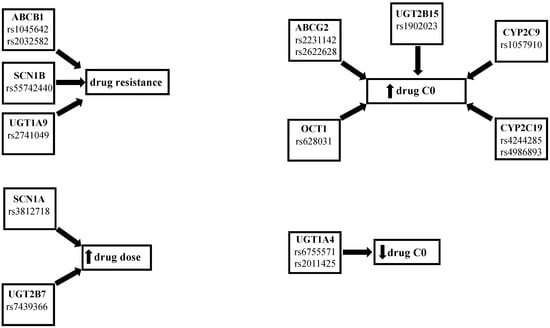
Figure 1.
Summary of the effects of the SNPs.

Table 2.
Summary of studies on SNPs affecting AED transporters.
3. Polymorphisms Affecting AED Brain-Targets
Regarding the brain targets of the AEDs, the most interesting data up until now have been reported on voltage-dependent Na+ channels (SCNs), which are responsible for the generation and propagation of action potential in neurons, and for this reason, many AEDs act by reducing the high-frequency firing of the voltage-dependent SCN that occurs during seizures [37]. SCN are formed by one alpha subunit and two beta subunits. The alpha subunits are encoded by the SCN (1–10)A genes and beta subunits by the SCN(1–4)B genes [38]. Exon 5 of the SCN1A gene encodes for the I-S4 voltage sensor domain, one of the sodium channels expressed in brain cells. Genetic variations in the α-subunit may affect the electro-physiological properties of sodium channels in drug-resistant epilepsy patients. For example, it has been suggested that the SCN1A-A3184G (p.Tr1067Ala) polymorphism may be involved in the gating of sodium channels, therefore making them insensitive to sodium-channel blockers [39].
In this regard, Lin et al. studied the influence of SCN1A, SCN1B, SCN2A, and SCN9A SNPs on the response to several sodium blocker AEDs in 214 Taiwanese patients. They observed that non-responder patients had a significantly higher frequency of allele C of rs55742440 in SCN1B. Moreover, they found no significant association between SNPs and AED response in the predominant sodium channel blocking AED group, which included oxcarbazepine, lamotrigine, and lacosamide [40]. Moreover, Angelopoulou and colleagues reported a similar distribution of SCN1A IVS5-91 rs3812718 G > A genotypes between drug-responsive and drug-resistant patients treated with sodium channel blocker AEDs. They also found that within the monotherapy-responsive group, G/G carriers needed lower doses than A/A or A/G carriers [41]. Furthermore, Gazhala et al. studied the SCN1A-A3184G SNP in 326 children with non-lesional epilepsy, 163 AEDs-resistant, 163 AEDs responders, and 163 healthy controls, and they did not find a significant influence of the studied SNPs between the AED responders and those resistant [42]. Pejanovic-Skobic found no significant difference of SCN2A 56G > A regarding G allele frequency between the responders or no responders to lamotrigine monotherapy, and between epileptic patients and healthy control subjects [43]. Ma et al. found that SCN1A IVS5-91G > A variant allele carriers needed significantly higher oxcarbazepine maintenance doses than non-carriers [22]. Studying the same polymorphism, other studies did not find a significant influence on lamotrigine and oxcarbazepine response or maintenance dosage. Thus, Kumari and colleagues did not observe an effect of the SCN1A IVS5-91G > A genotype on drug response in patients that showed drug resistance to oxcarbazepine [44]. This SNP was also investigated by Markovic et al., who did not find a significant influence of SCN1A IVS5-91 G > A SNPs on lamotrigine efficacy. However, they suggest that these SNPs may affect the average maintenance dose because in the responder group, the AA genotype needed higher doses than the GA and GG carriers, even if the difference was not statistically significant [45]. Furthermore, Manna et al. investigated the relationship between the c.603-91G > A polymorphism and response to oxcarbazepine, and they did not find a significant difference in the frequency of the presence of the 603-91G > A genotype between the drug-resistant and drug-responsive patients, and between this polymorphism and the response to oxcarbazepine. Moreover, the same authors found no influence of the 603-91G > A genotype on the drug dose in both the whole group of patients or within the drug-resistant compared to the drug-responsive patients [46].
In addition to SCNs, another important AED target is the synaptic vesicle protein SV2A, the prevalent of three isoforms (SV2A, SV2B, and SV2C) in the brain, and site of action of levetiracetam and brivaracetam. It has been reported that homozygous SV2A knockout mice are normal at birth but fail to grow, experience severe seizures, and die prematurely. In heterozygous knockout mice, the same study reported a likelihood to have seizure 10-fold higher than wild-type animals [47]. However, Lynch et al. found no association between the SV2A, SV2B, or SV2C polymorphisms and levetiracetam levels [48]. In summary, some studies found that SCN1A IVS5-91G > A GG carriers needed lower AED dose and a study found a higher presence of the SCN1B rs55742440 genotype in no-responders to sodium channel blockers (Figure 1 and Table 3); however, considering that few pharmacogenetic studies in epileptic patients treated with SCNs are present in the literature, the role of these SNPs on the PK, PD, and efficacy of AEDs should be further investigated.

Table 3.
Summary of studies on SNPs affecting AED brain-targets.
4. Polymorphisms Affecting AEDs Metabolizing Enzymes
Important SNPs in genes for enzymes responsible for drug metabolism are those concerning the UDP-glucuronosyltransferase (UGT) and CYP P450 isoenzymes. In fact, UGT isoenzymes metabolize oxcarbazepine in its main active metabolite 10-monohydroxy derivative (MHD) [49] and lamotrigine in lamotrigine-2-N-glucuronide (2-N-GLUC) [50], whilst the most important isoenzymes for the metabolization of AEDs are CYP3A4 and CYP2C19 [51]. In particular, UGT1A4*3 (142T > G, L48V) and UGT1A4*2 (70C > A, P24T) the most common SNPs involved in AED metabolism, caused a decreased glucuronidation, whereas the presence of one or two defective alleles CYP2C9*2, CYP2C9*3, CYP2C19*2, CYP2C19*3 in the CYP2C9 and CYP2C19 genes led to an increase in the drug concentrations [52,53]. In this regard, Lu et al. observed that UGT1A9 I399 C > T carriers in 124 Chinese patients treated with oxcarbazepine had significantly lower MHD plasma levels and worse seizure control compared to non-carriers [54]. Shen et al. found that UGT2B7 802T > C CC genotype carriers had a higher oxcarbazepine concentration than TT [21]. Opposite results were reported by Lin et al., who did not observe any influence on oxcarbazepine PK when the presence of UGT2B7 802T > C and UGT1A9 I399C > T SNPs was investigated [34]. One of these SNPs was also studied by Ma et al., who found that carriers of UGT2B7 802T > C variant alleles required significantly higher oxcarbazepine maintenance doses than noncarriers [22]. Moreover, Petrenaite et al. evaluated the impact of SNPs UGT1A4*2 70C > A, UGT1A4*3 142T > G, UGT2B7*2 802C > T, UGT2B15*2 253G > Ton lamotrigine PK. They observed lower lamotrigine ratios (LTG plasma concentration/LTG dose/weight) in wild-type UGT1A4*2 C, UGT2B7*2 TT, and UGT2B15*2 TT carriers compared with heterozygous C-carriers, CC, and GG carriers, respectively [35]. Another two studies observed a higher plasma lamotrigine concentration and better therapeutic efficacy in patients with the UGT1A4 142T > G TT polymorphism [55,56]. Moreover, Wang et al. found that carriers of the UGT1A4 -219C > T/-163G > A variant had a significantly higher adjusted lamotrigine levels compared to wild-type carriers. They also found no associations between the UGT1A4 142T > G SNPs and lamotrigine levels [52]. Concerning CYP P450 SNPs, Ahn et al. studied the effects of CYP2C19*2, CYPC19*3, and CYP2C9*3 genotypes on lacosamide PK. Based on CYPC19 genotypes, they stratified the patients in EMs (*1/*1), IMs (*1/*2 or *1/*3) as poor metabolizers (PMs) (*2/*2, *2/*3, *3/*3). They found significantly higher lacosamide concentrations and CDR in PMs, and the PM group presented the lowest proportion of lacosamide-resistant patients [57]. In a similar study, Okada et al. divided 99 Japanese patients in zonisamide extensive (EMs) and poor metabolizers (PMs) based on their CYP2C19 genotype. They observed lower zonisamide clearance in extensive metabolizers. They also found no influence of CYP3A5 SNPs on zonisamide clearance [58]. No influence of CYP3A4 and CYP3A5 SNPs on perampanel and oxcarbazebine was found by two other studies conducted in an Asian population [59]. Wang et al. did not find a significant correlation between the CYP3A4 and CYP3A5 genes and oxcarbazepine PK [33]. In summary, some studies demonstrated a correlation between the presence of SNPs in the UGT1A4, UGT1A9, UGT2B7, CYP2C9, 2CYP2C19 genes and the PK, PD, and efficacy of AEDs (Figure 1 and Table 4).

Table 4.
SNPs affecting the AED metabolizing enzymes.
5. Discussion and Conclusions
Epilepsy is one of the most diffuse neurological diseases; it can affect people within a wide age range, reaching its incidence peaks in the first few years of life and in the elderly [36]. Nowadays, thanks to an ample selection of available AEDs, most of the patients are able to achieve seizure freedom, but nevertheless, this result is not possible for almost 30% of the subjects, even if under polytherapy treatment. This pharmacological variability between and within patients can be due to various causes including sex, age, ethnicity, type of seizure, and genetic factors [12,13].
In this paper, we reviewed studies that analyzed the influence of genetic polymorphisms on the PK, PD, and efficacy of the new antiepileptic drugs. Our results confirmed an important role played by SNPs in the ABCB1 and ABCC2 genes. In fact, ABCB1 3435C > T CC has been observed to cause higher lacosamide and oxcarbazepine MHD levels, and a study observed a higher frequency of this polymorphism in pharmaco-resistant patients. Moreover, ABCB1 G2677T/A caused higher levetiracetam levels and lacosamide resistance. Regarding ABCC2 SNPs, we observed lower lacosamide levels in the ABCC2 -24C > T CT and TT carriers. When we assessed the role of SNPs in genes encoding for the most important targets of new antiepileptic drugs, all but one of our results did not find a significant correlation between the SNPs in SCNs and SVA2 and the studied parameters. Finally, an influence of genetic polymorphisms in metabolizing enzymes has been observed. In fact, the UGT1A9 I399 C > T carriers had significantly lower MHD plasma levels and worse seizure control, the UGT2B7 802T > C CC genotype had a higher oxcarbazepine concentration, and the UGT2B7 802T > C variant alleles needed higher OXC maintenance doses than non-carriers. Our results also showed lower lamotrigine ratios in the wild-type UGT1A4*2 C, UGT2B7*2 TT, UGT2B15*2 TT, and UGT1A4 -219C > T/-163G > A carriers. In this regard, it is important to underline that several studies found a significant influence of UGT SNPs on the lamotrigine levels only after including other variables in their statistical analysis, and from the data observed in these studies, the most important factor influencing lamotrigine levels is the co-administration of enzyme inhibitors such as valproic acid [60,61,62,63,64]. Its use, in fact, can prolong lamotrigine half-life even by two-fold [65], while enzyme inducers like phenytoin can drastically shorten this parameter [66]. Regarding CYP P450 SNPs, we overall observed an influence of CYP2C9 and CYPSC19 but not of SNPs in the CYP3A4 and CYP3A5 genes on the PK of AEDs.
In light of all of these SNPs that could affect the AED plasma levels and efficacy, genotyping is a fascinating option when choosing therapy for epileptic patients. In fact, recent developments in understanding the genetic and neurobiological basis of epilepsy are prospecting a new era for the treatment of this disease, where testing for gene variations might help to improve the efficacy and safety of epilepsy therapies, allowing for specialists to identify the best drug and dose-adjustment for each patient, reducing the frequency of therapeutic drug monitoring [67].
In conclusion, our work showed that SNPs in enzymes and transporters influence the pharmacokinetics of AEDs, and therefore, the use of population pharmacokinetic modelling incorporating the genotypes of drug-metabolizing enzymes and transporters can be one of the most useful tools to facilitate the determination of individualized dosing regimens in AED therapy, and genetic and non-genetic factors affecting enzyme activity can be used reliably under such models by clinicians in selecting the best AED and dose-adjustment for their patients. Nonetheless, further pharmacogenetic studies in large populations and including various ethnic groups should be conducted to consolidate the importance of genotyping as a standard practice in epileptic patients.
Author Contributions
Conceptualization, V.U.B. and T.P.V.; Writing—Original Draft Preparation, V.U.B. and T.P.V.; Writing—Review & Editing, G.P. and L.M.; Supervision, G.P. and L.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Duncan, J.S.; Sander, J.W.; Sisodiya, S.M.; Walker, M.C. Adult epilepsy. Lancet 2006, 367, 1087–1100. [Google Scholar] [CrossRef]
- Manford, M. Recent advances in epilepsy. J. Neurol. 2017, 264, 1811–1824. [Google Scholar] [CrossRef] [PubMed]
- Perucca, E.; Brodie, M.J.; Kwan, P.; Tomson, T. 30 years of second-generation antiseizure medications: Impact and future perspectives. Lancet Neurol. 2020, 19, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Guery, D.; Rheims, S. Is the mechanism of action of antiseizure drugs a key element in the choice of treatment? Fundam. Clin. Pharmacol. 2021, 35, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Hakami, T. Neuropharmacology of Antiseizure Drugs. Neuropsychopharmacol. Rep. 2021, 41, 336–351. [Google Scholar] [CrossRef] [PubMed]
- Johannessen, S.I.; Tomson, T. Pharmacokinetic variability of newer antiepileptic drugs: When is monitoring needed? Clin. Pharmacokinet. 2006, 45, 1061–1075. [Google Scholar] [CrossRef]
- Sills, G.J.; Rogawski, M.A. Mechanisms of action of currently used antiseizure drugs. Neuropharmacology 2020, 168, 107966. [Google Scholar] [CrossRef]
- Lynch, B.A.; Lambeng, N.; Nocka, K.; Kensel-Hammes, P.; Bajjalieh, S.M.; Matagne, A.; Fuks, B. The synaptic vesicle protein SV2A is the binding site for the antiepileptic drug levetiracetam. Proc. Natl. Acad. Sci. USA 2004, 101, 9861–9866. [Google Scholar] [CrossRef]
- Klitgaard, H.; Matagne, A.; Nicolas, J.M.; Gillard, M.; Lamberty, Y.; De Ryck, M.; Kaminski, R.M.; Leclercq, K.; Niespodziany, I.; Wolff, C.; et al. Brivaracetam: Rationale for discovery and preclinical profile of a selective SV2A ligand for epilepsy treatment. Epilepsia 2016, 57, 538–548. [Google Scholar] [CrossRef]
- Mendoza-Torreblanca, J.G.; Vanoye-Carlo, A.; Phillips-Farfan, B.V.; Carmona-Aparicio, L.; Gomez-Lira, G. Synaptic vesicle protein 2A: Basic facts and role in synaptic function. Eur. J. Neurosci. 2013, 38, 3529–3539. [Google Scholar] [CrossRef]
- Rheims, S.; Ryvlin, P. Profile of perampanel and its potential in the treatment of partial onset seizures. Neuropsychiatr. Dis. Treat. 2013, 9, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Sisodiya, S.M.; Marini, C. Genetics of antiepileptic drug resistance. Curr. Opin. Neurol. 2009, 22, 150–156. [Google Scholar] [CrossRef]
- MacDonald, B.K.; Johnson, A.L.; Goodridge, D.M.; Cockerell, O.C.; Sander, J.W.; Shorvon, S.D. Factors predicting prognosis of epilepsy after presentation with seizures. Ann. Neurol. 2000, 48, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Franco, V.; Perucca, E. The pharmacogenomics of epilepsy. Expert Rev. Neurother. 2015, 15, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Löscher, W.; Klotz, U.; Zimprich, F.; Schmidt, D. The clinical impact of pharmacogenetics on the treatment of epilepsy. Epilepsia 2009, 50, 1–23. [Google Scholar] [CrossRef]
- Loscher, W.; Potschka, H. Role of multidrug transporters in pharmacoresistance to antiepileptic drugs. J. Pharmacol. Exp. Ther. 2002, 301, 7–14. [Google Scholar] [CrossRef]
- Białecka, M.; Hnatyszyn, G.; Bielicka-Cymerman, J.; Droździk, M. Znaczenie polimorfizmu genu MDR-1 w patogenezie i leczeniu padaczki lekoopornej. The effect of MDR1 gene polymorphism in the pathogenesis and the treatment of drug-resistant epilepsy. Neurol. I Neurochir. Pol. 2005, 39, 476–481. [Google Scholar]
- Siddiqui, A.; Kerb, R.; Weale, M.E.; Brinkmann, U.; Smith, A.; Goldstein, D.B.; Wood, N.W.; Sisodiya, S.M. Association of multidrug resistance in epilepsy with a polymorphism in the drug-transporter gene ABCB1. New Engl. J. Med. 2003, 348, 1442–1448. [Google Scholar] [CrossRef]
- Hoffmeyer, S.; Burk, O.; von Richter, O.; Arnold, H.P.; Brockmoller, J.; Johne, A.; Cascorbi, I.; Gerloff, T.; Roots, I.; Eichelbaum, M.; et al. Functional polymorphisms of the human multidrug-resistance gene: Multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 3473–3478. [Google Scholar] [CrossRef]
- Lazarowski, A.; Czornyj, L. Potential role of multidrug resistant proteins in refractory epilepsy and antiepileptic drugs interactions. Drug Metab. Drug Interact. 2011, 26, 21–26. [Google Scholar] [CrossRef]
- Shen, C.; Zhang, B.; Liu, Z.; Tang, Y.; Zhang, Y.; Wang, S.; Guo, Y.; Ding, Y.; Wang, S.; Ding, M. Effects of ABCB1, ABCC2, UGT2B7 and HNF4α genetic polymorphisms on oxcarbazepine concentrations and therapeutic efficacy in patients with epilepsy. Seizure 2017, 51, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.L.; Wu, X.Y.; Jiao, Z.; Hong, Z.; Wu, Z.Y.; Zhong, M.K. SCN1A, ABCC2 and UGT2B7 gene polymorphisms in association with individualized oxcarbazepine therapy. Pharmacogenomics 2015, 16, 347–360. [Google Scholar] [CrossRef]
- Yao, N.; Huang, S.; Huang, A.; Song, H. Analysis of influencing factors on monohydroxylated derivative of oxcarbazepine plasma concentration in children with epilepsy. Eur. J. Clin. Pharmacol. 2022, 78, 1667–1675. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Li, H.J.; Feng, J.; Zhang, H.L.; Yu, J.; Sun, Y.; Yu, L.H. Impact of ABCC2 1249G>A and -24C>T polymorphisms on lacosamide efficacy and plasma concentrations in Uygur pediatric epilepsy patients in China. Ther. Drug Monit. 2022, 45, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Li, H.J.; Feng, J.; Zhang, H.L.; Ting-Ting, W.; Ma, L.; Yu, J.; Zhao, W.B.; Sun, L.; Yu, L.H.; et al. Impact of ABCB1 Polymorphisms on Lacosamide Serum Concentrations in Uygur Pediatric Patients With Epilepsy in China. Ther. Drug Monit. 2022, 44, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, X.; Li, H.; Zhang, J.; Chen, Z.; Xie, W.; Zhang, J.; Li, J.; Zhou, L.; Huang, M. Polymorphisms of ABCG2, ABCB1 and HNF4α are associated with Lamotrigine trough concentrations in epilepsy patients. Drug Metab. Pharmacokinet. 2015, 30, 282–287. [Google Scholar] [CrossRef]
- Babeu, J.P.; Boudreau, F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J. Gastroenterol. 2014, 20, 22–30. [Google Scholar] [CrossRef]
- Kamiyama, Y.; Matsubara, T.; Yoshinari, K.; Nagata, K.; Kamimura, H.; Yamazoe, Y. Role of human hepatocyte nuclear factor 4alpha in the expression of drug-metabolizing enzymes and transporters in human hepatocytes assessed by use of small interfering RNA. Drug Metab. Pharmacokinet. 2007, 22, 287–298. [Google Scholar] [CrossRef]
- Shen, C.H.; Zhang, Y.X.; Lu, R.Y.; Jin, B.; Wang, S.; Liu, Z.R.; Tang, Y.L.; Ding, M.P. Specific OCT1 and ABCG2 polymorphisms are associated with Lamotrigine concentrations in Chinese patients with epilepsy. Epilepsy Res. 2016, 127, 186–190. [Google Scholar] [CrossRef]
- Lovrić, M.; Božina, N.; Hajnšek, S.; Kuzman, M.R.; Sporiš, D.; Lalić, Z.; Božina, T.; Granić, P. Association between lamotrigine concentrations and ABCB1 polymorphisms in patients with epilepsy. Ther. Drug Monit. 2012, 34, 518–525. [Google Scholar] [CrossRef]
- Stasiołek, M.; Romanowicz, H.; Połatyńska, K.; Chamielec, M.; Skalski, D.; Makowska, M.; Smolarz, B. Association between C3435T polymorphism of MDR1 gene and the incidence of drug-resistant epilepsy in the population of Polish children. Behav. Brain Funct. BBF 2016, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Ufer, M.; von Stülpnagel, C.; Muhle, H.; Haenisch, S.; Remmler, C.; Majed, A.; Plischke, H.; Stephani, U.; Kluger, G.; Cascorbi, I. Impact of ABCC2 genotype on antiepileptic drug response in Caucasian patients with childhood epilepsy. Pharm. Genom. 2011, 21, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yin, T.; Ma, H.Y.; Liu, D.Q.; Sheng, Y.A.; Zhou, B.T. First Analysis of the Association Between CYP3A4/5, ABCB1 Genetic Polymorphisms and Oxcarbazepine Metabolism and Transport in Chinese Epileptic Patients with Oxcarbazepine Monotherapy and Bitherapy. J. Pharm. Pharm. Sci. A Publ. Can. Soc. Pharm. Sci. Soc. Can. Des Sci. Pharm. 2015, 18, 256–265. [Google Scholar]
- Lin, W.W.; Li, X.W.; Jiao, Z.; Zhang, J.; Rao, X.; Zeng, D.Y.; Lin, X.H.; Wang, C.L. Population pharmacokinetics of oxcarbazepine active metabolite in Chinese paediatric epilepsy patients and its application in individualised dosage regimens. Eur. J. Clin. Pharmacol. 2019, 75, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Petrenaite, V.; Öhman, I.; Jantzen, F.; Ekström, L. Effect of UGT1A4, UGT2B7, UGT2B15, UGT2B17 and ABC1B polymorphisms on lamotrigine metabolism in Danish patients. Epilepsy Res. 2022, 182, 106897. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Yu, J.; Wang, T.T.; Feng, J.; Zhao, W.B.; Sun, L.; Yu, L.H.; Li, H.J.; Sun, Y. Impact of ABCB1 Polymorphism on Levetiracetam Serum Concentrations in Epileptic Uygur Children in China. Ther. Drug Monit. 2020, 42, 886–892. [Google Scholar] [CrossRef]
- Abou-Khalil, B.W. Antiepileptic Drugs. Contin. (Minneap. Minn.) 2016, 22, 132–156. [Google Scholar] [CrossRef] [PubMed]
- Meisler, M.H.; Kearney, J.A. Sodium channel mutations in epilepsy and other neurological disorders. J. Clin. Investig. 2005, 11, 2010–2017. [Google Scholar] [CrossRef] [PubMed]
- El Fotoh, W.M.M.A.; Habib, M.S.E.D.; ALrefai, A.A.; Kasemy, Z.A. The potential implication of SCN1A and CYP3A5 genetic variants on antiepileptic drug resistance among Egyptian epileptic children. Seizure 2016, 41, 75–80. [Google Scholar] [CrossRef]
- Lin, C.H.; Ho, C.J.; Lu, Y.T.; Tsai, M.H. Response to Sodium Channel blocking Antiseizure medications and coding polymorphisms of Sodium Channel genes in Taiwanese epilepsy patients. BMC Neurol. 2021, 21, 367. [Google Scholar] [CrossRef]
- Angelopoulou, C.; Veletza, S.; Heliopoulos, I.; Vadikolias, K.; Tripsianis, G.; Stathi, C.; Piperidou, C. Association of SCN1A gene polymorphism with antiepileptic drug responsiveness in the population of Thrace, Greece. Arch. Med. Sci. AMS 2017, 13, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Ghazala, E.; Shahin, D.A.; Wahba, Y. Polymorphisms of the sodium voltage-gated channel, alpha subunit 1 (SCN1A -A3184G) gene among children with non-lesional epilepsy: A case-control study. Ital. J. Pediatr. 2022, 48, 157. [Google Scholar] [CrossRef]
- Pejanovic-Skobic, N.; Markovic, I.; Bozina, N.; Basic, S. Lack of association of SCN2A rs17183814 polymorphism with the efficacy of lamotrigine monotherapy in patients with focal epilepsy from Herzegovina area, Bosnia and Herzegovina. Epilepsy Res. 2019, 158, 106221. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Lakhan, R.; Kumar, S.; Garg, R.K.; Misra, U.K.; Kalita, J.; Mittal, B. SCN1AIVS5-91G>A polymorphism is associated with susceptibility to epilepsy but not with drug responsiveness. Biochimie 2013, 95, 1350–1353. [Google Scholar] [CrossRef] [PubMed]
- Markovic, I.; Pejanovic-Skobic, N.; Bozina, N.; Susak Sporis, I.; Sporis, D.; Basic, S. The lack of influence of IVS5-91 G>A polymorphism of the SCN1A gene on efficacy of lamotrigine in patients with focal epilepsy. Neurol. Res. 2019, 41, 930–935. [Google Scholar] [CrossRef]
- Manna, I.; Gambardella, A.; Bianchi, A.; Striano, P.; Tozzi, R.; Aguglia, U.; Beccaria, F.; Benna, P.; Campostrini, R.; Canevini, M.P.; et al. A functional polymorphism in the SCN1A gene does not influence antiepileptic drug responsiveness in Italian patients with focal epilepsy. Epilepsia 2011, 52, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Crowder, K.M.; Gunther, J.M.; Jones, T.A.; Hale, B.D.; Zhang, H.Z.; Peterson, M.R.; Scheller, R.H.; Chavkin, C.; Bajjalieh, S.M. Abnormal neurotransmission in mice lacking synaptic vesicle protein 2A (SV2A). Proc. Natl. Acad. Sci. USA 1999, 96, 15268–15273. [Google Scholar] [CrossRef]
- Lynch, J.M.; Tate, S.K.; Kinirons, P.; Weale, M.E.; Cavalleri, G.L.; Depondt, C.; Murphy, K.; O’Rourke, D.; Doherty, C.P.; Shianna, K.V.; et al. No major role of common SV2A variation for predisposition or levetiracetam response in epilepsy. Epilepsy Res. 2009, 83, 44–51. [Google Scholar] [CrossRef]
- Antunes, N.J.; Wichert-Ana, L.; Coelho, E.B.; Della Pasqua, O.; Junior, V.A.; Takayanagui, O.M.; Marques, M.P.; Lanchote, V.L. Analysis of unbound plasma concentration of oxcarbazepine and the 10-hydroxycarbazepine enantiomers by liquid chromatography with tandem mass spectrometry in healthy volunteers. J. Pharm. Biomed. Anal. 2018, 149, 442–447. [Google Scholar] [CrossRef]
- Argikar, U.A.; Remmel, R.P. Variation in glucuronidation of lamotrigine in human liver microsomes. Xenobiotica Fate Foreign Compd. Biol. Syst. 2009, 39, 355–363. [Google Scholar] [CrossRef]
- Ingelman-Sundberg, M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: The past, present and future. Trends Pharmacol. Sci. 2004, 25, 193–200. [Google Scholar] [CrossRef]
- Wang, Q.; Liang, M.; Dong, Y.; Yun, W.; Qiu, F.; Zhao, L.; Guo, Y. Effects of UGT1A4 genetic polymorphisms on serum lamotrigine concentrations in Chinese children with epilepsy. Drug Metab. Pharmacokinet. 2015, 30, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Garcia, M.A.; Feria-Romero, I.A.; Fernando-Serrano, H.; Escalante-Santiago, D.; Grijalva, I.; Orozco-Suarez, S. Genetic polymorphisms associated with antiepileptic metabolism. Front. Biosci. (Elite Ed.) 2014, 6, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Fang, Y.; Wu, X.; Ma, C.; Wang, Y.; Xu, L. Effects of UGT1A9 genetic polymorphisms on monohydroxylated derivative of oxcarbazepine concentrations and oxcarbazepine monotherapeutic efficacy in Chinese patients with epilepsy. Eur. J. Clin. Pharmacol. 2017, 73, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Yang, L.Y.; Zhang, M.C.; Liu, S.Y. Correlation of the UGT1A4 gene polymorphism with serum concentration and therapeutic efficacy of lamotrigine in Han Chinese of Northern China. Eur. J. Clin. Pharmacol. 2014, 70, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Jiao, Y.; Shi, L. Association of UGT2B7 and UGT1A4 Polymorphisms with Serum Concentration of Antiepileptic Drugs in Children. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016, 22, 4107–4113. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.J.; Oh, J.; Kim, D.Y.; Son, H.; Hwang, S.; Shin, H.R.; Kim, E.Y.; Lee, H.S.; Lee, W.J.; Moon, J.; et al. Effects of CYP2C19 genetic polymorphisms on the pharmacokinetics of lacosamide in Korean patients with epilepsy. Epilepsia 2022, 63, 2958–2969. [Google Scholar] [CrossRef]
- Okada, Y.; Seo, T.; Ishitsu, T.; Wanibuchi, A.; Hashimoto, N.; Higa, Y.; Nakagawa, K. Population estimation regarding the effects of cytochrome P450 2C19 and 3A5 polymorphisms on zonisamide clearance. Ther. Drug Monit. 2008, 30, 540–543. [Google Scholar] [CrossRef]
- Ohkubo, S.; Akamine, Y.; Ohkubo, T.; Kikuchi, Y.; Miura, M. Quantification of the Plasma Concentrations of Perampanel Using High-Performance Liquid Chromatography and Effects of the CYP3A4*1G Polymorphism in Japanese Patients. J. Chromatogr. Sci. 2020, 58, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Vázquez, A.; Fricke-Galindo, I.; Dorado, P.; Jung-Cook, H.; Martínez-Juárez, I.E.; Monroy-Jaramillo, N.; Rojas-Tomé, I.S.; Peñas-Lledó, E.; Llerena, A.; López-López, M. Influence of genetic variants and antiepileptic drug co-treatment on lamotrigine plasma concentration in Mexican Mestizo patients with epilepsy. Pharm. J. 2020, 20, 845–856. [Google Scholar] [CrossRef]
- Reimers, A.; Sjursen, W.; Helde, G.; Brodtkorb, E. Frequencies of UGT1A4*2 (P24T) and *3 (L48V) and their effects on serum concentrations of lamotrigine. Eur. J. Drug Metab. Pharmacokinet. 2016, 41, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Singkham, N.; Towanabut, S.; Lertkachatarn, S.; Punyawudho, B. Influence of the UGT2B7 -161C>T polymorphism on the population pharmacokinetics of lamotrigine in Thai patients. Eur. J. Clin. Pharmacol. 2013, 69, 1285–1291. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Yamamoto, Y.; Suzuki, E.; Takahashi, T.; Umemura, A.; Takahashi, Y.; Imai, K.; Inoue, Y.; Hirai, K.; Tsuji, D.; et al. Factors that influence the pharmacokinetics of lamotrigine in Japanese patients with epilepsy. Eur. J. Clin. Pharmacol. 2016, 72, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Z.; Zhang, Y.F.; Huang, W.C.; Wang, X.P.; Ni, X.J.; Lu, H.Y.; Hu, J.Q.; Deng, S.H.; Zhu, X.Q.; Xie, H.S.; et al. Effects of Comedication and Genetic Factors on the Population Pharmacokinetics of Lamotrigine: A Prospective Analysis in Chinese Patients With Epilepsy. Front. Pharmacol. 2019, 10, 832. [Google Scholar] [CrossRef] [PubMed]
- Rivas, N.; Buelga, D.S.; Christian, E.; Santos-Borbujo, J.; Otero, M.J.; Domínguez-Gil, A.; García, M.J. Population pharmacokinetics of lamotrigine with data from therapeutic drug monitoring in German and Spanish patients with epilepsy. Ther. Drug Monit. 2008, 30, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Brzaković, B.; Vučićević, K.; Kovačević, S.V.; Miljković, B.; Prostran, M.; Martinović, Ž.; Pokrajac, M. Pharmacokinetics of lamotrigine in paediatric and young adult epileptic patients-nonlinear mixed effects modelling approach. Eur. J. Clin. Pharmacol. 2014, 70, 179–185. [Google Scholar] [CrossRef]
- Balestrini, S.; Sisodiya, S.M. Pharmacogenomics in epilepsy. Neurosci. Lett. 2018, 667, 27–39. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).