The Switch and Reciprocating Models for the Function of ABC Multidrug Exporters: Perspectives on Recent Research
Abstract
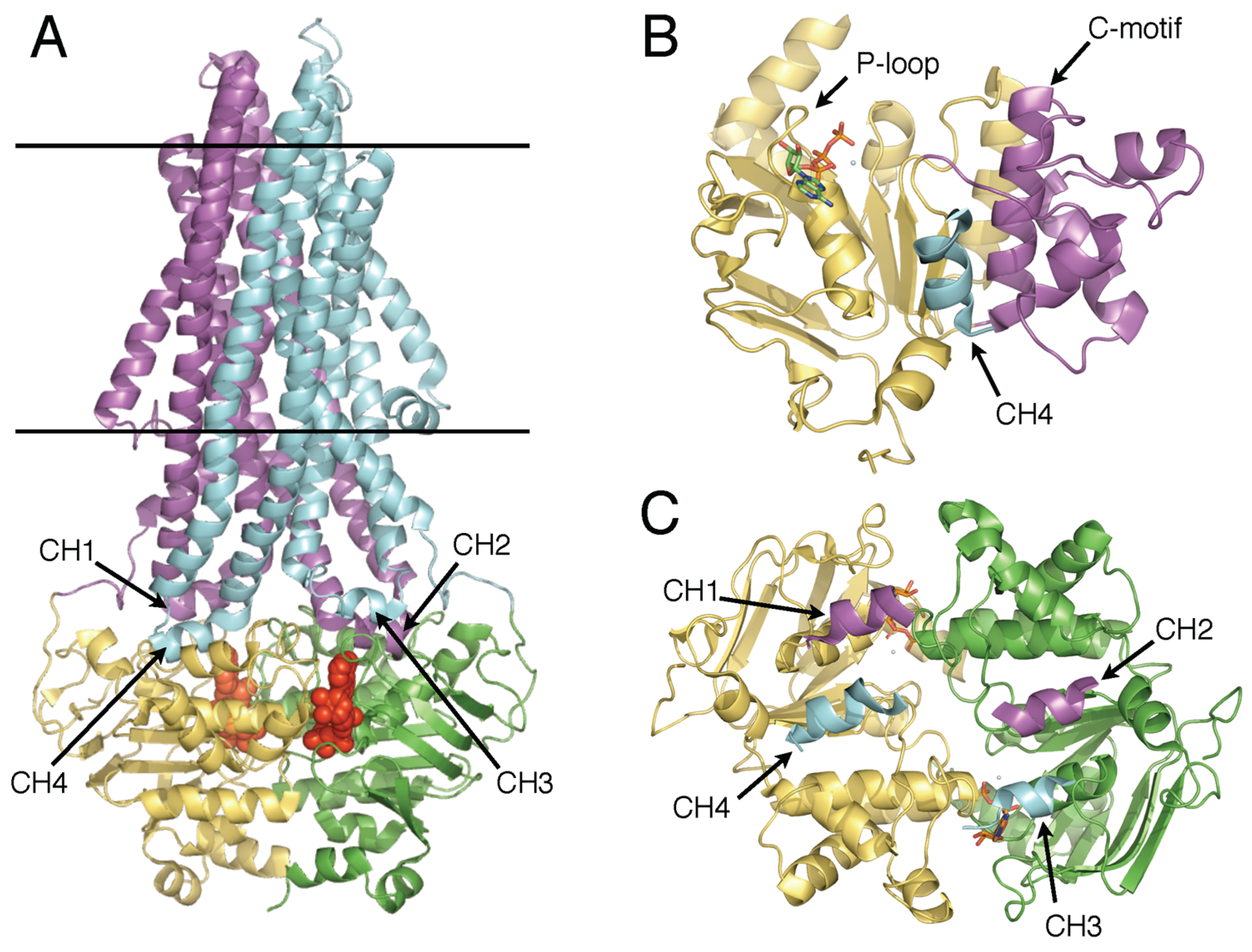
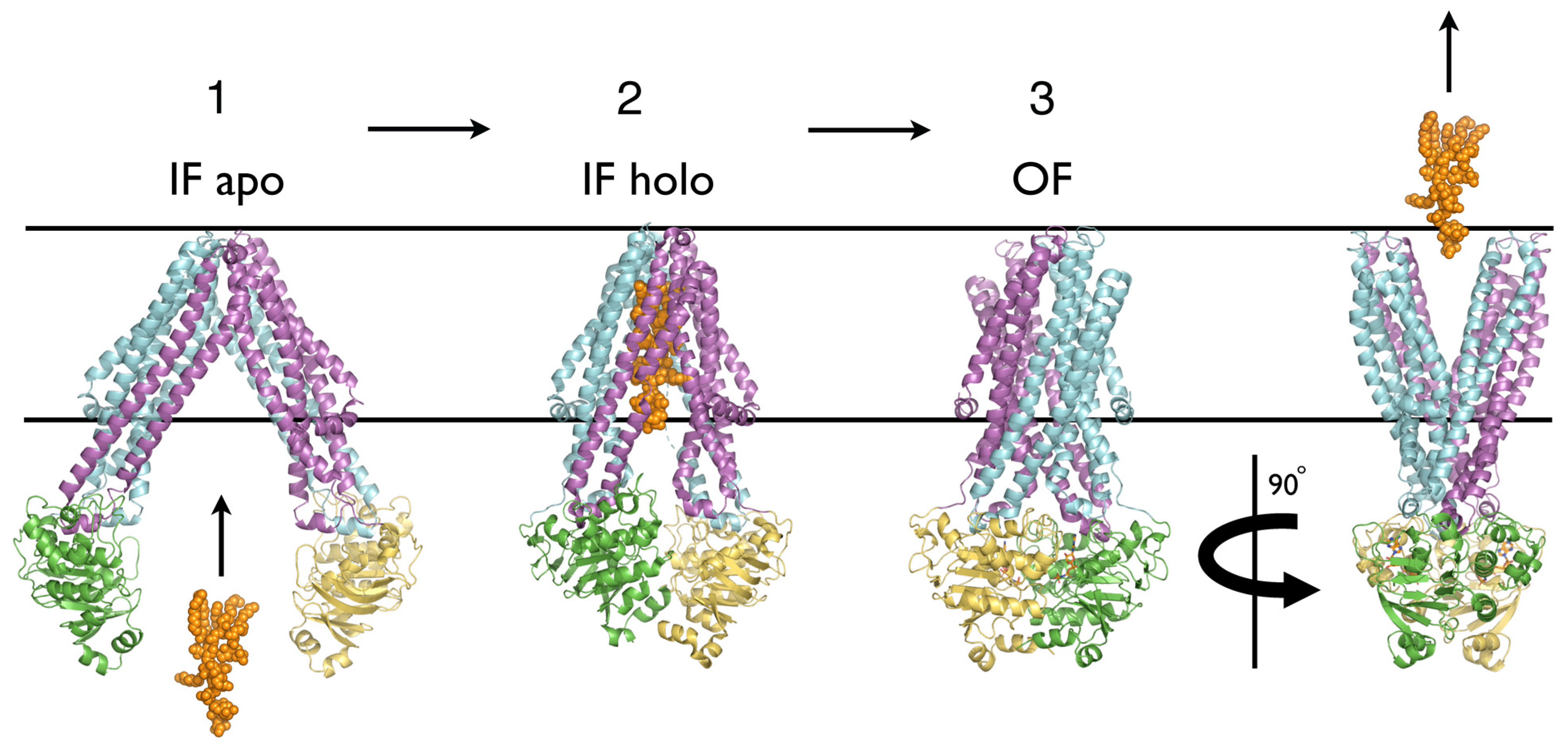
1. The Ubiquity of ABC Transporters: Architecture and Function
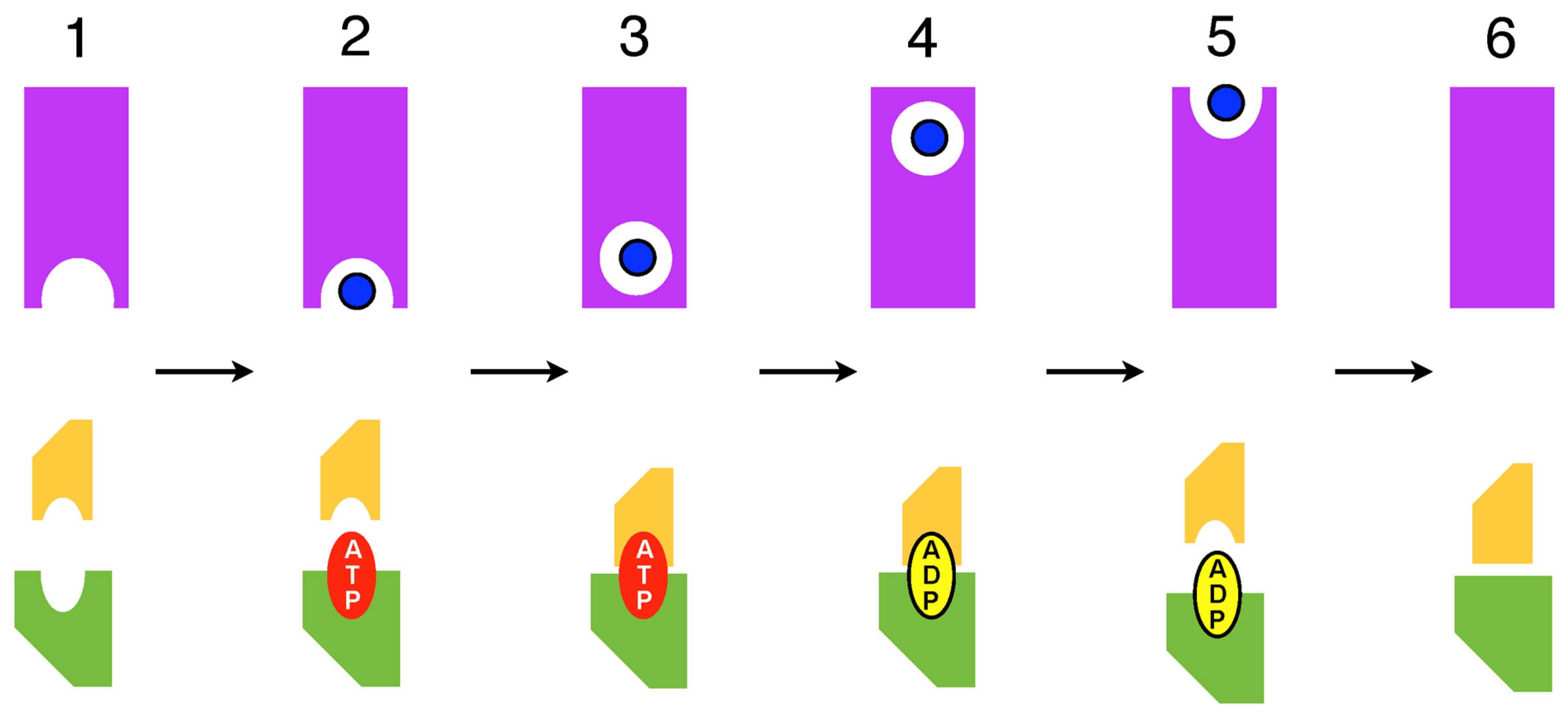
2. Do Recent NMR and HDX Studies of ABC Exporters Depart from the Reciprocating Model?
3. Summary and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ames, G.F.; Mimura, C.S.; Holbrook, S.R.; Shyamala, V. Traffic ATPases: A superfamily of transport proteins operating from Escherichia coli to humans. Adv. Enzymol. Relat. Areas Mol. Biol. 1992, 65, 1–47. [Google Scholar]
- Saurin, W.; Hofnung, M.; Dassa, E. Getting in or out: Early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J. Mol. Evol. 1999, 48, 22–41. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.; Rzhetsky, A.; Allikmets, R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001, 11, 1156–1166. [Google Scholar] [CrossRef]
- Jones, P.M.; George, A.M. The ABC transporter structure and mechanism: Perspectives on recent research. Cell Mol. Life Sci. 2004, 61, 682–699. [Google Scholar] [CrossRef]
- Jones, P.M.; George, A.M. Opening of the ADP-bound active site in the ABC transporter ATPase dimer: Evidence for a constant contact, alternating sites model for the catalytic cycle. Proteins 2009, 75, 387–396. [Google Scholar] [CrossRef]
- Locher, K.P. Review. Structure and mechanism of ATP-binding cassette transporters. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 239–245. [Google Scholar] [CrossRef]
- Rees, D.C.; Johnson, E.; Lewinson, O. ABC transporters: The power to change. Nat. Rev. Mol. Cell Biol. 2009, 10, 218–227. [Google Scholar] [CrossRef]
- Higgins, C.F. ABC transporters; from microorganisms to man. Annu. Rev. Cell Biol. 1992, 8, 67–113. [Google Scholar]
- Huls, M.; Russel, F.G.; Masereeuw, R. The role of ATP binding cassette transporters in tissue defense and organ regeneration. J. Pharmacol. Exp Ther. 2009, 328, 3–9. [Google Scholar] [CrossRef]
- Kotlyarov, S.; Kotlyarova, A. The Role of ABC Transporters in Lipid Metabolism and the Comorbid Course of Chronic Obstructive Pulmonary Disease and Atherosclerosis. Int. J. Mol. Sci. 2021, 22, 6711. [Google Scholar] [CrossRef] [PubMed]
- Abuznait, A.H.; Qosa, H.; Busnena, B.A.; El Sayed, K.A.; Kaddoumi, A. Olive-oil-derived oleocanthal enhances beta-amyloid clearance as a potential neuroprotective mechanism against Alzheimer’s disease: In vitro and in vivo studies. ACS Chem. Neurosci. 2013, 4, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Pahnke, J.; Frohlich, C.; Krohn, M.; Schumacher, T.; Paarmann, K. Impaired mitochondrial energy production and ABC transporter function-A crucial interconnection in dementing proteopathies of the brain. Mech. Ageing Dev. 2013, 134, 506–515. [Google Scholar] [CrossRef]
- Walker, J.E.; Saraste, M.; Runswick, M.J.; Gay, N.J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982, 1, 945–951. [Google Scholar] [CrossRef]
- Dawson, R.J.; Locher, K.P. Structure of a bacterial multidrug ABC transporter. Nature 2006, 443, 180–185. [Google Scholar] [CrossRef]
- Hunke, S.; Mourez, M.; Jehanno, M.; Dassa, E.; Schneider, E. ATP modulates subunit-subunit interactions in an ATP-binding cassette transporter (MalFGK2) determined by site-directed chemical cross-linking. J. Biol. Chem. 2000, 275, 15526–15534. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, D.A.; Ward, A.; Urbatsch, I.L.; Chang, G.; van Veen, H.W. Understanding polyspecificity of multidrug ABC transporters: Closing in on the gaps in ABCB1. Trends Biochem. Sci. 2010, 35, 36–42. [Google Scholar] [CrossRef]
- Locher, K.P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 2016, 23, 487–493. [Google Scholar] [CrossRef]
- Alam, A.; Kowal, J.; Broude, E.; Roninson, I.; Locher, K.P. Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 2019, 363, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Osborne, A.R.; Groll, M.; Rapoport, T.A. RecA-like motor ATPases--lessons from structures. Biochim. Biophys. Acta 2004, 1659, 1–18. [Google Scholar] [CrossRef]
- Bianchet, M.A.; Ko, Y.H.; Amzel, L.M.; Pedersen, P.L. Modeling of nucleotide binding domains of ABC transporter proteins based on a F1-ATPase/recA topology: Structural model of the nucleotide binding domains of the cystic fibrosis transmembrane conductance regulator (CFTR). J. Bioenerg. Biomembr. 1997, 29, 503–524. [Google Scholar] [CrossRef]
- Jones, P.M.; George, A.M. Subunit interactions in ABC transporters: Towards a functional architecture. FEMS Microbiol. Lett. 1999, 179, 187–202. [Google Scholar] [CrossRef]
- Smith, P.C.; Karpowich, N.; Millen, L.; Moody, J.E.; Rosen, J.; Thomas, P.J.; Hunt, J.F. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 2002, 10, 139–149. [Google Scholar] [CrossRef]
- Jones, P.M.; George, A.M. Mechanism of the ABC transporter ATPase domains: Catalytic models and the biochemical and biophysical record. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 39–50. [Google Scholar] [CrossRef]
- Lewinson, O.; Livnat-Levanon, N. Mechanism of Action of ABC Importers: Conservation, Divergence, and Physiological Adaptations. J. Mol. Biol. 2017, 429, 606–619. [Google Scholar] [CrossRef]
- Szollosi, D.; Szakacs, G.; Chiba, P.; Stockner, T. Dissecting the Forces that Dominate Dimerization of the Nucleotide Binding Domains of ABCB1. Biophys. J. 2018, 114, 331–342. [Google Scholar] [CrossRef]
- Ford, R.C.; Beis, K. Learning the ABCs one at a time: Structure and mechanism of ABC transporters. Biochem. Soc. Trans. 2019, 47, 23–36. [Google Scholar] [CrossRef]
- Shaikh, S.; Wen, P.C.; Enkavi, G.; Huang, Z.; Tajkhorshid, E. Capturing Functional Motions of Membrane Channels and Transporters with Molecular Dynamics Simulation. J. Comput. Theor. Nanosci. 2010, 7, 2481–2500. [Google Scholar] [CrossRef]
- Jardetzky, O. Simple allosteric model for membrane pumps. Nature 1966, 211, 969–970. [Google Scholar] [CrossRef]
- Higgins, C.F.; Linton, K.J. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 2004, 11, 918–926. [Google Scholar] [CrossRef]
- Dawson, R.J.; Locher, K.P. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 2007, 81, 935–938. [Google Scholar] [CrossRef]
- Reyes, C.L.; Chang, G. Structure of the ABC transporter MsbA in complex with ADP.vanadate and lipopolysaccharide. Science 2005, 308, 1028–1031. [Google Scholar] [CrossRef] [PubMed]
- Padayatti, P.S.; Lee, S.C.; Stanfield, R.L.; Wen, P.C.; Tajkhorshid, E.; Wilson, I.A.; Zhang, Q. Structural Insights into the Lipid A Transport Pathway in MsbA. Structure 2019, 27, 1114–1123.e3. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.; Miu, A.; Alexander, M.K.; Garcia, N.K.; Oh, A.; Zilberleyb, I.; Reichelt, M.; Austin, C.D.; Tam, C.; Shriver, S.; et al. Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nature 2018, 557, 196–201. [Google Scholar] [CrossRef]
- Ward, A.; Reyes, C.L.; Yu, J.; Roth, C.B.; Chang, G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. USA 2007, 104, 19005–19010. [Google Scholar] [CrossRef]
- Jones, P.M.; George, A.M. A reciprocating twin-channel model for ABC transporters. Q. Rev. Biophys. 2014, 47, 189–220. [Google Scholar] [CrossRef]
- Spadaccini, R.; Kaur, H.; Becker-Baldus, J.; Glaubitz, C. The effect of drug binding on specific sites in transmembrane helices 4 and 6 of the ABC exporter MsbA studied by DNP-enhanced solid-state NMR. Biochim. Biophys. Acta Biomembr. 2018, 1860, 833–840. [Google Scholar] [CrossRef]
- Kaur, H.; Lakatos, A.; Spadaccini, R.; Vogel, R.; Hoffmann, C.; Becker-Baldus, J.; Ouari, O.; Tordo, P.; Mchaourab, H.; Glaubitz, C. The ABC exporter MsbA probed by solid state NMR—Challenges and opportunities. Biol. Chem. 2015, 396, 1135–1149. [Google Scholar] [CrossRef]
- Szabo, K.; Welker, E.; Bakos Muller, M.; Roninson, I.; Varadi, A.; Sarkadi, B. Drug-stimulated nucleotide trapping in the human multidrug transporter MDR1. Cooperation of the nucleotide binding domains. J. Biol. Chem. 1998, 273, 10132–101388. [Google Scholar] [CrossRef]
- Kerr, K.M.; Sauna, Z.E.; Ambudkar, S.V. Correlation between steady-state ATP hydrolysis and vanadate-induced ADP trapping in Human P-glycoprotein. Evidence for ADP release as the rate-limiting step in the catalytic cycle and its modulation by substrates. J. Biol. Chem. 2001, 276, 8657–8664. [Google Scholar] [CrossRef]
- Clouser, A.F.; Atkins, W.M. Long Range Communication between the Drug-Binding Sites and Nucleotide Binding Domains of the Efflux Transporter ABCB1. Biochemistry 2022, 61, 730–740. [Google Scholar] [CrossRef]
- Jones, P.M.; George, A.M. How Intrinsic Dynamics Mediates the Allosteric Mechanism in the ABC Transporter Nucleotide Binding Domain Dimer. J. Chem. Theory Comput. 2017, 13, 1712–1722. [Google Scholar] [CrossRef] [PubMed]
- Lacabanne, D.; Orelle, C.; Lecoq, L.; Kunert, B.; Chuilon, C.; Wiegand, T.; Ravaud, S.; Jault, J.-M.; Meier, B.H.; Böckmann, A. Flexible-to-rigid transition is central for substrate transport in the ABC transporter BmrA from Bacillus subtilis. Commun. Biol. 2019, 2, 149. [Google Scholar] [CrossRef]
- Lacabanne, D.; Wiegand, T.; Di Cesare, M.; Orelle, C.; Ernst, M.; Jault, J.M.; Meier, B.H.; Böckmann, A. Solid-State NMR Reveals Asymmetric ATP Hydrolysis in the Multidrug ABC Transporter BmrA. J. Am. Chem. Soc. 2022, 144, 12431–12442. [Google Scholar] [CrossRef]
- Javed, W.; Vallet, S.; Clement, M.P.; Le Roy, A.; Moulin, M.; Härtlein, M.; Breyton, C.; Burlet-Schiltz, O.; Marcoux, J.; Orelle, C.; et al. Structural Insights into the Catalytic Cycle of a Bacterial Multidrug ABC Efflux Pump. J. Mol. Biol. 2022, 434, 167541. [Google Scholar] [CrossRef] [PubMed]
- Chaptal, V.; Zampieri, V.; Wiseman, B.; Orelle, C.; Martin, J.; Nguyen, K.A.; Gobet, A.; Di Cesare, M.; Magnard, S.; Javed, W.; et al. Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter. Sci. Adv. 2022, 8, eabg9215. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.E.; Bhagat, S. P-glycoprotein shows strong catalytic cooperativity between the two nucleotide sites. Biochemistry 1998, 37, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.E.; Gros, P.; Urbatsch, I.L. Residues in P-glycoprotein catalytic sites that react with the inhibitor 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole. Arch. Biochem. Biophys. 1998, 357, 121–125. [Google Scholar] [CrossRef]
- Urbatsch, I.L.; Sankaran, B.; Bhagat, S.; Senior, A.E. Both P-glycoprotein nucleotide-binding sites are catalytically active. J. Biol. Chem. 1995, 270, 26956–26961. [Google Scholar] [CrossRef]
- Russell, P.L.; Sharom, F.J. Conformational and functional characterization of trapped complexes of the P-glycoprotein multidrug transporter. Biochem. J. 2006, 399, 315–323. [Google Scholar] [CrossRef]
- Oldham, M.L.; Chen, J. Snapshots of the maltose transporter during ATP hydrolysis. Proc. Natl. Acad. Sci. USA 2011, 108, 15152–15156. [Google Scholar] [CrossRef]
- Kehlenbeck, D.M.; Traore, D.A.; Josts, I.; Sander, S.; Moulin, M.; Haertlein, M.; Prevost, S.; Forsyth, V.T.; Tidow, H. Cryo-EM structure of MsbA in saposin-lipid nanoparticles (Salipro) provides insights into nucleotide coordination. FEBS J. 2022, 289, 2959–2970. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jones, P.M.; George, A.M. The Switch and Reciprocating Models for the Function of ABC Multidrug Exporters: Perspectives on Recent Research. Int. J. Mol. Sci. 2023, 24, 2624. https://doi.org/10.3390/ijms24032624
Jones PM, George AM. The Switch and Reciprocating Models for the Function of ABC Multidrug Exporters: Perspectives on Recent Research. International Journal of Molecular Sciences. 2023; 24(3):2624. https://doi.org/10.3390/ijms24032624
Chicago/Turabian StyleJones, Peter M., and Anthony M. George. 2023. "The Switch and Reciprocating Models for the Function of ABC Multidrug Exporters: Perspectives on Recent Research" International Journal of Molecular Sciences 24, no. 3: 2624. https://doi.org/10.3390/ijms24032624
APA StyleJones, P. M., & George, A. M. (2023). The Switch and Reciprocating Models for the Function of ABC Multidrug Exporters: Perspectives on Recent Research. International Journal of Molecular Sciences, 24(3), 2624. https://doi.org/10.3390/ijms24032624




