Roles of Tryptophan and Charged Residues on the Polymorphisms of Amyloids Formed by K-Peptides of Hen Egg White Lysozyme Investigated through Molecular Dynamics Simulations
Abstract
1. Introduction
2. Results and Discussions
2.1. Disruption of the Antiparallel Beta Amyloid with a Negative Charge Addition
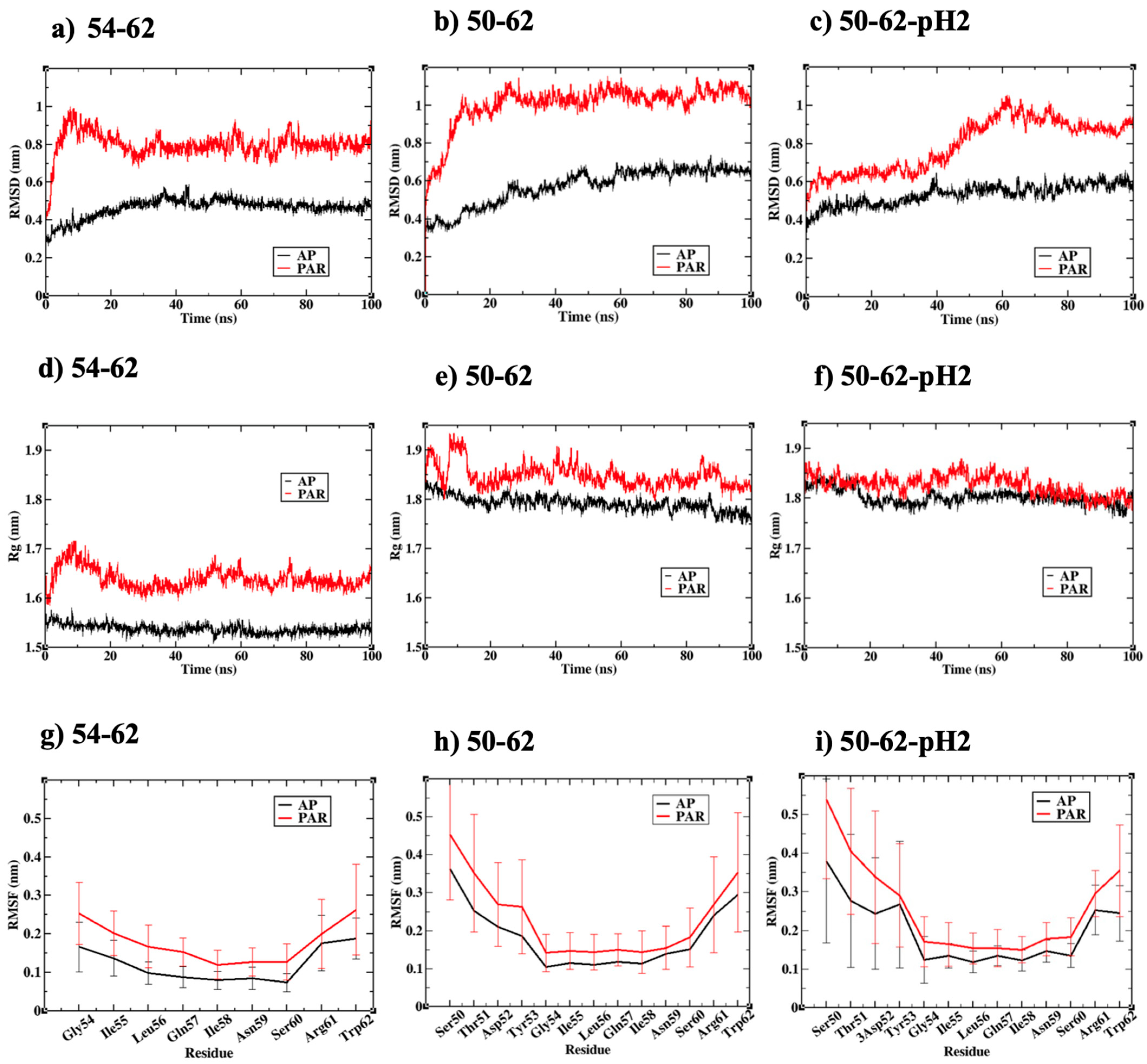
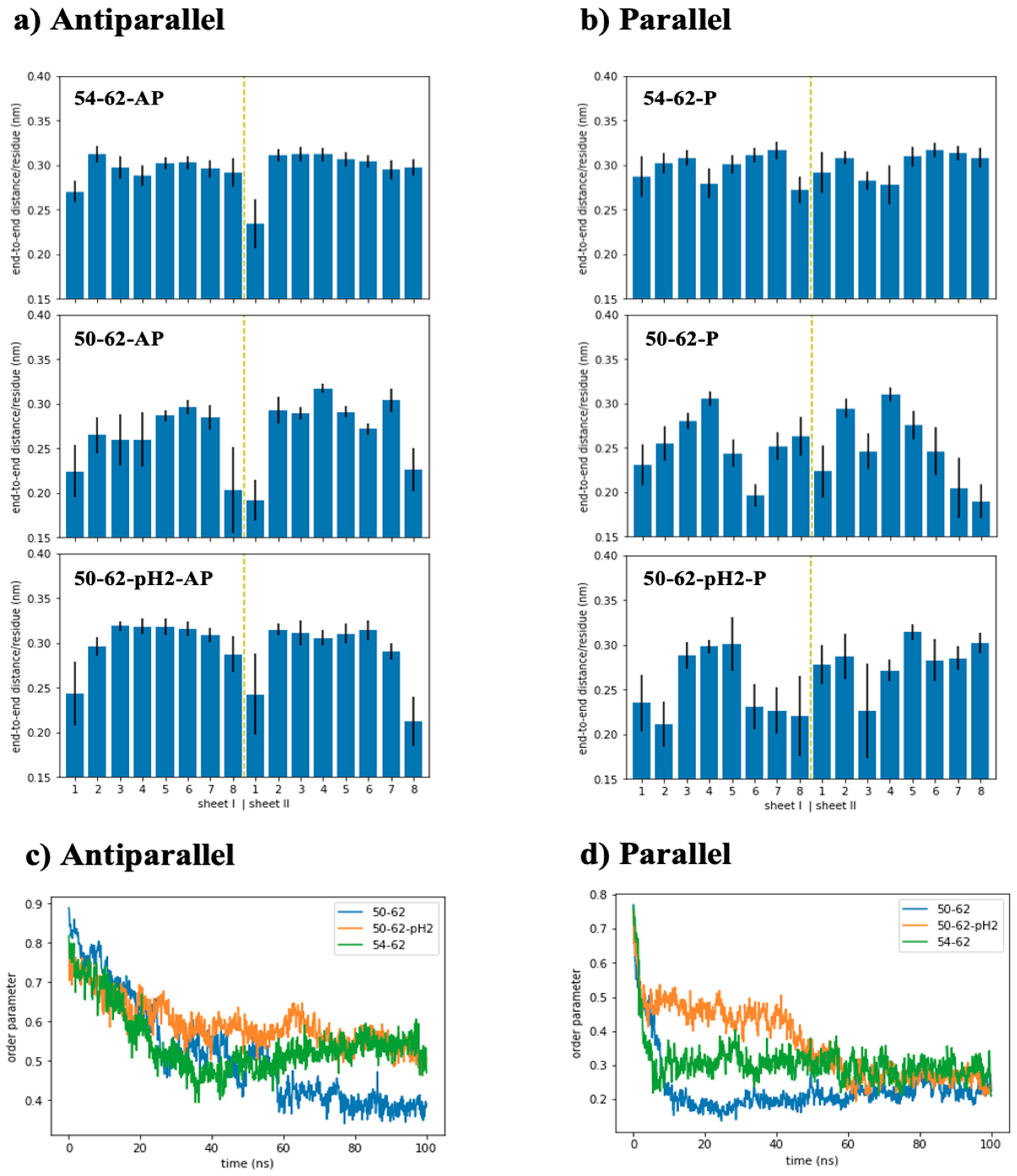
2.2. Antiparallel Beta Amyloid Models Displayed Greater Compactness and Stability
2.3. Antiparallel Amyloid Models of K-Peptides and STDY-K-Peptide at pH 2 Displayed Stretched and Ordered Chains
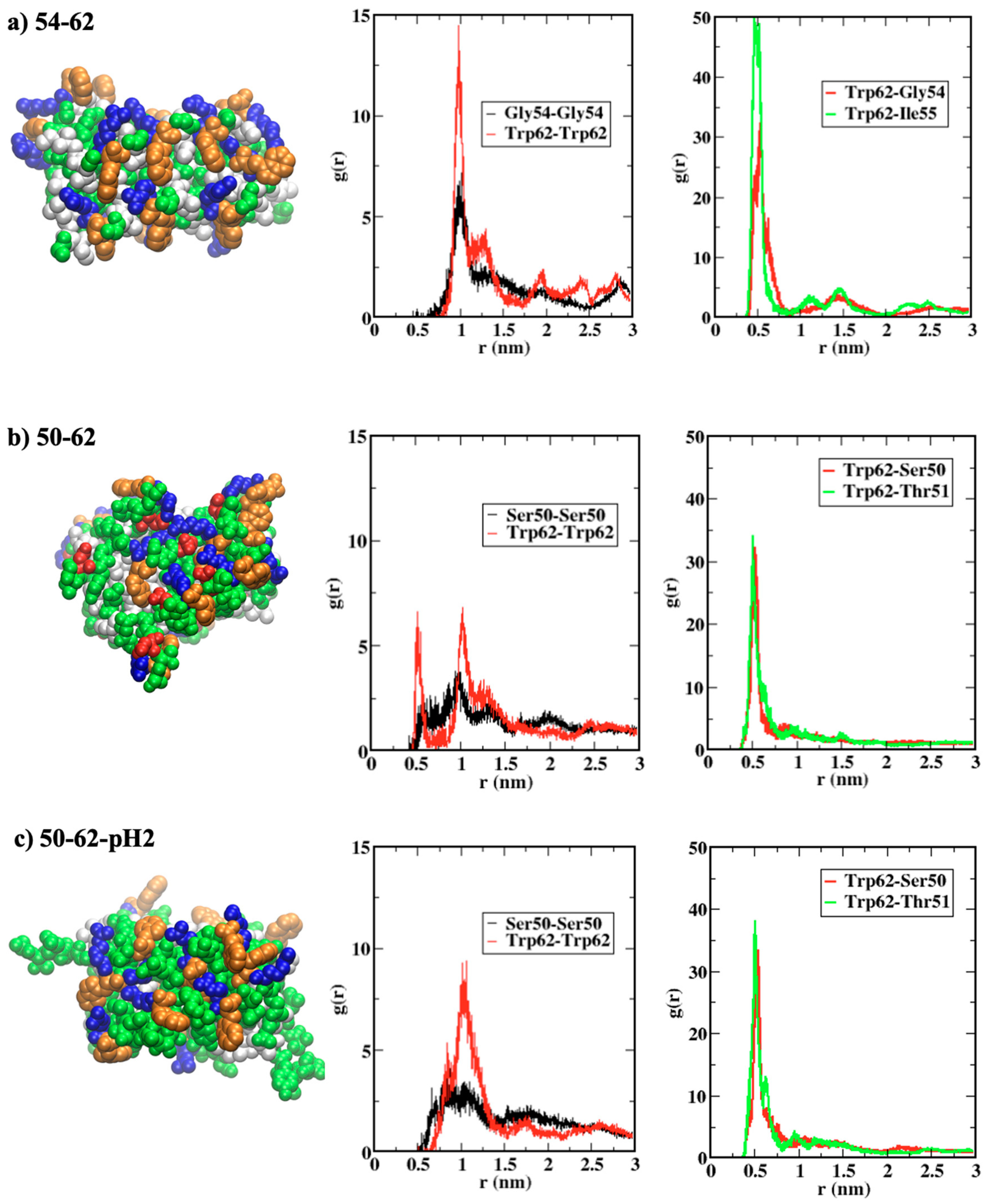
2.4. Roles of Trp62 Capping the Ends of Highly Ordered Amyloid Models
3. Materials and Methods
3.1. Beta Amyloid Model Creation
3.2. Atomistic Molecular Dynamics Simulations
3.3. Conformational and Interaction Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chiti, F.; Dobson, C.M. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M. Protein Folding and Misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Folding Proteins in Fatal Ways. Nature 2003, 426, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.M. Peptide Aggregation in Neurodegenerative Disease. Annu. Rev. Biomed. Eng. 2002, 4, 155–174. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, M.K.; Alam, P.; Malik, S.; Majid, N.; Chaturvedi, S.K.; Rajan, S.; Ajmal, M.R.; Khan, M.V.; Uversky, V.N.; Khan, R.H. Stabilizing Proteins to Prevent Conformational Changes Required for Amyloid Fibril Formation. J. Cell. Biochem. 2019, 120, 2642–2656. [Google Scholar] [CrossRef] [PubMed]
- Gharibyan, A.L.; Wasana Jayaweera, S.; Lehmann, M.; Anan, I.; Olofsson, A. Endogenous Human Proteins Interfering with Amyloid Formation. Biomolecules 2022, 12, 446. [Google Scholar] [CrossRef]
- Seraj, Z.; Seyedarabi, A.; Saboury, A.A.; Habibi-Rezaei, M.; Ahmadian, S.; Ghasemi, A. Unraveling the Novel Effects of Aroma from Small Molecules in Preventing Hen Egg White Lysozyme Amyloid Fibril Formation. PLoS ONE 2018, 13, e0189754. [Google Scholar] [CrossRef]
- Ariesandi, W.; Chang, C.-F.; Chen, T.-E.; Chen, Y.-R. Temperature-Dependent Structural Changes of Parkinson’s Alpha-Synuclein Reveal the Role of Pre-Existing Oligomers in Alpha-Synuclein Fibrillization. PLoS ONE 2013, 8, e53487. [Google Scholar] [CrossRef]
- Hwang, W.; Zhang, S.; Kamm, R.D.; Karplus, M. Kinetic Control of Dimer Structure Formation in Amyloid Fibrillogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 12916–12921. [Google Scholar] [CrossRef]
- Khurana, R.; Ionescu-Zanetti, C.; Pope, M.; Li, J.; Nielson, L.; Ramírez-Alvarado, M.; Regan, L.; Fink, A.L.; Carter, S.A. A General Model for Amyloid Fibril Assembly Based on Morphological Studies Using Atomic Force Microscopy. Biophys. J. 2003, 85, 1135–1144. [Google Scholar] [CrossRef]
- DeBenedictis, E.P.; Liu, J.; Keten, S. Adhesion Mechanisms of Curli Subunit CsgA to Abiotic Surfaces. Sci. Adv. 2016, 2, e1600998. [Google Scholar] [CrossRef] [PubMed]
- Zeng, G.; Vad, B.S.; Dueholm, M.S.; Christiansen, G.; Nilsson, M.; Tolker-Nielsen, T.; Nielsen, P.H.; Meyer, R.L.; Otzen, D.E. Functional Bacterial Amyloid Increases Pseudomonas Biofilm Hydrophobicity and Stiffness. Front. Microbiol. 2015, 6, 1099. [Google Scholar] [CrossRef] [PubMed]
- Iconomidou, V.A.; Vriend, G.; Hamodrakas, S.J. Amyloids Protect the Silkmoth Oocyte and Embryo. FEBS Lett. 2000, 479, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.M.; Koulov, A.V.; Alory-Jost, C.; Marks, M.S.; Balch, W.E.; Kelly, J.W. Functional Amyloid Formation within Mammalian Tissue. PLoS Biol. 2005, 4, e6. [Google Scholar] [CrossRef]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.R.; Simon, R.; Schubert, D.; et al. Functional Amyloids As Natural Storage of Peptide Hormones in Pituitary Secretory Granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef]
- Edwards, S.J.; Kjellerup, B.V. Applications of Biofilms in Bioremediation and Biotransformation of Persistent Organic Pollutants, Pharmaceuticals/Personal Care Products, and Heavy Metals. Appl. Microbiol. Biotechnol. 2013, 97, 9909–9921. [Google Scholar] [CrossRef]
- Li, J.; Zhang, F. Amyloids as Building Blocks for Macroscopic Functional Materials: Designs, Applications and Challenges. IJMS 2021, 22, 10698. [Google Scholar] [CrossRef]
- Catalini, S.; Perinelli, D.R.; Sassi, P.; Comez, L.; Palmieri, G.F.; Morresi, A.; Bonacucina, G.; Foggi, P.; Pucciarelli, S.; Paolantoni, M. Amyloid Self-Assembly of Lysozyme in Self-Crowded Conditions: The Formation of a Protein Oligomer Hydrogel. Biomacromolecules 2021, 22, 1147–1158. [Google Scholar] [CrossRef]
- Knowles, T.P.J.; Oppenheim, T.W.; Buell, A.K.; Chirgadze, D.Y.; Welland, M.E. Nanostructured Films from Hierarchical Self-Assembly of Amyloidogenic Proteins. Nature Nanotech. 2010, 5, 204–207. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, P. Amyloid-Mediated Fabrication of Organic–Inorganic Hybrid Materials and Their Biomedical Applications. Adv. Mater. Interfaces 2020, 7, 2001060. [Google Scholar] [CrossRef]
- Walker, D.J.; Adhikari, R.Y.; Holmes, D.E.; Ward, J.E.; Woodard, T.L.; Nevin, K.P.; Lovley, D.R. Electrically Conductive Pili from Pilin Genes of Phylogenetically Diverse Microorganisms. ISME J. 2018, 12, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, D.J.; Abdali, Z.; Modafferi, D.; Janfeshan, B.; Dorval Courchesne, N.-M. Fabrication of Fluorescent PH-Responsive Protein–Textile Composites. Sci. Rep. 2020, 10, 13052. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Mezzenga, R. Functionalization of Multiwalled Carbon Nanotubes and Their PH-Responsive Hydrogels with Amyloid Fibrils. Langmuir 2012, 28, 10142–10146. [Google Scholar] [CrossRef]
- Bolisetty, S.; Vallooran, J.J.; Adamcik, J.; Mezzenga, R. Magnetic-Responsive Hybrids of Fe3O4 Nanoparticles with β-Lactoglobulin Amyloid Fibrils and Nanoclusters. ACS Nano. 2013, 7, 6146–6155. [Google Scholar] [CrossRef] [PubMed]
- Lutz-Bueno, V.; Bolisetty, S.; Azzari, P.; Handschin, S.; Mezzenga, R. Self-Winding Gelatin–Amyloid Wires for Soft Actuators and Sensors. Adv. Mater. 2020, 32, 2004941. [Google Scholar] [CrossRef]
- Alam, I.; Lertanantawong, B.; Sutthibutpong, T.; Punnakitikashem, P.; Asanithi, P. Molecularly Imprinted Polymer-Amyloid Fibril-Based Electrochemical Biosensor for Ultrasensitive Detection of Tryptophan. Biosensors 2022, 12, 291. [Google Scholar] [CrossRef]
- Duncan, K.L.; Ulijn, R.V. Short Peptides in Minimalistic Biocatalyst Design. Biocatalysis 2015, 1, 67–81. [Google Scholar] [CrossRef]
- Díaz-Caballero, M.; Navarro, S.; Nuez-Martínez, M.; Peccati, F.; Rodríguez-Santiago, L.; Sodupe, M.; Teixidor, F.; Ventura, S. PH-Responsive Self-Assembly of Amyloid Fibrils for Dual Hydrolase-Oxidase Reactions. ACS Catal. 2021, 11, 595–607. [Google Scholar] [CrossRef]
- Garcia, A.M.; Kurbasic, M.; Kralj, S.; Melchionna, M.; Marchesan, S. A Biocatalytic and Thermoreversible Hydrogel from a Histidine-Containing Tripeptide. Chem. Commun. 2017, 53, 8110–8113. [Google Scholar] [CrossRef]
- Das, S.; Jacob, R.S.; Patel, K.; Singh, N.; Maji, S.K. Amyloid Fibrils: Versatile Biomaterials for Cell Adhesion and Tissue Engineering Applications. Biomacromolecules 2018, 19, 1826–1839. [Google Scholar] [CrossRef]
- Nyström, G.; Fong, W.-K.; Mezzenga, R. Ice-Templated and Cross-Linked Amyloid Fibril Aerogel Scaffolds for Cell Growth. Biomacromolecules 2017, 18, 2858–2865. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.R.; Vekilov, P.G.; Rosenberger, F. Heterogeneity Determination and Purification of Commercial Hen Egg-White Lysozyme. Acta Crystallogr. D Biol. Crystallogr. 1996, 52, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Yang, H.; Miao, X.; Wang, W.; Wang, J. Macromolecular Protein Crystallisation with Biotemplate of Live Cells. Sci. Rep. 2022, 12, 3005. [Google Scholar] [CrossRef] [PubMed]
- Ndlovu, H.; Ashcroft, A.E.; Radford, S.E.; Harris, S.A. Effect of Sequence Variation on the Mechanical Response of Amyloid Fibrils Probed by Steered Molecular Dynamics Simulation. Biophys. J. 2012, 102, 587–596. [Google Scholar] [CrossRef]
- Ndlovu, H.; Ashcroft, A.E.; Radford, S.E.; Harris, S.A. Molecular Dynamics Simulations of Mechanical Failure in Polymorphic Arrangements of Amyloid Fibrils Containing Structural Defects. Beilstein J. Nanotechnol. 2013, 4, 429–440. [Google Scholar] [CrossRef]
- Wang, Y.; Jia, B.; You, M.; Fan, H.; Cao, S.; Li, H.; Zhang, W.; Ma, G. Modulation of Surface-Catalyzed Secondary Nucleation during Amyloid Fibrillation of Hen Egg White Lysozyme by Two Common Surfactants. J. Phys. Chem. B 2019, 123, 6200–6211. [Google Scholar] [CrossRef]
- Nitani, A.; Muta, H.; Adachi, M.; So, M.; Sasahara, K.; Sakurai, K.; Chatani, E.; Naoe, K.; Ogi, H.; Hall, D.; et al. Heparin-Dependent Aggregation of Hen Egg White Lysozyme Reveals Two Distinct Mechanisms of Amyloid Fibrillation. J. Biol. Chem. 2017, 292, 21219–21230. [Google Scholar] [CrossRef]
- Jafari, M.; Mehrnejad, F. Molecular Insight into Human Lysozyme and Its Ability to Form Amyloid Fibrils in High Concentrations of Sodium Dodecyl Sulfate: A View from Molecular Dynamics Simulations. PLoS ONE 2016, 11, e0165213. [Google Scholar] [CrossRef]
- Xing, L.; Fan, W.; Chen, N.; Li, M.; Zhou, X.; Liu, S. Amyloid Formation Kinetics of Hen Egg White Lysozyme under Heat and Acidic Conditions Revealed by Raman Spectroscopy. J. Raman Spectrosc. 2019, 50, 629–640. [Google Scholar] [CrossRef]
- Alam, I.; Lertanantawong, B.; Prongmanee, W.; Lertvanithphol, T.; Horprathum, M.; Sutthibutpong, T.; Asanithi, P. Investigating Lysozyme Amyloid Fibrillization by Electrochemical Impedance Spectroscopy for Application in Lysozyme Sensor. J. Electroanal. Chem. 2021, 901, 115799. [Google Scholar] [CrossRef]
- Zein, H.F.; Alam, I.; Asanithi, P.; Sutthibutpong, T. Molecular Dynamics Study on the Effects of Charged Amino Acid Distribution under Low PH Condition to the Unfolding of Hen Egg White Lysozyme and Formation of Beta Strands. PLoS ONE 2022, 17, e0249742. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Sörgjerd, K.; Nyström, S.; Nordigården, A.; Yu, Y.-C.; Hammarström, P. Lysozyme Amyloidogenesis Is Accelerated by Specific Nicking and Fragmentation but Decelerated by Intact Protein Binding and Conversion. J. Mol. Biol. 2007, 366, 1029–1044. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Kamada, Y.; Tokunaga, Y.; Shinohara, H.; Matsumoto, M.; Kusakabe, T.; Ohkuri, T.; Ueda, T. Aggregates with Lysozyme and Ovalbumin Show Features of Amyloid-like Fibrils. Biochem. Cell Biol. 2011, 89, 533–544. [Google Scholar] [CrossRef]
- Tokunaga, Y.; Sakakibara, Y.; Kamada, Y.; Watanabe, K.; Sugimoto, Y. Analysis of Core Region from Egg White Lysozyme Forming Amyloid Fibrils. Int. J. Biol. Sci. 2013, 9, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Rahimi Araghi, L.; Dee, D.R. Cross-Species and Cross-Polymorph Seeding of Lysozyme Amyloid Reveals a Dominant Polymorph. Front. Mol. Biosci. 2020, 7, 206. [Google Scholar] [CrossRef]
- Reynolds, N.P.; Adamcik, J.; Berryman, J.T.; Handschin, S.; Zanjani, A.A.H.; Li, W.; Liu, K.; Zhang, A.; Mezzenga, R. Competition between Crystal and Fibril Formation in Molecular Mutations of Amyloidogenic Peptides. Nat. Commun. 2017, 8, 1338. [Google Scholar] [CrossRef]
- Ohkuri, T.; Shioi, S.; Imoto, T.; Ueda, T. Effect of the Structure of the Denatured State of Lysozyme on the Aggregation Reaction at the Early Stages of Folding from the Reduced Form. J. Mol. Biol. 2005, 347, 159–168. [Google Scholar] [CrossRef]
- Mishima, T.; Ohkuri, T.; Monji, A.; Imoto, T.; Ueda, T. A Particular Hydrophobic Cluster in the Residual Structure of Reduced Lysozyme Drastically Affects the Amyloid Fibrils Formation. Biochem. Biophys. Res. Commun. 2007, 356, 769–772. [Google Scholar] [CrossRef]
- Zhou, R.; Eleftheriou, M.; Royyuru, A.K.; Berne, B.J. Destruction of Long-Range Interactions by a Single Mutation in Lysozyme. Proc. Natl. Acad. Sci. USA 2007, 104, 5824–5829. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK Server for Integrated Protein–Protein Docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Bjelkmar, P.; Larsson, P.; Cuendet, M.A.; Hess, B.; Lindahl, E. Implementation of the CHARMM Force Field in GROMACS: Analysis of Protein Stability Effects from Correction Maps, Virtual Interaction Sites, and Water Models. J. Chem. Theory Comput. 2010, 6, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Price, D.J.; Brooks, C.L. A Modified TIP3P Water Potential for Simulation with Ewald Summation. J. Chem. Phys. 2004, 121, 10096–10103. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N⋅log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.; Fraaije, J. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1998, 18, 1463–1472. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A.; Open Source Drug Discovery Consortium. G_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]


| Name | Sequence | Asp Protonation | Orientation | No. of Strands | No. of Sheets |
|---|---|---|---|---|---|
| 54–62-AP | GILQINSRW | N/A | Antiparallel | 8 | 2 |
| 50–62-AP | STDYGILQINSRW | No | Antiparallel | 8 | 2 |
| 50–62-pH2-AP | STDYGILQINSRW | Yes | Antiparallel | 8 | 2 |
| 54–62-P | GILQINSRW | N/A | Parallel | 8 | 2 |
| 50–62-P | STDYGILQINSRW | No | Parallel | 8 | 2 |
| 50–62-pH2-P | STDYGILQINSRW | Yes | Parallel | 8 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zein, H.F.; Sutthibutpong, T. Roles of Tryptophan and Charged Residues on the Polymorphisms of Amyloids Formed by K-Peptides of Hen Egg White Lysozyme Investigated through Molecular Dynamics Simulations. Int. J. Mol. Sci. 2023, 24, 2626. https://doi.org/10.3390/ijms24032626
Zein HF, Sutthibutpong T. Roles of Tryptophan and Charged Residues on the Polymorphisms of Amyloids Formed by K-Peptides of Hen Egg White Lysozyme Investigated through Molecular Dynamics Simulations. International Journal of Molecular Sciences. 2023; 24(3):2626. https://doi.org/10.3390/ijms24032626
Chicago/Turabian StyleZein, Husnul Fuad, and Thana Sutthibutpong. 2023. "Roles of Tryptophan and Charged Residues on the Polymorphisms of Amyloids Formed by K-Peptides of Hen Egg White Lysozyme Investigated through Molecular Dynamics Simulations" International Journal of Molecular Sciences 24, no. 3: 2626. https://doi.org/10.3390/ijms24032626
APA StyleZein, H. F., & Sutthibutpong, T. (2023). Roles of Tryptophan and Charged Residues on the Polymorphisms of Amyloids Formed by K-Peptides of Hen Egg White Lysozyme Investigated through Molecular Dynamics Simulations. International Journal of Molecular Sciences, 24(3), 2626. https://doi.org/10.3390/ijms24032626





