MRPL12 Acts as A Novel Prognostic Biomarker Involved in Immune Cell Infiltration and Tumor Progression of Lung Adenocarcinoma
Abstract
:1. Introduction
2. Results
2.1. MRPL12 Expression Level in LUAD
2.2. Prognostic Value of MRPL12 in LUAD
2.3. Correlation between MRPL12 Expression and Survival of LUAD Patients in TMA Cohort
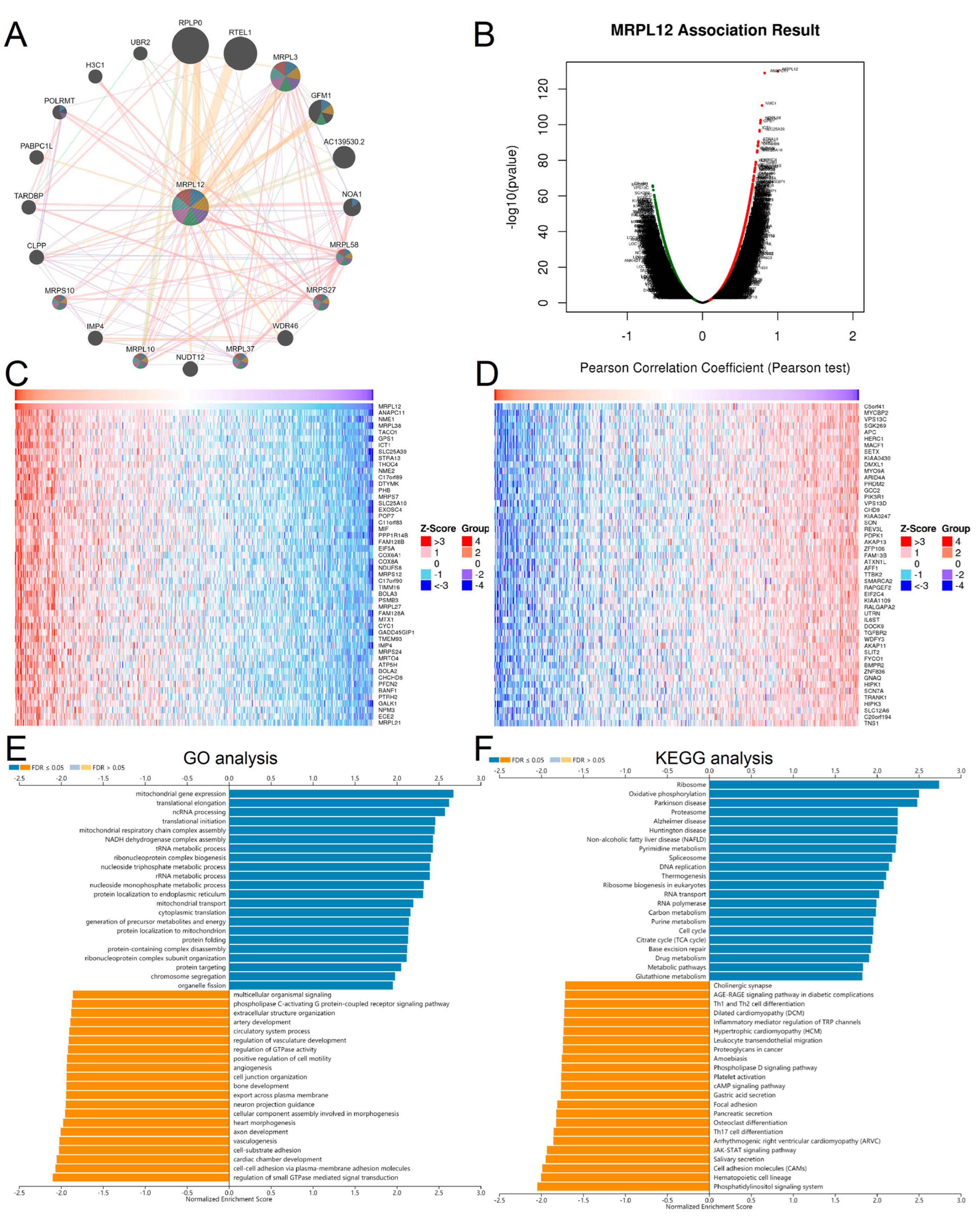
2.4. GO and KEGG Pathway Enrichment Analysis of MRPL12
2.5. MRPL12 Expression in TME at the Single-Cell Level
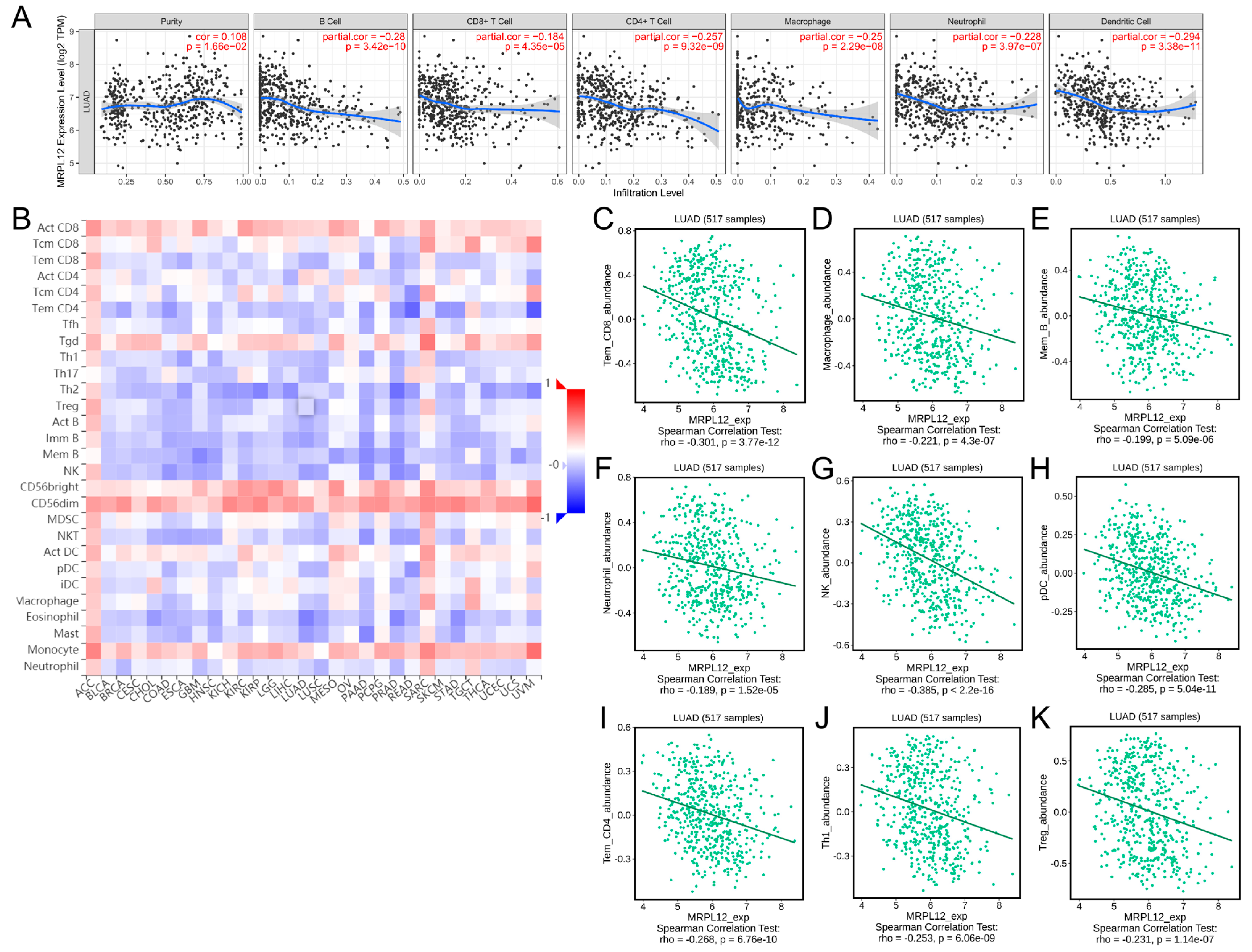
2.6. Correlation between MRPL12 Expression and Immune Infiltrates
2.7. Correlation between MRPL12 Expression and Gene Markers of Immune Cells
2.8. Correlation between MRPL12 Expression and Immunomodulators
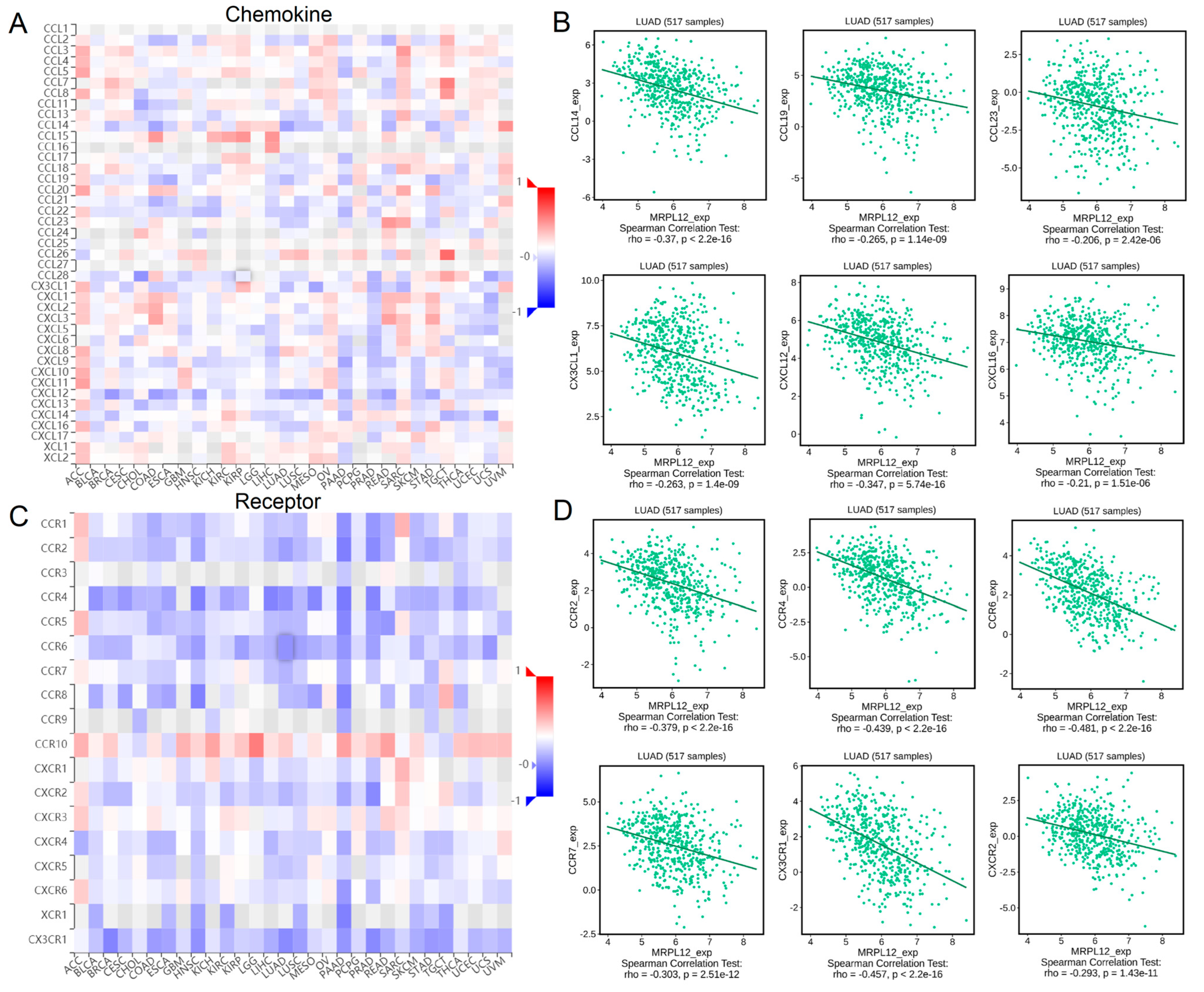
2.9. Correlation between MRPL12 Expression and Chemokines
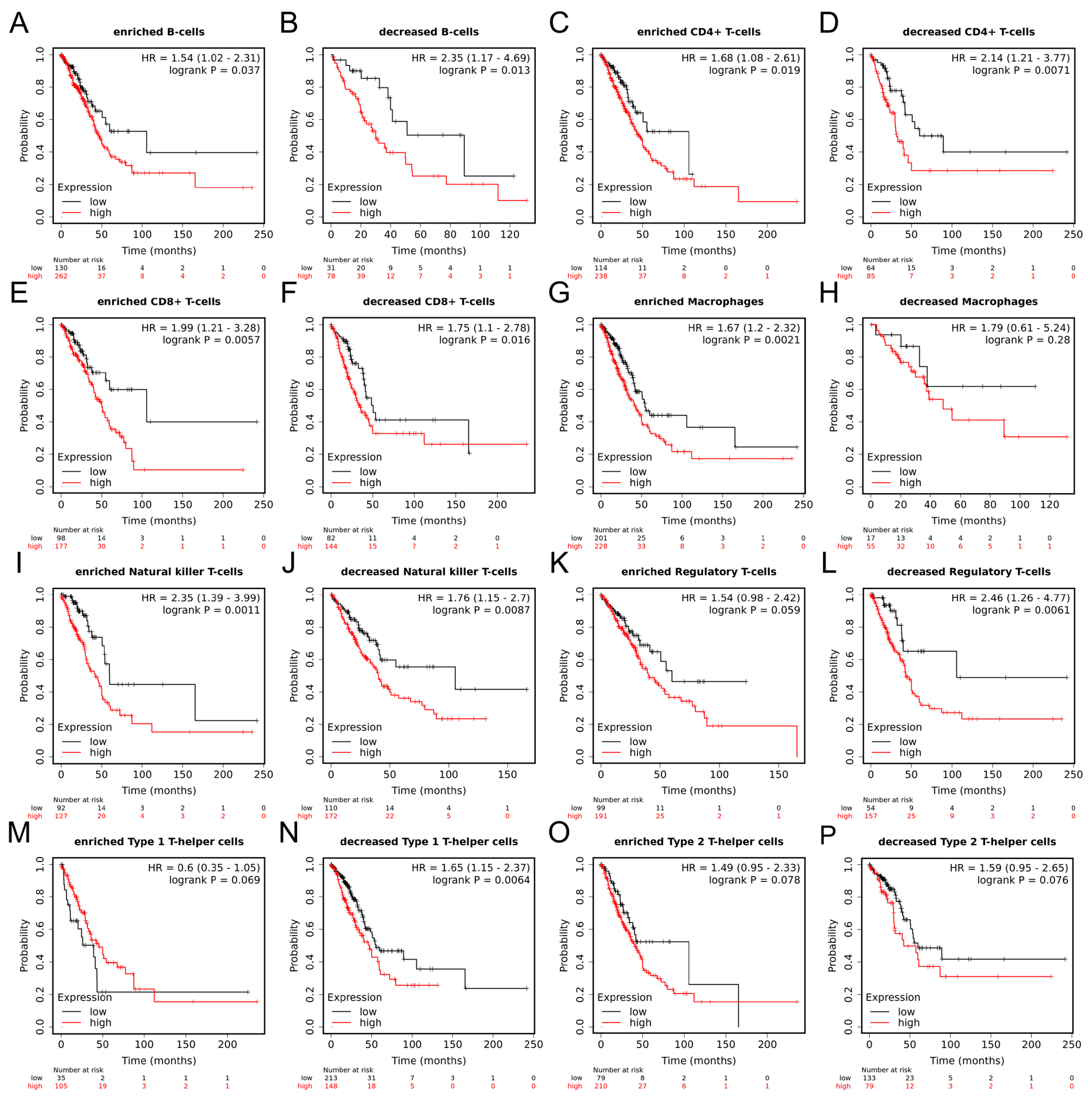
2.10. Prognostic Value of MRPL12 in LUAD Based on Immune Cells
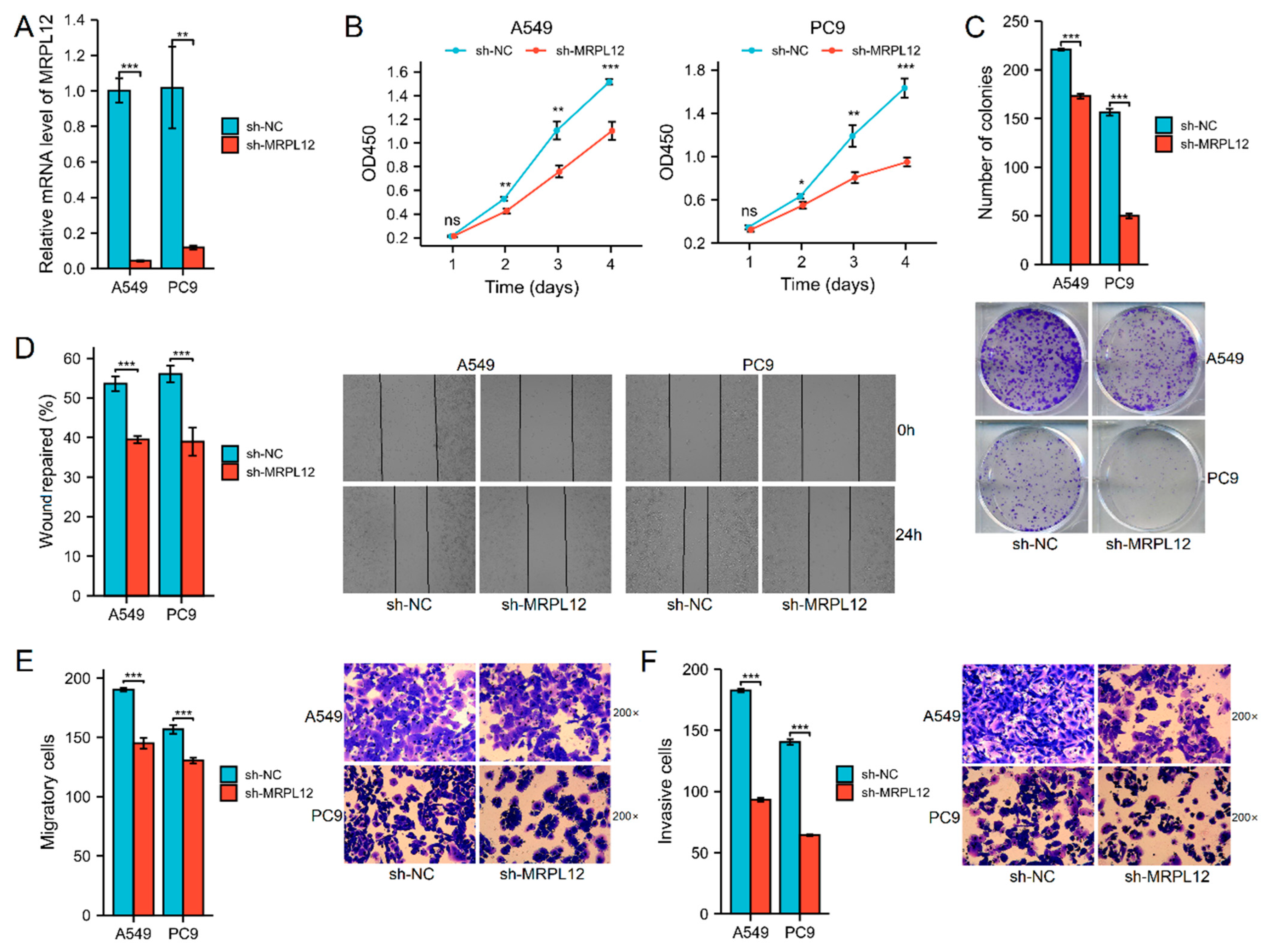
2.11. MRPL12 Knockdown Inhibited Proliferation and Invasion of LUAD Cells
3. Discussion
4. Materials and Methods
4.1. UALCAN Analysis
4.2. HPA Analysis
4.3. TMA Analysis
4.4. TIMER Analysis
4.5. GEPIA Analysis
4.6. Kaplan–Meier Plotter Analysis
4.7. LinkedOmics Analysis
4.8. PPI Network Analysis
4.9. TISIDB Analysis
4.10. TISCH Analysis
4.11. Cell Transfection
4.12. Quantitative Real-Time PCR Assay
4.13. Cell Proliferation Assay
4.14. Colony Formation Assay
4.15. Wound-Healing Assay
4.16. Transwell Assay
4.17. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.K. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar] [CrossRef]
- Denisenko, T.V.; Budkevich, I.N.; Zhivotovsky, B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018, 9, 117. [Google Scholar]
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Niksic, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Estève, J.; et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): Analysis of individual records for 37,513,025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [PubMed]
- Cheong, A.; Lingutla, R.; Mager, J. Expression analysis of mammalian mitochondrial ribosomal protein genes. Gene Expr. Patterns 2020, 38, 119147. [Google Scholar] [CrossRef] [PubMed]
- Surovtseva, Y.V.; Shutt, T.E.; Cotney, J.; Cimen, H.; Chen, S.Y.; Koc, E.C.; Shadel, G.S. Mitochondrial ribosomal protein L12 selectively associates with human mitochondrial RNA polymerase to activate transcription. Proc. Natl. Acad. Sci. USA 2011, 108, 17921–17926. [Google Scholar] [CrossRef]
- Kim, H.J.; Maiti, P.; Barrientos, A. Mitochondrial ribosomes in cancer. Semin. Cancer Biol. 2017, 47, 67–81. [Google Scholar]
- Min, S.; Lee, Y.K.; Hong, J.; Park, T.J.; Woo, H.G.; Kwon, S.M.; Yoon, G. MRPS31 loss is a key driver of mitochondrial deregulation and hepatocellular carcinoma aggressiveness. Cell Death Dis. 2021, 12, 1076. [Google Scholar] [CrossRef]
- Gu, X.; Liu, Y.; Wang, N.; Zhen, J.; Zhang, B.; Hou, S.; Cui, Z.; Wan, Q.; Feng, H. Transcription of MRPL12 regulated by Nrf2 contributes to the mitochondrial dysfunction in diabetic kidney disease. Free Radic. Biol. Med. 2021, 164, 329–340. [Google Scholar] [CrossRef]
- Chen, Y.; Cairns, R.; Papandreou, I.; Koong, A.; Denko, N.C. Oxygen consumption can regulate the growth of tumors, a new perspective on the Warburg effect. PLoS ONE 2009, 4, e7033. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, H.; Li, X.; Liu, Q.; Zhao, Y.; Li, L.; Xu, B.; Hou, Y.; Jin, W. Identification of a Three-RNA Binding Proteins (RBPs) Signature Predicting Prognosis for Breast Cancer. Front. Oncol. 2021, 11, 663556. [Google Scholar] [CrossRef]
- Ye, L.; Zhang, T.; Kang, Z.; Guo, G.; Sun, Y.; Lin, K.; Huang, Q.; Shi, X.; Ni, Z.; Ding, N.; et al. Tumor-Infiltrating Immune Cells Act as a Marker for Prognosis in Colorectal Cancer. Front. Immunol. 2019, 10, 2368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Yang, W.; Zhang, H.; Ren, Y.; Fang, Z.; Yuan, C.; Yao, Z. The Prognostic Landscape of Tumor-Infiltrating Immune Cells and Immune Checkpoints in Glioblastoma. Technol. Cancer Res. Treat. 2019, 18, 1533033819869949. [Google Scholar] [CrossRef]
- Liu, X.; Shang, X.; Li, J.; Zhang, S. The Prognosis and Immune Checkpoint Blockade Efficacy Prediction of Tumor-Infiltrating Immune Cells in Lung Cancer. Front. Cell Dev. Biol. 2021, 9, 707143. [Google Scholar] [CrossRef]
- Zeng, Y.; Shi, Y.; Xu, L.; Zeng, Y.; Cui, X.; Wang, Y.; Yang, N.; Zhou, F.; Zhou, Y. Prognostic Value and Related Regulatory Networks of MRPL15 in Non-Small-Cell Lung Cancer. Front. Oncol. 2021, 11, 656172. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Zhang, N. Identification of the Upregulation of MRPL13 as a Novel Prognostic Marker Associated with Overall Survival Time and Immunotherapy Response in Breast Cancer. Comput. Math. Methods Med. 2021, 2021, 1498924. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, P.; Yan, L.; Yang, L.; Wang, Y.; Chen, J.; Dai, J.; Li, Y.; Kang, Z.; Bai, T.; et al. MRPL35 Is Up-Regulated in Colorectal Cancer and Regulates Colorectal Cancer Cell Growth and Apoptosis. Am. J. Pathol. 2019, 189, 1105–1120. [Google Scholar] [CrossRef]
- Ma, Y.; Zhu, S.; Lv, T.; Gu, X.; Feng, H.; Zhen, J.; Xin, W.; Wan, Q. SQSTM1/p62 Controls mtDNA Expression and Participates in Mitochondrial Energetic Adaption via MRPL12. iScience 2020, 23, 101428. [Google Scholar] [CrossRef] [PubMed]
- Ashton, T.M.; McKenna, W.G.; Kunz-Schughart, L.A.; Higgins, G.S. Oxidative Phosphorylation as an Emerging Target in Cancer Therapy. Clin. Cancer Res. 2018, 24, 2482–2490. [Google Scholar] [CrossRef]
- Chen, D.; Barsoumian, H.B.; Fischer, G.; Yang, L.; Verma, V.; Younes, A.I.; Hu, Y.; Masropour, F.; Klein, K.; Vellano, C.; et al. Combination treatment with radiotherapy and a novel oxidative phosphorylation inhibitor overcomes PD-1 resistance and enhances antitumor immunity. J. Immunother. Cancer 2020, 8, e000289. [Google Scholar] [CrossRef]
- Gao, Y.; Luo, L.; Xie, Y.; Zhao, Y.; Yao, J.; Liu, X. PYCR1 knockdown inhibits the proliferation, migration, and invasion by affecting JAK/STAT signaling pathway in lung adenocarcinoma. Mol. Carcinog. 2020, 59, 503–511. [Google Scholar] [CrossRef]
- Prabhu, K.S.; Bhat, A.A.; Siveen, K.S.; Kuttikrishnan, S.; Raza, S.S.; Raheed, T.; Jochebeth, A.; Khan, A.Q.; Chawdhery, M.; Haris, M.; et al. Sanguinarine mediated apoptosis in Non-Small Cell Lung Cancer via generation of reactive oxygen species and suppression of JAK/STAT pathway. Biomed. Pharmacother. 2021, 144, 112358. [Google Scholar] [CrossRef]
- Potter, S.S. Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 2018, 14, 479–492. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Wang, D.; Lu, P.; Yang, N.; Xue, Z.; Zhu, X.; Zhang, P.; Fan, G. Single-cell RNA sequencing reveals heterogeneous tumor and immune cell populations in early-stage lung adenocarcinomas harboring EGFR mutations. Oncogene 2021, 40, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yin, C.; Geng, L.; Cai, W. Immune Infiltration Landscape in Clear Cell Renal Cell Carcinoma Implications. Front. Oncol. 2020, 10, 491621. [Google Scholar] [CrossRef] [PubMed]
- de Winde, C.M.; Munday, C.; Acton, S.E. Molecular mechanisms of dendritic cell migration in immunity and cancer. Med. Microbiol. Immunol. 2020, 209, 515–529. [Google Scholar] [CrossRef]
- Fu, C.; Jiang, A. Dendritic Cells and CD8 T Cell Immunity in Tumor Microenvironment. Front. Immunol. 2018, 9, 3059. [Google Scholar] [CrossRef]
- Kravtsov, D.S.; Erbe, A.K.; Sondel, P.M.; Rakhmilevich, A.L. Roles of CD4+ T cells as mediators of antitumor immunity. Front. Immunol. 2022, 13, 972021. [Google Scholar] [CrossRef] [PubMed]
- Aktas, O.N.; Ozturk, A.B.; Erman, B.; Erus, S.; Tanju, S.; Dilege, S. Role of natural killer cells in lung cancer. J. Cancer Res. Clin. Oncol. 2018, 144, 997–1003. [Google Scholar] [CrossRef]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef]
- Ozga, A.J.; Chow, M.T.; Luster, A.D. Chemokines and the immune response to cancer. Immunity 2021, 54, 859–874. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.W.; Onder, L.; Cupovic, J.; Boesch, M.; Novkovic, M.; Pikor, N.; Tarantino, I.; Rodriguez, R.; Schneider, T.; Jochum, W.; et al. CCL19-producing fibroblastic stromal cells restrain lung carcinoma growth by promoting local antitumor T-cell responses. J. Allergy Clin. Immunol. 2018, 142, 1257–1271.e4. [Google Scholar] [CrossRef] [Green Version]
- Itakura, M.; Terashima, Y.; Shingyoji, M.; Yokoi, S.; Ohira, M.; Kageyama, H.; Matui, Y.; Yoshida, Y.; Ashinuma, H.; Moriya, Y.; et al. High CC chemokine receptor 7 expression improves postoperative prognosis of lung adenocarcinoma patients. Br. J. Cancer 2013, 109, 1100–1108. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, C.; Yang, Z.; Bai, Y.; Shukuya, T.; Poh, M.E.; Ekman, S.; Li, J.; Xu, Y.; Deng, S. Identification of GPX4 as a therapeutic target for lung adenocarcinoma after EGFR-TKI resistance. Transl. Lung Cancer Res. 2022, 11, 786–801. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Yu, T.; Li, C.; Li, J.; Liang, Y.; Wang, X.; Chen, Y.; Wang, X. Expression of CAMK1 and its association with clinicopathologic characteristics in pancreatic cancer. J. Cell. Mol. Med. 2021, 25, 1198–1206. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Wang, J.; Han, Y.; Dong, X.; Ge, J.; Zheng, R.; Shi, X.; Wang, B.; Li, Z.; Ren, P.; et al. TISCH: A comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021, 49, D1420–D1430. [Google Scholar] [CrossRef] [PubMed]



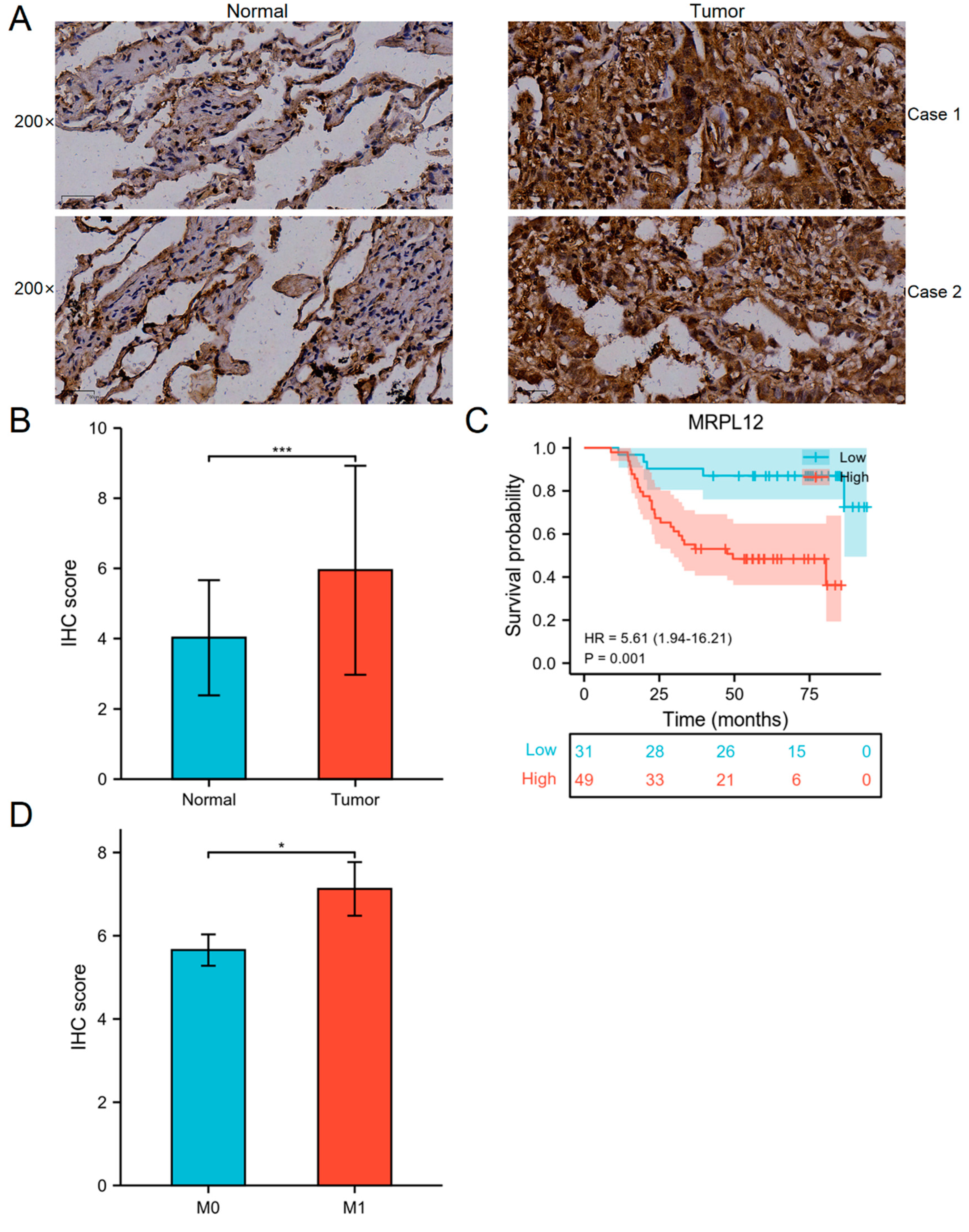


| Variables | N | MRPL12 Expression | p Value | |
|---|---|---|---|---|
| Low (n = 31) | High (n = 49) | |||
| Age (years) | 0.910 | |||
| ≤60 | 29 | 11 | 18 | |
| >60 | 51 | 20 | 31 | |
| Gender | 0.050 | |||
| Female | 33 | 17 | 16 | |
| Male | 47 | 14 | 33 | |
| Tumor size (cm) | 0.185 | |||
| ≤3 | 39 | 18 | 21 | |
| >3 | 41 | 13 | 28 | |
| T stage | 0.102 | |||
| T1/T2 | 59 | 26 | 33 | |
| T3/T4 | 21 | 5 | 16 | |
| N stage | 0.433 | |||
| N0 | 58 | 24 | 34 | |
| N1/N2 | 22 | 7 | 15 | |
| Pathologic stage | 0.003 | |||
| I/II | 51 | 26 | 25 | |
| III/IV | 29 | 5 | 24 | |
| Chemotherapy | 0.009 | |||
| No | 42 | 22 | 20 | |
| Yes | 38 | 9 | 29 | |
| Survival status | 0.001 | |||
| Alive | 49 | 26 | 23 | |
| Dead | 31 | 5 | 26 | |
| Survival time (months) | 68.22 ± 22.02 | 43.56 ± 23.21 | <0.001 | |
| Variables | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| OR | 95%CI | p | OR | 95%CI | p | |
| Age (years) | ||||||
| ≤60 | ref | |||||
| >60 | 0.703 | 0.344–1.437 | 0.334 | |||
| Gender | ||||||
| Female | ref | |||||
| Male | 1.576 | 0.737–3.369 | 0.240 | |||
| Tumor size (cm) | ||||||
| ≤3 | ref | ref | ||||
| >3 | 3.677 | 1.631–8.292 | 0.001 | 0.72 | 0.289–1.793 | 0.481 |
| T stage | ||||||
| T1/T2 | ref | ref | ||||
| T3/T4 | 13.87 | 6.139–31.32 | <0.001 | 5.455 | 1.713–17.368 | 0.004 |
| N stage | ||||||
| N0 | ref | ref | ||||
| N1/N2 | 3.315 | 1.633–6.728 | <0.001 | 1.144 | 0.418–3.131 | 0.792 |
| Pathologic stage | ||||||
| I/II | ref | ref | ||||
| III/IV | 32.17 | 10.95–94.47 | <0.001 | 9.477 | 1.537–58.42 | 0.015 |
| Chemotherapy | ||||||
| No | ref | ref | ||||
| Yes | 12.85 | 4.47–36.96 | <0.001 | 2.685 | 0.687–10.485 | 0.155 |
| MRPL12 expression | ||||||
| Low | ref | ref | ||||
| High | 5.611 | 1.942–16.21 | 0.001 | 3.861 | 1.187–12.562 | 0.024 |
| Description | Gene Markers | None Cor | p Value | Purity Cor | p Value |
|---|---|---|---|---|---|
| CD8+ T cell | CD8A | −0.058 | 0.187 | −0.016 | 0.718 |
| CD8B | 0.007 | 0.872 | 0.044 | 0.325 | |
| T cell (general) | CD3D | −0.121 | ** | −0.077 | 0.087 |
| CD3E | −0.222 | *** | −0.196 | *** | |
| CD2 | −0.224 | *** | −0.2 | *** | |
| B cell | CD19 | −0.194 | *** | −0.165 | *** |
| CD79A | −0.201 | *** | −0.172 | *** | |
| Monocyte | CD86 | −0.213 | *** | −0.191 | *** |
| CD115 (CSF1R) | −0.272 | *** | −0.251 | *** | |
| TAM | CCL2 | −0.126 | *** | −0.096 | * |
| CD68 | −0.191 | *** | −0.169 | *** | |
| IL10 | −0.181 | *** | −0.151 | *** | |
| M1 macrophage | INOS (NOS2) | 0.002 | 0.964 | 0.009 | 0.835 |
| IRF5 | −0.067 | 0.13 | −0.033 | 0.467 | |
| COX2(PTGS2) | −0.026 | 0.562 | −0.039 | 0.392 | |
| M2 macrophage | CD163 | −0.209 | *** | −0.191 | *** |
| VSIG4 | −0.193 | *** | −0.175 | *** | |
| MS4A4A | −0.25 | *** | −0.23 | *** | |
| Neutrophils | CD66b (CEACAM8) | −0.35 | *** | −0.35 | *** |
| CD11b (ITGAM) | −0.305 | *** | −0.281 | *** | |
| CCR7 | −0.308 | *** | −0.287 | *** | |
| NK cell | KIR2DL1 | 0 | 0.993 | 0.012 | 0.785 |
| KIR2DL3 | −0.002 | 0.97 | 0.022 | 0.629 | |
| KIR2DL4 | 0.185 | *** | 0.21 | *** | |
| KIR3DL1 | −0.031 | 0.481 | −0.013 | 0.772 | |
| KIR3DL2 | 0.017 | 0.705 | 0.053 | 0.245 | |
| KIR3DL3 | 0.068 | 0.123 | 0.082 | 0.068 | |
| KIR2DS4 | −0.063 | 0.152 | −0.038 | 0.397 | |
| DC | HLA-DPB1 | −0.409 | *** | −0.401 | *** |
| HLA-DQB1 | −0.258 | *** | −0.24 | *** | |
| HLA-DRA | −0.37 | *** | −0.362 | *** | |
| HLA-DPA1 | −0.408 | *** | −0.398 | *** | |
| BCDA-1 (CD1C) | −0.439 | *** | −0.42 | *** | |
| BDCA-4 (NRP1) | −0.144 | ** | −0.136 | ** | |
| CD11c (ITGAX) | −0.238 | *** | −0.218 | *** | |
| Th1 cell | T-bet (TBX21) | −0.158 | *** | −0.133 | ** |
| STAT4 | −0.21 | *** | −0.196 | *** | |
| STAT1 | 0.029 | 0.506 | 0.049 | 0.278 | |
| IFN-γ (IFNG) | 0.074 | 0.093 | 0.114 | * | |
| TNF-α (TNF) | −0.162 | *** | −0.122 | ** | |
| Th2 cell | GATA3 | −0.198 | *** | −0.169 | *** |
| STAT6 | −0.302 | *** | −0.326 | *** | |
| STAT5A | −0.288 | *** | −0.263 | *** | |
| IL13 | −0.065 | 0.139 | −0.051 | 0.257 | |
| Th17 cell | STAT3 | −0.242 | *** | −0.256 | *** |
| IL17A | 0.022 | 0.617 | 0.048 | 0.289 | |
| Treg | FOXP3 | −0.115 | ** | −0.086 | 0.057 |
| CCR8 | −0.219 | *** | −0.196 | *** | |
| STAT5B | −0.241 | *** | −0.233 | *** | |
| TGFβ (TGFB1) | −0.201 | *** | −0.178 | *** | |
| T-cell exhaustion | PD-1 (PDCD1) | 0.042 | 0.341 | 0.101 | * |
| PDL1 (PDCD1LG2) | −0.148 | *** | −0.11 | * | |
| CTLA4 | −0.114 | ** | −0.076 | 0.093 | |
| LAG3 | 0.047 | 0.283 | 0.09 | * | |
| TIM-3 (HAVCR2) | −0.189 | *** | −0.164 | *** | |
| GZMB | 0.176 | *** | 0.232 | *** |
| Description | Gene Markers | Tumor Cor | p Value | Normal Cor | p Value |
|---|---|---|---|---|---|
| T cell (general) | CD3D | −0.17 | *** | 0.13 | 0.31 |
| CD3E | −0.28 | *** | −0.11 | 0.4 | |
| CD2 | −0.28 | *** | −0.044 | 0.74 | |
| B cell | CD19 | −0.29 | *** | 0.07 | 0.6 |
| CD79A | −0.32 | *** | −0.1 | 0.45 | |
| TAM | CCL2 | −0.13 | ** | −0.26 | * |
| CD68 | −0.2 | *** | 0.042 | 0.75 | |
| IL10 | −0.23 | *** | −0.029 | 0.83 | |
| M2 macrophage | CD163 | −0.16 | *** | 0.13 | 0.32 |
| VSIG4 | −0.2 | *** | 0.17 | 0.19 | |
| MS4A4A | −0.27 | *** | 0.16 | 0.23 | |
| DC | HLA-DPB1 | −0.39 | *** | 0.12 | 0.35 |
| HLA-DQB1 | −0.18 | *** | −0.044 | 0.74 | |
| HLA-DRA | −0.36 | *** | 0.21 | 0.11 | |
| HLA-DPA1 | −0.4 | *** | −0.22 | 0.096 | |
| BCDA-1 (CD1C) | −0.37 | *** | 0.083 | 0.53 | |
| BDCA-4 (NRP1) | −0.12 | ** | −0.44 | *** | |
| CD11c (ITGAX) | −0.26 | *** | −0.017 | 0.9 | |
| Th1 cell | T-bet (TBX21) | −0.23 | *** | −0.1 | 0.44 |
| STAT4 | −0.21 | *** | −0.27 | * | |
| STAT1 | −0.009 | 0.85 | −0.19 | 0.14 | |
| IFN-γ (IFNG) | −0.008 | 0.86 | 0.03 | 0.82 | |
| TNF-α (TNF) | −0.18 | *** | −0.034 | 0.8 | |
| Th2 cell | GATA3 | −0.21 | *** | −0.14 | 0.31 |
| STAT6 | −0.24 | *** | −0.37 | ** | |
| STAT5A | −0.3 | *** | −0.067 | 0.61 | |
| IL13 | −0.098 | * | −0.16 | 0.22 | |
| Treg | FOXP3 | −0.15 | ** | 0.029 | 0.83 |
| CCR8 | −0.26 | *** | −0.12 | 0.36 | |
| STAT5B | −0.22 | *** | −0.53 | *** | |
| TGFβ (TGFB1) | −0.15 | *** | 0.14 | 0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Liu, Y.; Ma, C.; Ai, K. MRPL12 Acts as A Novel Prognostic Biomarker Involved in Immune Cell Infiltration and Tumor Progression of Lung Adenocarcinoma. Int. J. Mol. Sci. 2023, 24, 2762. https://doi.org/10.3390/ijms24032762
Hu Y, Liu Y, Ma C, Ai K. MRPL12 Acts as A Novel Prognostic Biomarker Involved in Immune Cell Infiltration and Tumor Progression of Lung Adenocarcinoma. International Journal of Molecular Sciences. 2023; 24(3):2762. https://doi.org/10.3390/ijms24032762
Chicago/Turabian StyleHu, Yangyang, Yue Liu, Chenchao Ma, and Kaixing Ai. 2023. "MRPL12 Acts as A Novel Prognostic Biomarker Involved in Immune Cell Infiltration and Tumor Progression of Lung Adenocarcinoma" International Journal of Molecular Sciences 24, no. 3: 2762. https://doi.org/10.3390/ijms24032762






