Abstract
A series of 15 new derivatives of 6-acetyl-7-hydroxy-4-methylcoumarin containing a piperazine group were designed with the help of computational methods and were synthesized to study their affinity for the serotonin 5-HT1A and 5-HT2A receptors. Among them, 6-acetyl-7-{4-[4-(3-bromophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (4) and 6-acetyl-7-{4-[4-(2-chlorophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (7) exhibited excellent activity for 5-HT1A receptors with Ki values 0.78 (0.4–1.4) nM and 0.57 (0.2–1.3) nM, respectively, comparable to the Ki values of 8-OH-DPAT (0.25 (0.097–0.66) nM). The equilibrium dissociation constant values of the tested compounds showed differential intrinsic activities of the agonist and antagonist modes.
1. Introduction
5-HT receptors belong to the group of G-protein-coupled membrane receptors located on the cell membrane of neurons and selected other cells, including smooth muscle, pancreatic β-cells, hepatocytes and adipocytes. 5-HT receptors mediate the action of serotonin both in the central nervous system and the periphery nerves [1,2,3,4]. They are also an important target for a variety of drugs for the treatment of anorexia, schizophrenia, psychosis or depression [1,2,3,4].
N-arylpiperazine-containing ligands are one of the families of chemical compounds known to strongly interact with serotonin receptors [5,6]. This large group owes its biological properties to the presence of a highly basic nitrogen atom of the piperazine. This atom can form strong interactions with the acidic amino acids in the GPCR transmembrane domain of proteins [7]. Coumarin–piperazine ligands are also known for their cytotoxic activity, acting as antibacterial and antifungal agents, acetylcholinesterase inhibitors and as dual serotonin and dopamine receptor agents with antipsychotic and antiparkinsonian properties [8]. These compounds, often bearing arylpiperazines linked to a coumarin system via an alkyl linker, can modulate central nervous system (CNS) affective function by targeting serotoninergic, dopaminergic and adrenergic receptors. From a medical point of view, the importance of finding new biologically active compounds lies in the fact that serotonin and dopamine receptors are involved in the pathomechanisms of many psychiatric and neurological disorders, such as schizophrenia, depression, epilepsy or Alzheimer’s and Parkinson’s diseases [8].
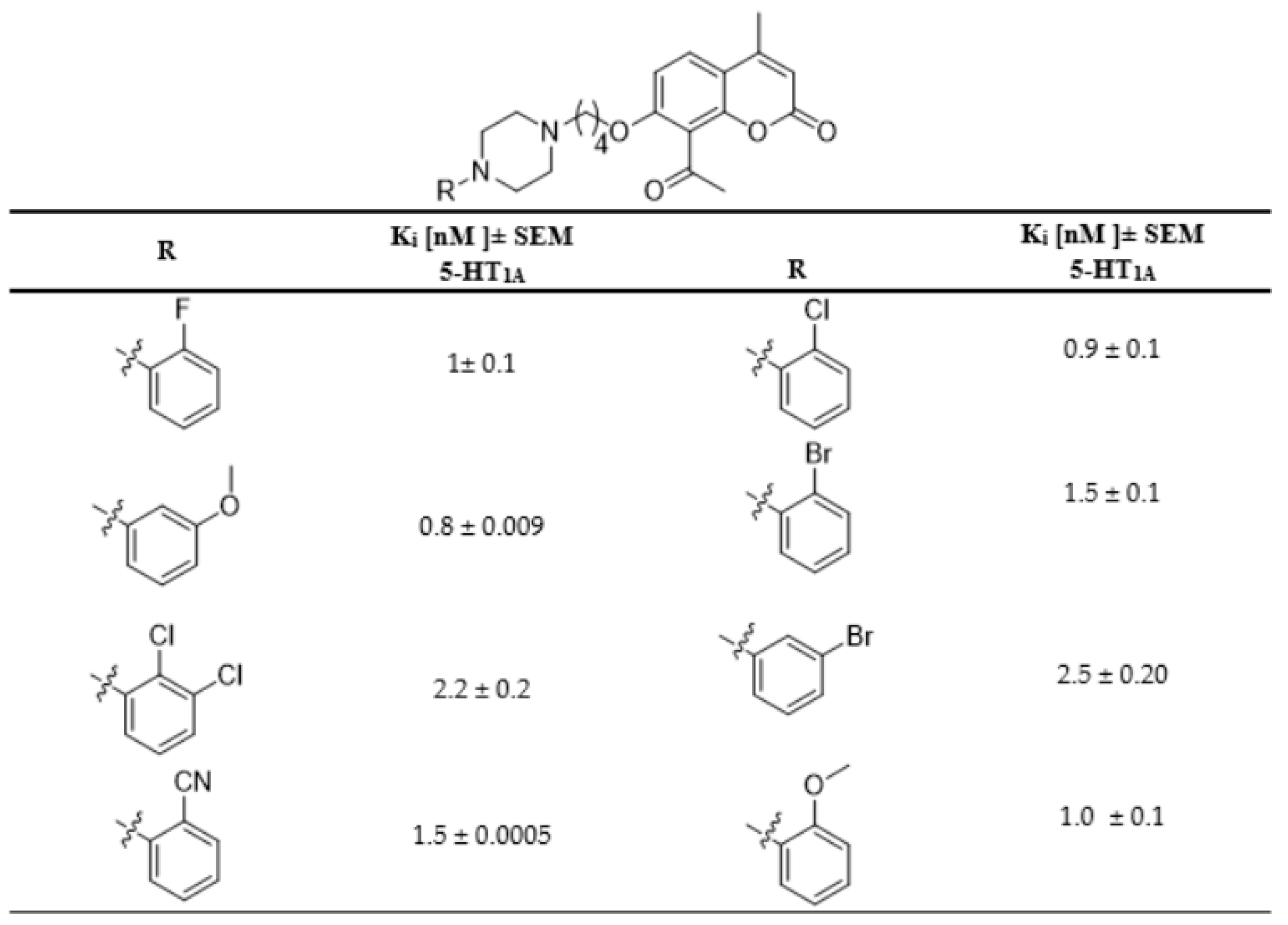
In this work, as a continuation of our previous research, 6-acetyl-7-hyroxy-4-methylcoumarin was used as the lead compound for further structural modifications [9,10,11,12,13,14,15,16]. Recently, we synthesized a series of aryl/heteroarylpiperazinyl derivatives of 8-acetyl-7-hydroxy-4-methylcoumarin and evaluated their antidepressant-like activity [9,10]. These compounds showed very high affinity for serotonin 5-HT1A receptors (Figure 1). The acetyl group in position 8 of the coumarin ring of these compounds increased the affinity for 5-HT1A and 5-HT2A receptors compared to their derivative bearing no acetyl group in position 8 [9]. Here, we decided to attach the acetyl group with its ability to form hydrogen bonds with residues in the 5-HT1A binding pocket, at the C-6 position of the coumarin ring. We designed a synthetic strategy for target compounds with different aryl and heteroarylpiperazine moieties and harboring a four-carbon linker between the coumarin and piperazine ring, which according to earlier studies, gave the most favorable binding features [9,10]. Following the design of new coumarin derivatives, we used molecular docking to multiple crystal structures of 5-HT1A and 5-HT2A receptors to estimate their affinities, followed by microwave-assisted protocols, which were used to synthesize all new compounds. Upon the successful synthesis all compounds, they have been evaluated for their binding affinities for 5-HT1A and 5-HT2A receptors and their agonistic/antagonistic properties at the 5-HT1A receptor.
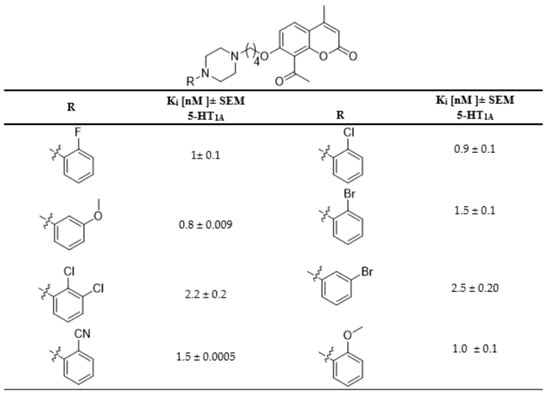
Figure 1.
Selected 5-HT1A receptor affinities of previously synthesized coumarin derivatives.
2. Results and Discussion
2.1. Chemistry
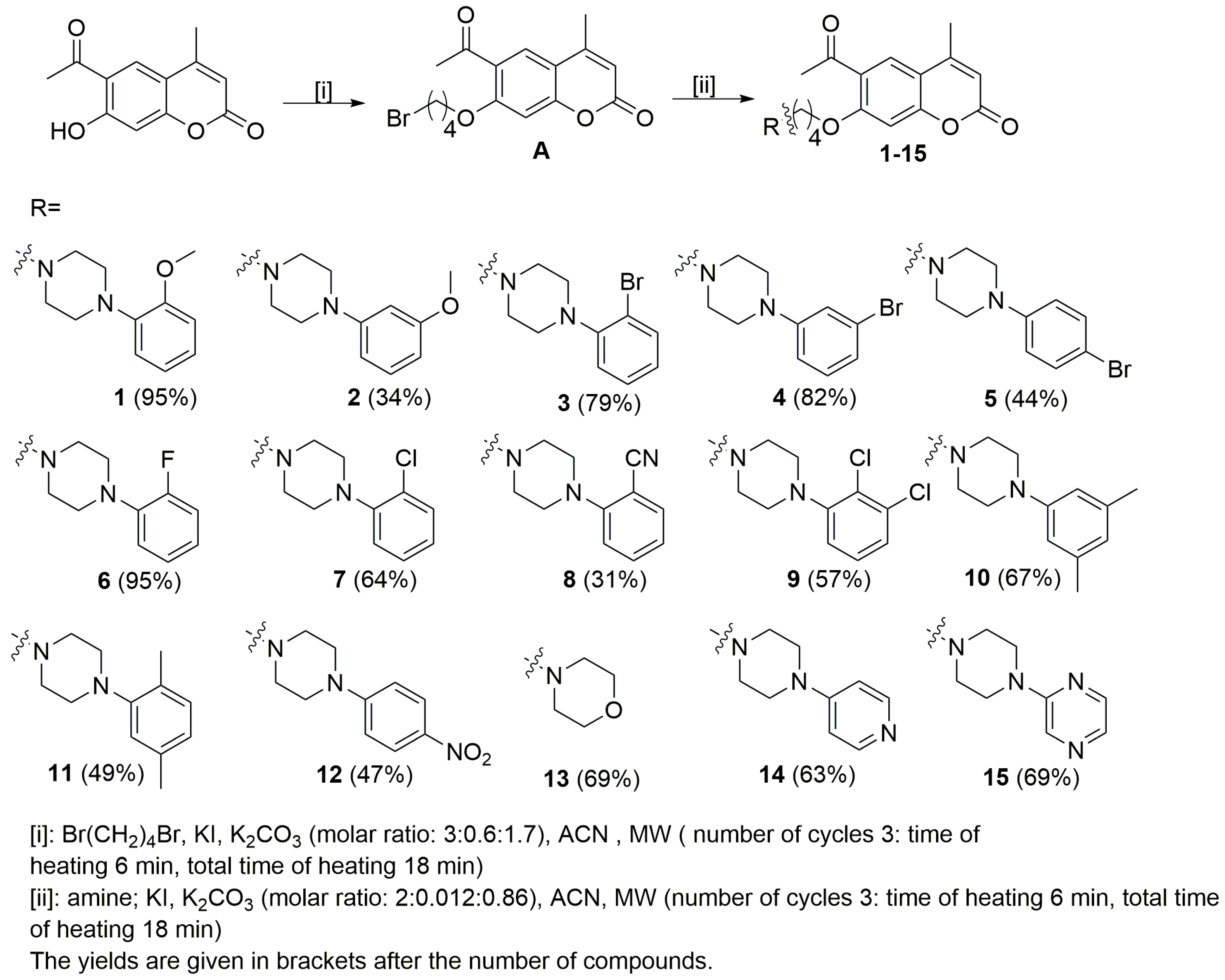
The starting coumarin 6-acetyl-7-hydroxy-4-methylchromen-2-one was resynthesized according to previously published studies [17]. The planned compounds were obtained in two steps. In the first step, 6-acetyl-7-(4-bromobutoxy)-4-methylchromen-2-one (A) was prepared by alkylation of the phenolic group with dibromobutane in acetonitrile in the presence of potassium iodide and potassium carbonate (Scheme 1). In the second step, the final compounds were obtained according to the previously published study [16]. Microwave irradiation was used in order to increase yield and to reduce the reaction time. The synthesis of compounds 1–15 was carried out by reacting the 6-acetyl-7-(4-bromobutoxy)-4-methylchromen-2-one (A) with appropriate arylpiperazine: 4-(2-methoxyphenyl)piperazine, 4-(3-methoxyphenyl)piperazine, 4-(2-bromophenyl)piperazine, 4-(3-bromophenyl)piperazine, 4-(4-bromophenyl)piperazine 4-(2-fluorophenyl)piperazine, 4-(2-chlorophenyl)piperazine, 4-(2-cyanophenyl)piperazine, 4-(2,3-dichlorophenyl)piperazine, 4-(3,5-dimethylphenyl)piperazine, 4-(2,5-dime thylphenyl)piperazine, 4-(4-nitrophenyl)piperazine, morpholine, 4-(pyrid-4-yl)pipera zine and 4-(pyrazin-2-yl)piperazine, in acetonitrile and in the presence of potassium iodide and potassium carbonate. The TLC method was used to monitor the progress of the reaction (silica gel plates, eluent: CHCl3: MeOH; 10: 0.25). We also used microwave irradiation in this step and column chromatography (silica gel, CHCl3:MeOH (100:1) as the eluent) to purify the final compounds 1–15. The final product yields were in the 31–95% range. All compounds were characterized using standard methods, 1H NMR, 13C NMR spectroscopy, and HRMS spectrometry. All NMR spectra are presented in the Supplementary Materials. To complete the structural characterization of compounds, we also report results of X-ray crystallographic studies for 6-acetyl-7-{4-[4-(3-methoxyphenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (2).
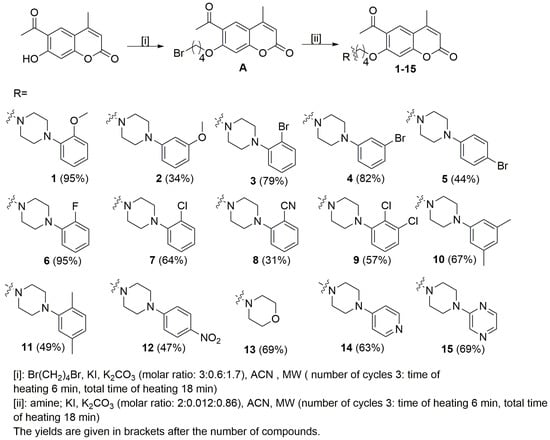
Scheme 1.
Synthesis of compounds 1–15 investigated in this work.
2.2. X-ray Crystallography
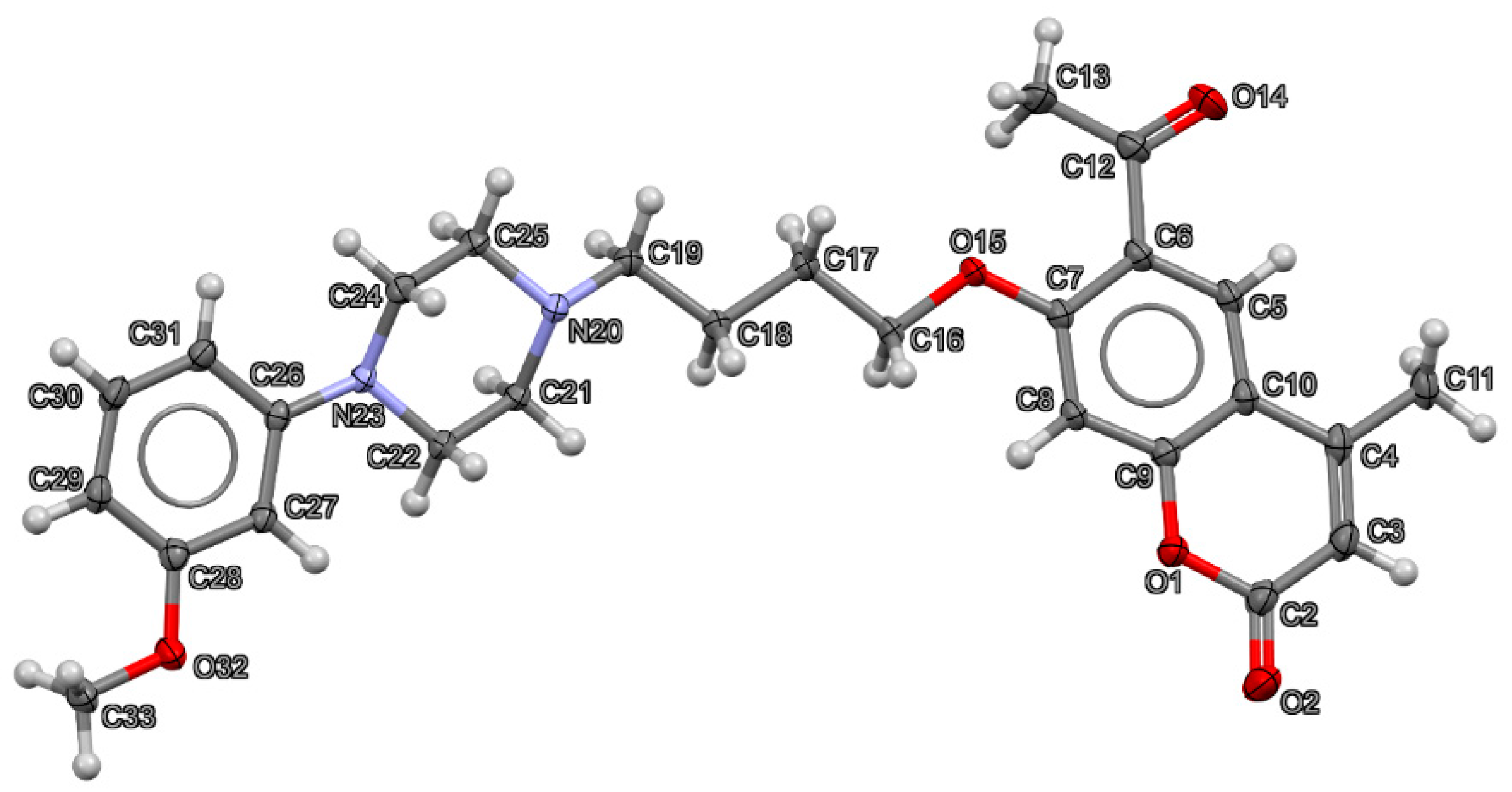
The crystal data and structure refinement parameters for 6-acetyl-7-{4-[4-(3-methoxyphenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (2) are collected in Table 1. Compound 2 crystallizes in P21/c space group. The structure of the crystal measured at 130 K is fully ordered, which is visible in Figure 2, displaying the asymmetric unit with anisotropic displacement parameters of all non-H atoms at a 50% probability level.

Table 1.
Data collection and structure refinement parameters for 2.
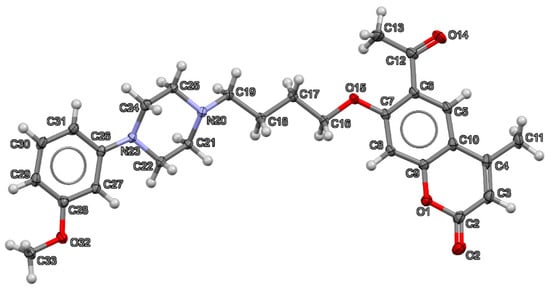
Figure 2.
Thermal ellipsoid plot at 50% probability level together with numbering scheme of all non-H atoms in the structure of 6-acetyl-7-{4-[4-(3-methoxyphenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (2). Oxygen atoms are shown in red, nitrogen in blue, carbon in dark grey and hydrogen in light grey.
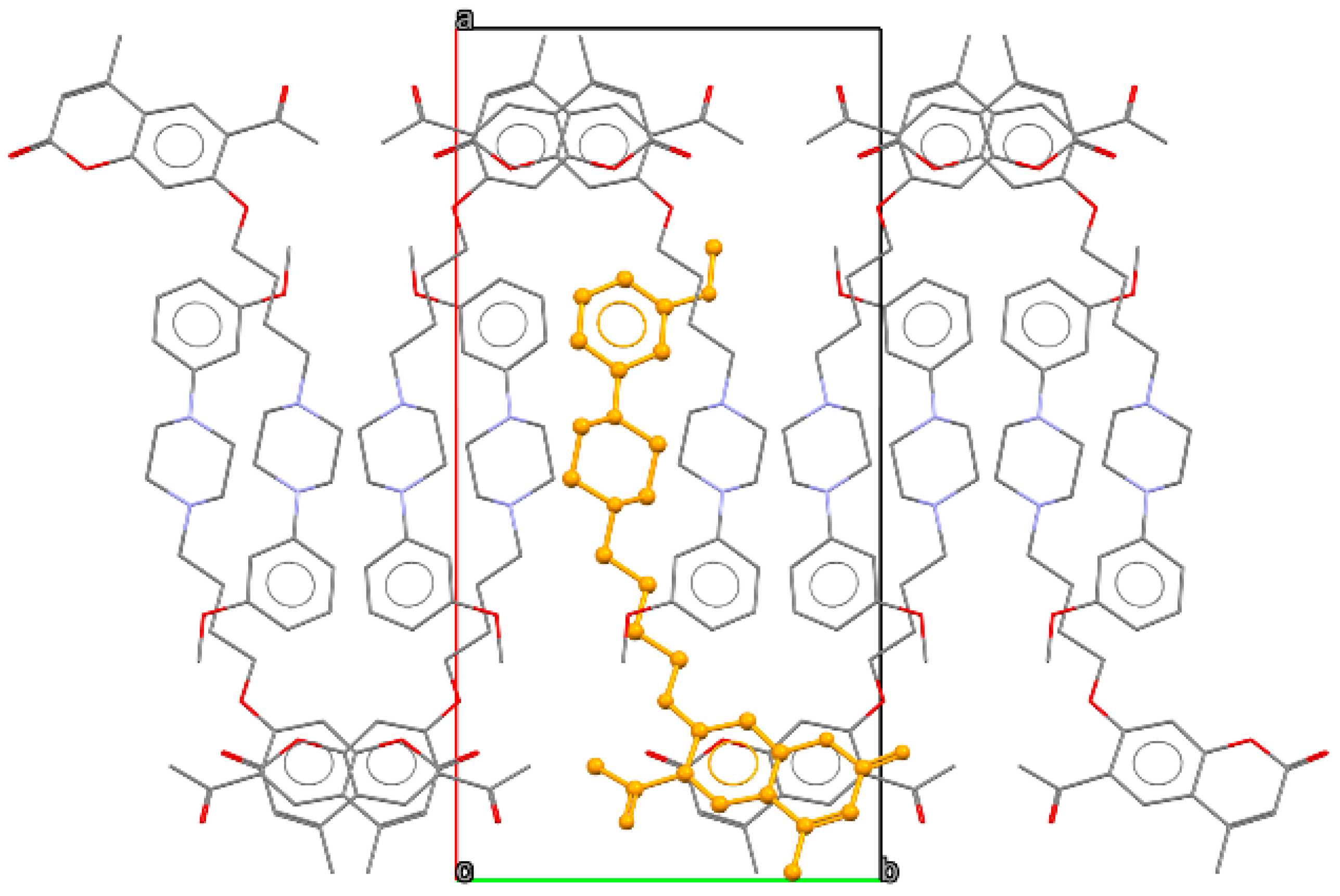
Because of the lack of strong hydrogen bond donors, the structure is supported by weak interactions only. However, such interactions slightly affect the geometry of the coumarin molecules, which deviate somewhat from planarity. The distance between the mean plane fitted to the coumarin C5–C10 atoms and O2 moiety is 0.15 Å. In addition, the acetyl group is slightly rotated with respect to the C5–C10 benzene moiety with the interplanar angle close to 15°. In the crystal lattice, the coumarin fragments are engaged in parallel π-π stacking, with the closest distances between the C5–C10 benzene plane and above/below coumarin moiety C atoms being ca. 3.52 Å. These stacks of moieties are parallel to the [001] direction. The side chains containing the piperazine unit are grouped in the central part of the unit cells, as presented in Figure 3. Here, some weak C H…N interactions (H…N distance of 2.64 Å) involving neighboring piperazine moieties can be observed.
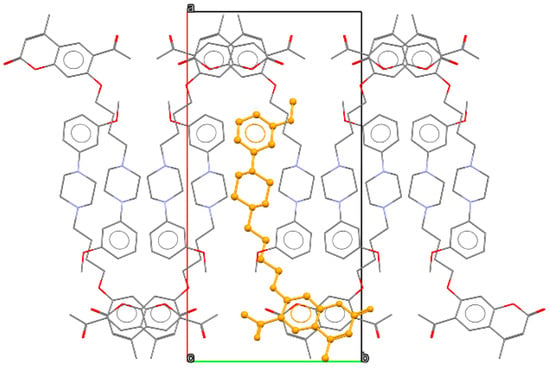
Figure 3.
Packing diagram of 2, view along the [001] direction. Orange color indicates single molecule, oxygen atoms are shown in red, nitrogen in blue, carbon in grey.
2.3. Computational Studies
2.3.1. ADME Properties
The major predicted ADME properties of all compounds studied are presented in Table 2. Some of the newly synthesized compounds exceed the desired common limit of 500 Da for systems with good oral bioavailability [18], but only slightly, and they all fall within the modern limit of <700 Da [19]. Compared to aripiprazole, there is a higher number of possible hydrogen bond acceptors due to the presence of one or two more oxygen atoms in the coumarin scaffold. As a result of the same structural feature, all investigated compounds were predicted to be more soluble in water. Finally, in all cases apart from 14, the nitrogen atom of the piperazine part of ligands is predicted to be basic and protonated, as in the case of aripiprazole (although, the expected accuracy of pKa estimate is of around 0.7–1.0 pH units).

Table 2.
Predicted ADME properties for compounds 1–15.
2.3.2. Molecular Docking
The results of the Ki estimates obtained from the local search are presented in Table 3. For both the 5-HT1A and 5-HT2A receptors, we were able to find several compounds with expected higher affinity for these receptors than aripiprazole. However, in most cases, the difference between compounds with the highest estimated affinity and aripiprazole is below 1 kcal/mol or one order of magnitude in the Ki estimates, which is likely below the accuracy of the docking method [20]; on the other hand, since all poses in the local search are very similar, it is likely that error cancelation occurs. This is also clear when comparing the computational Ki estimate of aripiprazole in the pose from the crystal structure (45.6 nM) with the experimental value of 1.5–5.6 nM [21,22,23].

Table 3.
Predicted affinities of compounds 1–15 to 5-HT receptors.
Comparing the results of the local search with the experimental Ki values presented in the next part of this study, the agreement between those two methods is only average. For example, compounds 12–15 were correctly predicted to have low affinities for the 5-HT1A receptor. Additionally, compounds 1–11 were predicted to have Ki values between 20 and 70 nM for the 5-HT1A receptor, which is in good agreement with the experimental values of 0.5–22 nM given the expected accuracy of the docking approach. To improve the computational results, after performing the biological affinity evaluation, we also conducted flexible docking with the Lamarckian genetic algorithm to multiple crystal structures of the 5-HT1A receptor (see Table 3). Here, the results were improved, and in many cases, the obtained affinity estimates were very close to the experimental values. On the other hand, for selected systems with relatively low affinities (e.g., 12), flexible docking predicts very low Ki values, which are clearly in disagreement with experimental data. In these cases, the poses of such docked complexes do not resemble the pose of aripiprazole with the crucial salt bridge to D116, but are stabilized by completely different intermolecular interactions.
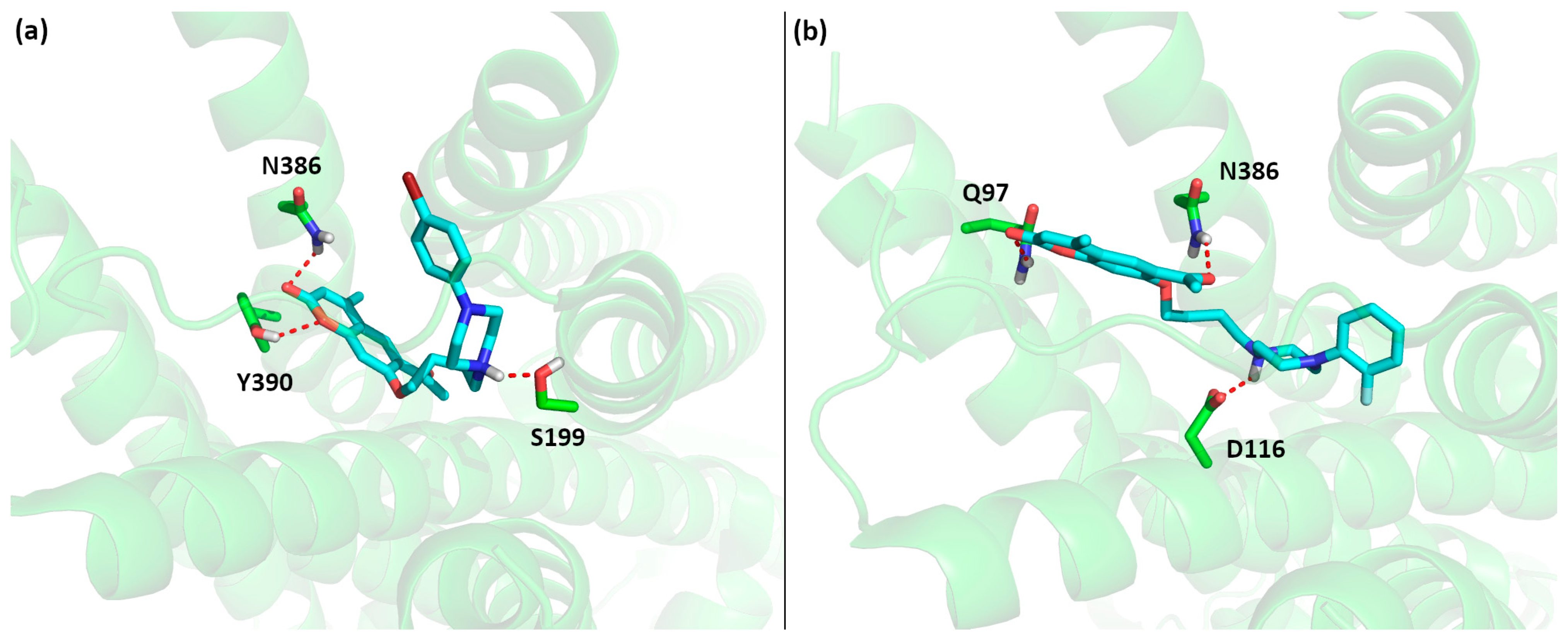
Based on the local search results for the 5-HT1A receptor combined with the experimental results presented in the next section, we can identify critical structural features in this family of ligands responsible for the high affinity of compounds 1–10 and low affinity of compounds 11–14 (see Figure 4). Clearly, the 6-acetyl-7-hydroxy-4-methylcoumarin scaffold fits well in the part of the 5-HT1A receptor binding site located close to transmembrane helices 2 and 7 (as in the case of aripiprazole), but making an additional strong hydrogen bond between the acetyl moiety and Q97. The middle part of the ligand anchors the ligand in the binding site due to a strong salt bridge to D116. The varied affinity results come from the variable part of the phenylpiperazine moiety. Here, small substituents in all possible positions (ortho, meta and para) of the phenyl ring likely do not form any additional interactions, as this part of the binding site (helices 3, 5 and 6) is mostly hydrophobic. On the other hand the presence of two substituents (10–11) lowers the affinity, likely due to the too large size of the entire ligand, causing steric hindrances inside the binding pocket. A similar cause of lowering the affinity to the 5-HT1A receptor can be attributed to compound 12 with a larger –NO2 substituent in the para position; on the other hand, compound 13 is likely to bind weakly due to the missing favorable interactions between the missing phenyl moiety and the hydrophobic pocket of helices 3, 5 and 6. In the case of compounds 12–14, the low affinity may also be an effect of electron-withdrawing properties of substituents, which diminish the basic character of the anchoring nitrogen atom of the piperazine.
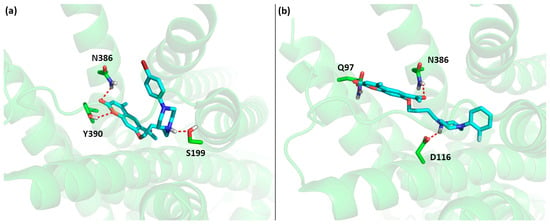
Figure 4.
Predicted pose of (a) 7 and (b) experimental pose aripiprazole inside the binding pocket of the 5-HT1A receptor, highlighting the most important interactions between ligands and the receptor (salt bridge between the ligand and D116 and hydrogen bond between the ligand and Q97). Oxygen atoms are shown in red, nitrogen in blue, carbon in cyan or green and hydrogen in light grey.
Finally, we performed analogous calculations to estimate affinities for 5-HT2A receptor affinity; see Table 4. Here, the computational results are in disagreement with the experimental data, as molecular docking predicts very high affinities, but the experimental values are rather low. The computational estimate of Ki of aripiprazole (0.85 nM) is very close to the experimental value of 3.4–3.5 nM [21,22,23]; thus, one could expect similar values for the studied compounds due to the structural similarities to aripiprazole. This is clearly not the case, and for now, we do not have a good explanation for this discrepancy between computational estimates and experimental values, although the most likely cause is the overestimation of the non-covalent interactions by our docking approach. This is a common problem in all docking protocols, as most available software is rather accurate in predicting correct ligand poses but fail in accurate estimation of the binding affinities [24,25].

Table 4.
Affinities of compounds 1–15 for 5-HT1A and 5-HT2A receptor.
2.4. Biological Evaluation
All arylpiperazinyl derivatives of 6-acetyl-7-hydroxy-4-methylchromen-2-one described in this study (1–15) were tested for their affinity for the 5-HT1A and 5-HT2A receptors, as shown in Table 4. The results show that some of the synthesized systems have affinities in the nanomolar range toward 5-HT1A and the low micromolar range toward 5-HT2A receptors.
As in our previous studies, we clearly see the influence of different substituents on the affinities of coumarin derivatives [9,10]. Compounds 3–7 with (2-bromophenyl)piperazinyl, (3-bromophenyl)piperazinyl, (4-bromophenyl)piperazinyl, (2-fluorophenyl)piperazinyl and (2-chlorophenyl)piperazinyl moieties showed high affinities for the 5-HT1A receptors. Among these derivatives, compounds 6-acetyl-7-{4-[4-(3-bromophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (4) and 6-acetyl-7-{4-[4-(2-chlorophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (7) displayed the highest affinities for the 5-HT1A receptor, with Ki values of 0.78 (0.4–1.4) nM and 0.57 (0.2–1.3) nM, respectively, nearby to the Ki values of 8-OH-DPAT (0.25 (0.097–0.66) nM). The introduction of the chloro substituents in the ortho position, or bromo substituents in the meta position of the phenyl ring increases affinities for the 5-HT1A receptor. The replacement of the chlorine group in the 2-postion of the phenyl ring with the bromine or fluorine moieties displayed a decrease in affinities for the 5-HT1A receptor form Ki = 0.57 nM to Ki = 1.96 nM or 1.04 nM for compounds 7, 3 and 6, respectively. Replacing the 2-position halogen with a methoxy group results in similar affinities. The situation is analogous when the bromine in position 3 of the phenyl ring of piperazine is replaced with a methoxy group, lowering the affinity from Ki = 0.78 nM for compound 4 to Ki = 12.9 nM for compound 2. This relation is interesting due to the fact that for derivatives containing an acetyl group in the 8-position of the coumarin ring and a methoxy group in the 2- or 3-position of the piperazine phenyl ring, we have previously demonstrated excellent affinity for the 5-HT1A receptor [9,10]. Changing the position of the acetyl group from 8 to 6 in the coumarin ring lowers the affinity of the derivatives with the methoxy substituent from Ki = 1.0 nM to Ki = 5.75 nM for the 2-position of the methoxy moiety and from Ki = 0.8 nM to Ki = 12.9 for the 3-position of methoxy moiety. The affinity for the 5-HT1A receptor for derivatives containing bromo, fluoro or chloro substituents in the ortho position of the phenyl ring of the piperazine is analogous for both 6-acetyl and 8-acetyl coumarin. In the case of (3-bromophenyl)piperazine, 5-HT1A receptor affinity increases by threefold for 6-acetylcoumarin compared to 8-acetylcoumarin (Ki = 2.5 nM to Ki = 0.78 nM). In the case of (2-fluorophenyl)piperazine, the affinity remains similar regardless of the position of the acetyl group in the coumarin ring (Ki = 1 nM for 8-acetylcoumarin and Ki = 1.04 nM for 6-acetylcoumarin). Surprisingly, we also obtained high affinity for compound 5 with bromine in the para position of the piperazine phenyl ring (Ki = 1.40 nM). An introduction of the nitro-substituents in the para position or the replacement of the phenyl ring with a heterocyclic moiety produced a decrease in 5-HT1A receptor affinity (Ki = 1658–9617 nM), in line with the trend described in our previous works.
For the 5-HT1A receptor, we also determined the agonist or antagonist properties of all of the new compounds, as shown in Table 5. Our results confirmed the 5-HT1A antagonistic properties for derivatives 1, 3, 5, and 11–14. The strongest antagonist in this group was 6-acetyl-7-{4-[4-(2-methoxyphenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (1) with an IC50 value of 301 nM, which is twenty times higher than for WAY 100636, the antagonist of the 5-HT1A receptor used as a reference substance. Curiously, the 6-acetyl-7-{4-[4-(3-methoxyphenyl)piperazin-1-yl] butoxy}-4-methylchromen-2-one (2) derivative, which differs only in the position of the methoxy group in the phenyl ring of piperazine, showed agonistic activity at the 5-HT1A receptor, although it was almost five times weaker than 8-OH-DPAT used as the reference compound (EC50 = 181 nM for 2 and EC50 = 38 for 8-OH-DPAT). The agonistic profile was also observed for compounds 4, 6–10 and 15. 6-Acetyl-7-{4-[4-(2-chlorophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (7), which showed the highest affinity for the 5-HT1A receptor, also turned out to be the strongest agonist among the group of compounds tested with an EC50 = 49 nM. Furthermore, we verified the relationship between the Ki and IC50/EC50 values for compounds 1–15 and found that there is a positive moderate correlation for both quantities (correlation coefficients R2 = 0.58 for Ki/IC50 and R2 = 0.66 for Ki/EC50, respectively). This result shows that Ki is a good preliminary indicator of the biological activity of these compounds, but functional tests are necessary to verify the functional nature of this activity.

Table 5.
Functional data for compounds 1–15.
All compounds studied showed low, micromolar affinities for the 5-HT2A receptor. Compounds bearing the (3,5-dimethylphenyl) and (2-cyanophenyl)piperazinyl piperazinyl moieties (10 and 8, respectively) showed the highest affinities in the whole series (Ki values of 516 nM or 705 nM, respectively). However, their affinities for the 5-HT2A receptor were more than two orders of magnitude weaker than ketanserin, which served as the reference compound (Ki value of 1.33 nM). In both cases of 10 and 8, the position of the acetyl group in the coumarin ring had a significant influence on the action at the 5-HT2A receptor, as the Ki values of analogous compounds with the acetyl group in position 8 of the coumarin ring were respectively found to be much lower at 58 and 91 nM, respectively [10,11].
3. Materials and Methods
3.1. Chemistry
All starting materials were purchased from Aldrich or Merck and were used without further purification. The Plazmatronika 1000 microwave oven was used (http://www.plazmatronika.com.pl (accessed on 27 December 2020). The melting points were determined with ElectroThermal 9001 Digital Melting Point apparatus and are uncorrected. High resolution mass spectra were recorded on a Micromsass LCT (ESI-TOF). 1H NMR, 13C NMR spectra in solution were recorded at 25 °C with a Varian NMRS-300 spectrometer, and standard Varian VnmrJ 2.1B software was employed. The calculated shielding constants were used as an aid in assigning resonances of 13C atoms. Chemical shifts δ (ppm) were referenced to TMS. TLC was carried out using Kieselgel 60 F254 sheets, eluent: CHCl3: MeOH; 10:0.25 and spots were visualized by UV e 254 and 365 nm.
Compounds A and 1–15 were prepared in accordance with the previously reported procedures [9]. Atom numbering, 1H NMR and 13C NMR spectra of all synthesized compounds are available in the ESI.
- 6-Acetyl-7-(4-bromobutoxy)-4-methylchromen-2-one (A)
M.p.: 114 °C, Rf = 0.32, yield 81%, 1H NMR (300 MHz, CHCl3) δ ppm: 8.07 (s, 1H, H-5), 6.86 (s, 1H, H-8), 6.20 (s, 1H, H-3), 4.19 (t, J = 6.0 Hz, 2H, H-1′), 3.52 (t, J = 6.0 Hz, 2H, H-4′), 2.67 (s, 3H, H-11), 2.45 (s, 3H, H-9), 2.12 (q, 4H, H-2′, H-3′); 13C NMR (75 MHz, CHCl3) δ ppm: 197.7 (C-10), 161.0 (C-7), 160.5 (C-2), 157.7 (C-8a), 152.9 (C-4), 128.1 (C-6), 124.9 (C-5), 113.6 (C-3), 112.9 (C-4a), 100.5 (C-8), 68.6 (C-1′), 33.1 (C-4′), 32.3 (C-3′), 29.5 (C-2′), 27.8 (C-11), 18.9 (C-9); TOF MS ES+: [M + Na]+ calcd for C16H17O4NaBr: 375.0208 found 375.0196.
- 6-Acetyl-7-{4-[4-(2-methoxyphenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (1)
M.p.: 126–127 °C, Rf = 0.38, yield 95%, 1H NMR (300 MHz, CDCl3) δ ppm: 8.08 (s, 1H, H-5), 6.98 (m, 5H, H-8, H-3”-H-6”), 6.19 (s, 1H, H-3), 4.19 (t, J = 6.0 Hz, 2H, H-1′), 3.86 (s, 3H, H-7”), 3.13 (br. s, 4H, H-3p, H-5p), 2.69 (br. s, 7H, H-11, H-2p, H-6p), 2.54 (t, J = 6.0 Hz, 2H, H-4′), 2.45 (s, 3H, H-9), 2.00 (m, 2H, H-3′), 1,81 (m, 2H, H-2′); 13C NMR (75 MHz, CDCL3) δ ppm: 197.9 (C-10), 161.3 (C-6”), 160.6 (C-7), 157.7 (C-2), 153.0 (C-8a), 152.4 (C-4), 141.3 (C-2”), 128.2 (C-6), 125.0 (C-5), 123.2 (C-6”), 121.2 (C-4”), 118.4 (C-5”), 113.5 (C-3”), 112.9 (C-4a), 111.3 (C-3), 100.6 (C-8), 69.5 (C-1′), 58.3 (C-3p, C-5p), 55.6 (C-4′), 53.6 (C-7”), 50.8 (C-2p, C-6p), 32.4 (C-2′), 29.9 (C-11), 23.6 (C-3′), 18.9 (C-9); TOF MS ES+: [M + H]+ calcd for C27H33O5N2: 465.2389 found 465.2401
- 6-Acetyl-7-{4-[4-(3-methoxyphenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (2)
M.p.: 83–85 °C, Rf = 0.27, yield 34%, 1H NMR (300 MHz, CDCl3) δ ppm: 8.07 (s, 1H, H-5), 7.20 (t, J = 7.5 Hz, 1H, H-5”), 6.87 (s, 1H, H-8), 6.56 (d, J = 9.0 Hz, 1H, H-4”), 6.47 (m, 2H, H-2”, H-6”), 6.20 (s, 1H, H-3), 4.19 (t, J = 6.0 Hz, 2H, H-1′), 3.81 (s, 3H, H-7”), 3.31 (br., s, 4H, H-3p, H-5p), 2.74 (br., s, 4H, H-2p, H-6p), 2.68 (s, 3H, H-11), 2.62 (br., s, 2H, H-4”), 2.45 (s, 3H, H-9), 2.01 (m, 2H, H-2′), 1.87 (br., s, 2H, H-3′); 13C NMR (75 MHz, CDCl3) δ ppm: 197.8 (C-10), 161.1 (C-7), 160.8 (C-2, C-3”), 160.6 (C-8a), 157.7 (C-4), 152.9 (C-1”), 130.2 (C-5”), 128.1 (C-6), 125.1 (C-5), 113.6 (C-4a), 113.1 (C-3), 109.4 (C-4”), 105.4 (C-6”), 103.1 (C-8), 100.6 (C-2”), 69.2 (C-1′), 57.9 (C-3p, C-5p), 55.3 (C-2p, C-6p), 52.9 (C-4′), 48.5 (C-2′), 48.3 (C-7”), 32.3 (C-3′), 26.9 (C-11), 18.9 (C-9); TOF MS ES+: [M + H]+ calcd for C27H33O5N2: 465.2389 found 465.2381.
- 6-Acetyl-7-{4-[4-(2-bromophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (3)
M.p.: 121–122 °C, Rf = 0.15, yield 79%, 1H NMR (300 MHz, CDCl3) δ ppm: 8.08 (s, 1H, H-5), 7.57 (dd, J1 = 12.0 Hz, J2 = 3.0 Hz, 1H, H-6”), 7.33 (s, 1H, H-8), 7.12 (d, J = 9.0 Hz, 1H, H-3”), 6.96 (br.t, J = 9.0 Hz, 1H, H-4”), 6.88 (s, 1H, H-5”), 6.21 (s, 1H, H-3), 4.21 (t, J = 4.5 Hz, 2H, H-1′), 3.20 (br. s, 4H, H-3p, H-5p), 2.69 (s, 3H, H-11), 2.46 (s, 3H, H-9), 2.03 (br. s, 2H, H-4′), 1.59 (br.s, 6H, H-2p, H-6p, H-2′), 1.27 (br.s., 2H, H-3′); 13C NMR (75 MHz, CDCl3) δ ppm: 197.7 (C-10), 160.8 (C-7, C-2), 160.5 (C-4), 157.7 (C-8a), 152.9 (C-1”), 134.0 (C-3”), 128.8 (C-6, C-5), 128.1 (C-4), 125.1 (C-5”), 121.6 (C-6”), 120.0 (C-2”), 113.7 (C-3), 113.1 (C-4a), 100.6 (C-8), 68.8 (C-1′), 57.6 (C-4′), 53.0 (C-3p, C-5p), 52.9 (C-2p, C-6p), 32.3 (C-2′), 29.9 (C-3′), 26.8 (C-11), 18.9 (C-9); TOF MS ES+: [M + Na]+ calcd for C26H29BrO4N2Na: 535.1208 found 535.1195.
- 6-Acetyl-7-{4-[4-(3-bromophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (4)
M.p.: 109–110 °C, Rf = 0.16, yield 82%, 1H NMR (300 MHz, CDCl3) δ ppm: 8.05 (s, 1H, H-5), 7.10 (t, J = 9.0 Hz, 1H, H-5”), 7.03 (m, 1H, H-6”), 6.96 (m, 1H, H-4”), 6.81 (m, 2H, H-8, H-2”), 6.17 (s, 1H, H-3), 4.17 (t, J = 6.0 Hz, 2H, H-1′), 3.23 (t, J = 4.5 Hz, 4H, H-3p, H-5p), 2.64 (m, 7H, H-11, H-2p, H-6p), 2.53 (t, J = 7.5 Hz, 2H, H-4′), 2.42 (s, 3H, H-9), 1.99 (m, 2H, H-2′), 1.78 (m, 2H, H-3′); 13C NMR (75 MHz, CDCl3) δ ppm: 197.8 (C-10), 161.2 (C-7), 160.5 (C-2), 157.7 (C-4), 152.9 (C-8a), 152.3 (C-1”), 130.5 (C-5”), 128.1 (C-6), 125.0 (C-5), 123.4 (C-3”), 122.6 (C-4”), 118.9 (C-2”, C-6”), 114.6 (C-4a), 113.5 (C-3), 100.5 (C-8), 69.3 (C-1′), 57.9 (C-4′), 53.0 (C-3p, C-5p), 48.5 (C-2p, C-6p), 32.3 (C-2′), 27.0 (C-3′), 23.3 (C-11), 18.9 (C-9); TOF MS ES+: [M + Na]+ calcd for C26H29BrO4N2Na: 535.1208 found 535.1197.
- 6-Acetyl-7-{4-[4-(4-bromophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (5)
M.p.: 145–146 °C, Rf = 0.26, yield 44%, 1H NMR (300 MHz, CDCl3) δ ppm: 8.07 (s, 1H, H-5), 7.35 (d, J = 9.0 Hz, 2H, H-3”, H-5”), 6.87 (s, 1H, H-8), 6.82 (d, J = 9.0 Hz, 2H, H-2”, H-6”), 6.20 (s, 1H, H-3), 4.19 (t, J = 6.0 Hz, 2H, H-1′), 3.25 (br. s, 4H, H-3p, H-5p), 6.28 (br. s, 6H, H-2p, H-6p, H-4′), 2.45 (s, 3H, H-9), 2.02 (m, 2H, H-2′), 1.96 (m, 2H, H-3′); 13C NMR (75 MHz, CDCL3) δ ppm: 197,8 (C-10), 161,1 (C-7, C-2), 160.5 (C-8a), 157.7 (C-4), 152.9 (C-1”), 132.1 (C-3”, C-5”), 128.1 (C-6), 125.0 (C-5), 118.0 (C-4”), 113.5 (C-6”, C-2”), 113.0 (C-4a, C-3), 100.6 (C-8), 69.3 (C-1′), 58.0 (C-3p, C-5p), 53.0 (C-4′), 48.7 (C-2p, C-6p), 32.3 (C-2′, C-3′), 27.0 (C-11), 18.9 (C-9); TOF MS ES+: [M + H]+ calcd for C26H30BrO4N2: 513.1389 found 513.1398.
- 6-Acetyl-7-{4-[4-(2-fluorophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (6)
M.p.: 112–113 °C, Rf = 0.32, yield 95%, 1H NMR (300 MHz, CDCl3) δ ppm: 8.07 (s, 1H, H-5), 7.01 (m, 4H, H-3”, H-4”, H-5”, H-6”), 6.87 (s, 1H, H-8), 6.20 (s, 1H, H-3), 4.20 (t, J = 6.0 Hz, 2H, H-1′), 3.23 (br. s, 4H, H-3p, H-5p), 2.81 (br. s, 4H, H-2p, H-6p), 2.68 (s, 3H, H-11), 2.64 (br. s. 2H, H-4′), 2.45 (s, 3H, H-9), 2.03 (m, 2H, H-2′), 1.90 (m, 2H, H-3′); 13C NMR (75 MHz, CDCL3) δ ppm: 197.8 (C-10), 161.0 (C-3”), 160.6 (C-7), 157.7 (C-2), 157.5 (C-8a), 154.2 (C-4), 152.9 (C-1”), 128.1 (C-5”), 125.1 (C-6). 124.8 (C-5), 123.5 (C-4a), 119.4 (C-3), 116.5 (C-4”), 116.3 (C-6”), 113.6, 113.1 (C-8), 100.6 (C-2”), 69.1 (C-1′), 57.8 (C-4”), 53.0 (C-2p, C-6p), 49.2 (C-3p, C-5p), 32.3 (C-2′), 29.9 (C-3′), 26.9 (C-11), 18.9 (C-9); TOF MS ES+: [M + H]+ calcd for C26H30FO4N2: 453.2190 found 453.2198.
- 6-Acetyl-7-{4-[4-(2-chlorophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (7)
M.p.: 132–133 °C, Rf = 0.19, yield 64%,1H NMR (300 MHz, CDCl3) δ ppm: 8.07 (s, 1H, H-5), 7.37 (d, J = 9.0 Hz, 1H, H-6”), 7.27 (d, J = 9.0 Hz, 1H, H-3”), 7.03 (m, 2H, H-4”, H-5”),6.87 (s, 1H, H-8), 6.19 (s, 1H, H-3), 4.20 (t, J = 6.0 Hz, 2H, H-1′), 3.18 (br. s, 4H, H-3p, H-5p), 2.78 (br. s, 4H, H-2p, H-6p), 2.69 (s, 3H, H-11), 2.64 (br. s, 2H, H-4′), 2.01 (m, 2H, H-2′), 1.86 (m, 2H, H-3′); 13C NMR (75 MHz, CDCl3) δ ppm: 197.8 (C-10), 161.1 (C-7), 160.6 (C-2), 157.7 (C-8a), 152.9 (C-5, C-1”), 130.8 (C-3”), 128.9 (C-2”), 128.2 (C-5”), 127.9 (C-5), 125.1 (C-6), 124.3 (C-4”), 120.7 (C-6”), 113.6 (C-3), 113.0 (C-4a), 100.6 (C-8), 69.2 (C-1′), 58.0 (C-3p, C-5p), 53.4 (C-2p, C-6p), 50.5 (C-4′), 32.3 (C-2′), 27.0 (C-3′), 22.9 (C-11), 18.9 (C-9); TOF MS ES+: [M + H]+ calcd for C26H30 ClO4N2: 469.1894 found 469.1898.
- 6-Acetyl-7-{4-[4-(2-cyanophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (8)
M.p.: 117–118 °C, Rf = 0.22, yield 31%, 1H NMR (300 MHz, CDCl3) δ ppm: 8.05 (s, 1H, H-5), 7.57 (m, 2H, H-3”, H-5”), 7.10 (m, 2H, H-6”, H-4”), 6.85 (s, 1H, H-8), 6.20 (s, 1H, H-3), 4.20 (t, J = 4.5 Hz, 2H, H-1′), 3.47 (br. s, 10H, H-2p, H-3p, H-5p, H-6p, H-4′), 2.67 (s, 3H, H-11), 2.44 (s, 3H, H-9), 2.04 (br., s, 2H, H-2′), 1.61 (br., s, 2H, H-3′); 13C NMR (75 MHz, CDCl3) δ ppm: 197.7 (C-10), 160.5 (C-2, C-7), 157.7 (C-4, C-8a), 152.9 (C-1”), 134.4 (C-3”, C-4”), 128.1 (C-5, C-6), 125.1 (C-4”, C-6”, C-7”), 113.7 (C-3), 113.2 (C-4a), 100.6 (C-2”, C-8), 68.8 (C-1′), 57.5 (C-4′, C-2p, C-6p), 52.8 (C-3p, C-5p), 32.2 (C-2′), 29.9 (C-3′), 26.7 (C-9), 18.9 (C-11); TOF MS ES+: [M + H]+ calcd for C27H30O4N3: 460.2236 found 460.2233.
- 6-Acetyl-7-{4-[4-(2,3-dichlorophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (9)
M.p.: 111–112 °C, Rf = 0.37, yield 57%,1H NMR (300 MHz, CDCl3) δ ppm: 8.07 (s, 1H, H-5), 7.17 (m, 2H, H-6”, H-4”), 6.98 (m, 1H, H-5”), 6.87 (s, 1H, H-8), 6.19 (s, 1H, H-3), 4.20 (t, J = 7.5 Hz, 2H, H-1′), 3.13 (br. s, 4H, H-3p, H-5p), 2.72 (br.s, 4H, H-2p, H-6p), 2.68 (s, 3H, H-11), 2.58 (m, 2H, H-4′), 2.45 (s, 3H, H-9), 2.00 (m, 2H, H-2′), 1.83 (m, 2H, H-3′); 13C NMR (75 MHz, CDCl3) δ ppm: 197.8 (C-10), 161.2 (C-2, C-7), 160.6 (C-8a), 157.7 (C-4), 152.9 (C-2”), 134.2 (C-3”), 128.1 (C-5”), 127.8 (C-1”), 125.2 (C-5), 125.0 (C-6), 118.9 (C-4”), 113.5 (C-6”), 112.9 (C-3, C-4a), 100.6 (C-8), 69.2 (C-1′), 57.9 (C-4′), 53.3 (C-3p, C-5p), 50.7 (C-2p, C-6p), 32.3 (C-2′), 26.9 (C-3′), 23.0 (C-11), 18.9 (C-9); TOF MS ES+: [M + Na]+ calcd for C26H28Cl2O4N2Na: 525.1324 found 525.1313.
- 6-Acetyl-7-{4-[4-(3,5-dimethylphenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (10)
M.p.: 101–102 °C, Rf = 0.20, yield 67%,1H NMR (300 MHz, CDCl3) δ ppm: 8.07 (s, 1H, H-5), 6.86 (s, 1H, H-5), 6.58 (m, 3H, H-2”, H-4”, H-6”), 6.19 (s, 1H, H-3), 4.19 (t, J = 6.0 Hz, 2H, H-1′), 3.26 (br. s, 4H, H-3p, H-5p), 2.71 (br.s, 4H, H-2p, H-6p), 2.68 (br. s, 3H, H-11), 2.58 (m, 2H, H-4′), 2.45 (s, 3H, H-9), 2.29 (s, 6H, H-7”, H-8”), 2.00 (m, 2H, H-2′), 1.86 (m, 2H, H-3′); 13C NMR (75 MHz, CDCl3) δ ppm: 197.7 (C-10), 161.2 (C-7), 160.5 (C-2), 157.7 (C-8a), 152.9 (C-4), 151.3 (C-1”), 138.7 (C-3”, C-5”), 128.1 (C-5), 124.9 (C-6), 121.9 (C-4”), 114.2 (C-2”, C-6”), 113.3 (C-4a), 112.8 (C-3), 100.5 (C-8), 69.3 (C-1′), 58.1 (C-4′), 53.4 (C-3p, C-5p), 49.2 (C-2p, C-6p), 32.3 (C-2′), 29.8 (C-3′), 27.1 (C-11), 23.4 (C-7”), 21.8 (C-8”), 18.8 (C-9); TOF MS ES+: [M + H]+ calcd for C28H35O4N2: 436.2597 found 436.2604.
- 6-Acetyl-7-{4-[4-(2,5-dimethylphenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (11)
M.p.: 130–131 °C, Rf = 0.28, yield 49%, 1H NMR (300 MHz, CDCl3) δ ppm: 8.07 (s, 1H, H-5), 7.08 (d, J = 6.0 Hz, 1H, H-3”), 6.87 (s, 1H, H-8), 6.83 (d, J= 9.0 Hz, 2H, H-4”, H-6”), 6.20 (s, 1H, H-3), 4.20 (t, J= 6.0 Hz, 2H, H-1′), 3.07 (br. s, 4H, H-3p, H-5p), 2.69 (s, 3H, H-11), 2.45 (s, 3H, H-9), 2.32 (s, 3H, H-7”), 2.26 (s, 3H, H-8”), 2.02 (m, 2H, H-2′), 1.92 (m, 2H, H-3′); 13C NMR (75 MHz, CDCL3) δ ppm: 197.8 (C-10), 160.9 (C-7), 160.5 (C-2), 157.7 (C-8a), 152.9 (C-4), 136.6 (C-1”), 131.1 (C-5”), 129.4 (C-2”), 128.1 (C-3”), 125.1 (C-5), 124.8 (C-6), 120.4 (C-4”), 113.6 (C-6”), 113.1 (C-3, C-4a), 100.6 (C-8), 69.0 (C-1′), 57.8 (C-4′), 53.5 (C-3p, C-5p), 50.4 (C-2p, C-6p), 32.3 (C-2′), 29.9 (C-11), 26.9 (C-3′), 22.4 (C-7”), 18.9 (C-9), 17.5 (C-8”); TOF MS ES+: [M + H]+ calcd for C28H35O4N2: 463.2597 found 463.2599.
- 6-Acetyl-7-{4-[4-(4-nitrophenyl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (12)
M.p.: 138–139 °C, Rf = 0.44, yield 47%, 1H NMR (300 MHz, CDCl3) δ ppm: 8.14 (d, J = 9.0 Hz, 2H, H-3”, H-5”), 8.07 (s, 1H, H-5), 6.87 (s, 1H, H-8), 6.84 (d, J= 9.0 Hz, 2H, H-2”, H-6”), 6.20 (s, 1H, H-3), 4.20 (t, J = 6.0 Hz, 2H, H-1′), 3.47 (br. s, 4H, H-3p, H-5p), 2.68 (s, 3H, H-11), 2.62 (m, 6H, H-4′,H-2p, H-6p), 2.01 (m, 2H, H-2′), 1.79 (m, 2H, H-3′); 13C NMR (75 MHz, CDCL3) δ ppm: 197.9 (C-10), 160.6 (C-7, C-2), 157.7 (C-8a), 152.9 (C-1”), 152.9 (C-4), 128.1 (C-4”), 126.2 (C-5, C-6), 125.1 (C-3”, C-5”), 113.6 (C-3, C-4a), 113.1 (C-2”, C-6”), 100.6 (C-8), 69.2 (C-1′), 57.9 (C-4′), 52.7 (C-3p, C-5p), 46.9 (C-2p, C-6p), 32.4 (C-2′), 32.3 (C-3′), 26.9 (C-11), 18.9 (C-9); TOF MS ES+: [M + Na]+ calcd for C26H29O6N3Na: 502.1954 found 502.1942.
- 6-Acetyl-7-[4-(morpholin-4-yl)butoxy]-4-methylchromen-2-one (13)
M.p.: 115–116 °C, Rf = 0.29, yield 69%, 1H NMR (300 MHz, CDCl3) δ ppm: 8.07 (s, 1H, H-5), 6.86 (s, 1H, H-8), 6.19 (s, 1H, H-8), 4.17 (t, J = 7.5 Hz, 2H, H-1′), 3.79 (br. s, 4H, H-3m, H-5m), 2.67 (s, 3H, H-11), 2.54 (br. s, 6H, H-4′, H-2m, H-6m), 2.45 (s, 3H, H-9), 1.98 (m, 2H, H-2′), 1.79 (m, 2H, H-3′); 13C NMR (75 MHz, CDCL3) δ ppm:197.8 (C-10), 161.2 (C-7), 160.6 (C-2), 157.7 (C-8a), 152.9 (C-4), 128.1 (C-6), 125.1 (C-5), 113.6 (C-4a), 113.0 (C-3), 100.6 (C-8), 69.3 (C-1′), 66.5 (C-3m, C-5m), 58.4 (C-2m, C-6m), 53.6 (C-4′), 32.3 (C-2′), 26.9 (C-3′), 22.8 (C-11), 18.9 (C-9); TOF MS ES+: [M + Na]+ calcd for C20H25NO5Na: 382.1630 found 382.1618.
- 6-Acetyl-7-{4-(4-piridin)piperazin-1-yl}butoxy}-4-methylchromen-2-one (14)
M.p.: 107–108 °C, Rf = 0.19, yield 63%, 1H NMR (300 MHz, CDCl3) δ ppm: 8.21 (br. s, 2H, H-3”, H-5”), 8.07 (s, 1H, H-5), 6.88 (s, 1H, H-8), 6.84 (br. d, J = 9.0 Hz, 2H, H-2”, H-6”), 6.20 (s, 1H, H-3), 4.20 (t, J = 6.0 Hz, 2H, H-1′), 3.59 (t, J= 4,5 Hz, 4H, H-3p, H-5p), 2.68 (s, 3H, H-11), 2.64 (t, J = 6 Hz, 4H, H-2p, H-6p), 2.53 (t, J = 7,5 Hz, 2H, H-4′), 2.46 (s, 1H, H-9), 2.00 (m, 2H, H-2′), 1.77 (m, 2H, H-3′); 13C NMR (75 MHz, CDCL3) δ ppm:197.9 (C-10), 161.2 (C-7), 160.6 (C-2), 157.8 (C-8a), 156.5 (C-4), 153.0 (C-1”), 143.1 (C-3”, C-5”), 128.2 (C-6), 125.1 (C-5), 113.6 (C-3), 113.1 (C-4a), 107.6 (C-2”, C-6”), 100.6 (C-8), 69.3 (C-1′), 57.8 (C-3p, C-5p), 52.5 (C-2p, C-6p), 46.3 (C-4′), 32.3 (C-2′), 29.3 (C-3′), 23.5 (C-11), 18.9 (C-9); TOF MS ES+: [M + H]+ calcd for C25H30O4N3: 436.2236 found 435.2247.
- 6-Acetyl-7-{4-[4-(pyrazin-2-yl)piperazin-1-yl]butoxy}-4-methylchromen-2-one (15)
M.p.: 150–151 °C, Rf = 0.35, yield 69%,1H NMR (300 MHz, CDCl3) δ ppm: 8.16 (s, 1H, H-6”), 8.08 (s, 2H, H-3”, H-4”), 7.87 (s, 1H, H-5), 6.86 (s, 1H, H-8), 6.18 (s, 1H, H-3), 4.19 (t, J = 6.0 Hz, 2H, H-1′), 3.66 (br. s, 4H, H-3p, H-5p), 2.67 (s, 3H, H-11), 2.63 (br. s, 4H, H-2p, H-6p), 2.55 (br., s, 2H, H-4′), 2.00 (m, 2H, H-2′), 1.81 (m, 2H, H-3′); 13C NMR (75 MHz, CDCl3) δ ppm: 197.8 (C-10), 161.3 (C-7), 160.6 (C-2), 157.4 (C-8a), 155.1 (C-1”), 153.0 (C-4), 141.9 (C-3”), 133.2 (C-4”), 131.2 (C-6”), 128.2 (C-6), 125.0 (C-5), 113.5 (C-4a), 112.9 (C-3), 100.5 (C-8), 69.4 (C-1′), 58.2 (C-4′), 52.9 (C-3p, C-5p), 44.6 (C-2p, C-6p), 32.3 (C-2′), 27.0 (C-3′), 23.5 (C-11), 18.9 (C-9); TOF MS ES+: [M + Na]+ calcd for C24H28O4N4Na: 459.2008 found 459.2018.
3.2. X-ray Crystallography
The X-ray measurement of 2 was performed at 130.0(5) K on a Bruker D8 Venture PhotonII diffractometer equipped with a TRIUMPH monochromator and a MoKα fine focus sealed tube (λ = 0.71073 Å). A total of 2690 frames were collected with the Bruker APEX3 program [26]. The frames were integrated with the Bruker SAINT, V8.40A software package [27] using a narrow-frame algorithm. Integration of the data using a monoclinic unit cell yielded a total of 62369 reflections to a maximum θ angle of 28.50° (0.74 Å resolution), of which 5948 were independent (average redundancy 10.486, completeness = 99.9%, Rint = 2.78%, Rsig = 1.41%) and 5240 (88.10%) were greater than 2σ(F2). The final cell constants of a = 24.3110(12) Å, b = 12.0631(6) Å, c = 8.0498(4) Å, β = 96.000(2)°, V = 2347.8(2) Å3 are based upon the refinement of the XYZ-centroids of 5239 reflections above 20 σ(I) with 4.042° < 2θ < 60.45°. Data were corrected for absorption effects using the Multi-Scan method (SADABS) [28]. The ratio of minimum to maximum apparent transmission was 0.943. The calculated minimum and maximum transmission coefficients (based on crystal size) are 0.947 and 0.991.
The structure was solved and refined using the SHELXTL Software Package [29,30] using the space group P21/c, with Z = 4 for the formula unit, C27H32N2O5. The final anisotropic full-matrix least-squares refinement on F2 with 311 variables converged at R1 = 3.68% for the observed data and wR2 = 10.58% for all data. The goodness of fit was 1.031. The largest peak in the final difference electron density synthesis was 0.355 e−/Å3, and the largest hole was −0.217 e−/Å3 with an RMS deviation of 0.042 e−/Å3. Based on the final model, the calculated density was 1.314 g/cm3 and F(000), 992 e−. The details concerning the crystal data and structural parameters of 2 are collected in Table 1.
All non-hydrogen atoms were refined anisotropically. All hydrogen atoms were placed in calculated positions and refined within the riding model. The temperature factors of the hydrogen atoms were not refined and were set at 1.2 (Car-H, CH2 groups) or 1.5 (CH3 group) times higher than the Ueq of the corresponding heavy atom. The atomic scattering factors were taken from the International Tables [31]. Molecular graphics was prepared using the program Mercury 2020.2.0 [32].
CCDC 2213451 contains the supplementary crystallographic data for this study. The data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/structures.
3.3. Biological Evaluation
3.3.1. Membrane Preparation
Sprague–Dawley rats were sacrificed by isoflurane overdose. Brains were rapidly removed and placed on ice. Hippocampi (for 5-HT1A assays) and frontal cortices (for 5-HT2A assays) were dissected on a Petri dish. The tissue from 10 rats was homogenized in 30 vol. homogenization buffer (50 mM Tris-HCl, pH = 4.7, 1 mM EDTA, 1 mM dithiothreitol) with a hand-held Teflon-glass homogenizer. The homogenate was centrifuged at 48,000× g at 4 °C for 15 min. The pellet was suspended and homogenized in homogenization buffer and incubated for 10 min at 36 °C. The centrifugation and suspension steps were repeated twice. The final pellet was homogenized in 5 vol. 50 mM Tris-HCl, pH = 7.4 buffer and stored at −80 °C for not more than 6 months.
3.3.2. Competitive 5-HT1A and 5-HT2A Binding Assays
For the 5-HT1A assay, ten concentrations equally spaced on a logarithmic scale (10−14M−10−5M) of each compound were incubated in duplicate with 1 nM [3H]8-OH-DPAT (specific activity: 200 Ci/mmol, Perkin Elmer, Waltham, MA, USA) for 60 min at 36 °C in a 50 mM Tris-HCl buffer (pH 7.4), supplemented with 0.1% ascorbate, 5 mM MgCl2 and 80 µg of rat hippocampal membrane suspension. For the 5-HT2A assay, 160 µg of rat frontal cortex membrane suspension was incubated with 1 nM [3H]ketanserin (specific activity: 22.8 Ci/mmol, Perkin Elmer, Waltham, MA, USA) for 60 min at 36 °C in a 50 mM Tris-HCl (pH 7.4) buffer, supplemented with 0.1% ascorbate and 3 mM CaCl2. Non-specific binding was determined with 10 μM serotonin in both assays. The final DMSO concentration in the assay was 5%. After incubation, the reaction mixture was deposited with the FilterMate-96 Harvester (Perkin Elmer, Waltham, MA, USA) onto Unifilter® GF/C plates (Perkin Elmer, Waltham, MA, USA) presoaked in 0.4% PEI for 1h. Each well was washed with 2 mL of 50 mM Tris-HCl (pH 7.4) buffer to separate bound ligands from free ones. Plates were left to dry overnight. Then, 35 µL of Microscint-20 scintillation fluid (Perkin Elmer, Waltham, MA, USA) was added to each filter well and left to equilibrate for 2 h. Filter-bound radioactivity was counted in a MicroBeta2 LumiJet scintillation counter (Perkin Elmer, Waltham, MA, USA). Binding curves were fitted with one-site non-linear regression. Binding affinity (pKi ± SEM and Ki ± 95% confidence intervals) for each compound was calculated from EC50 values with the Cheng-Prusoff equation from two separate experiments.
3.3.3. 5-HT1A Receptor Activation in the [35S]GTP-γ-S Assay
Ten compound concentrations equally spaced on a log scale (10−4.5 M to 10−9 M) were incubated in duplicate with rat hippocampal membrane preparations (5 μg per well) in assay buffer (50 mM Tris-HCl, pH = 7.4, 1 mM EGTA, 3 mM MgCl2, 100 mM NaCl and 30 µM GDP) and 0.08 nM [35S]GTPγS (specific activity: 1250 Ci/mmole, Perkin Elmer, Waltham, MA, USA). Non-specific binding was determined with 100 µM of unlabeled GTPγS. The compounds were tested in both the agonist and antagonist mode. In the antagonist mode, 10−6.8 M of 8-OH-DPAT was used as a stimulating ligand. The final DMSO concentration in the assay was 5%. The reaction mixture was incubated for 90 min at 30 °C on an orbital shaker set at 250 rpm. The reaction mixture was then deposited under vacuum with the FilterMate Harvester® (Perkin Elmer, Waltham, MA, USA) onto Unifilter® GF/C Plates (Perkin Elmer, Waltham, MA, USA) presoaked with wash buffer (50 mM Tris-HCl, pH = 7.4). The wells were then rapidly washed with 2 mL of wash buffer. The filter plates were dried overnight at room temperature. Once completely dry, 35 µL of MicroScint PS (Perkin Elmer, Waltham, MA, USA) scintillation fluid was added to each well. Radioactivity was counted in a MicroBeta2 LumiJet scintillation counter (Perkin Elmer, Waltham, MA, USA). Data were analyzed with GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA, USA, www.graphpad.com, accessed on 4 April 2012). The curves were fitted with the three-parameter non-linear regression model. Potency (EC50 or IC50 ± 95% confidence intervals) and efficacy (Emax ± SEM) were calculated and expressed as means from two separate experiments.
3.4. Computational Methods
In the computational part of this study, we used a protocol similar to our previous investigation on this topic [10,11,14] but based on recently obtained crystal structures of both 5-HT1A and 5-HT2A receptors. In the case of the 5-HT1A receptor, we selected three crystal structures: apo-5-HT1A (PDB id: 7e2x), serotonin-bound 5-HT1A (PDB id: 7e2y) and aripiprazole-bound 5-HT1A (PDB id: 7e2z), all complexed to a G protein [33]. In the case of the 5-HT2A receptor, we selected two crystal structures: 5-HT2A in complex with serotonin (PDB id: 7wc4) and 5-HT2A in complex with aripiprazole (PDB id: 7voe) [34,35]. The choice of these particular structures was made on the basis of a very high similarity of compounds studies in this work to aripiprazole. We used two different docking protocols for all investigated coumarin derivatives. First, we manually superimposed all studied coumarin derivatives onto the aripiprazole poses from crystal structures of 5-HT1A (7e2z) and 5-HT2A (7voe) and performed a local search procedure using standard Autodock 4.2 parameters and 1000 independent hybrid genetic algorithm local search runs [36]. Second, we performed standard flexible docking with the Lamarckian genetic algorithm and 200 runs for each ligand–receptor pair for each of the five GPCR crystal structures. In the case of the 5-HT1A receptor, the following residues are described in a flexible manner: Y96, Q97, F112, D116, T121, S199, F361, N386, and Y390, while for 5-HT2A receptor flexible residues were: W151, D155, V156, F243, F332, W336, F339, F340, N363, and V366. In each case, we used 60 × 60 × 60 Å3 boxes centered on binding pockets of studied receptors. Additionally, we performed computational assessment of ADME properties using the QikProp 4.6 software and evaluated pKa values of basic nitrogen-containing functional groups using Epik 5.3 software [37].
4. Conclusions
Our studies on determining the influence of the acetyl group position in the coumarin ring on the affinity for the 5-HT1A and 5-HT2A receptors allowed us to draw clear and interesting conclusions regarding the structure–activity relationship for the new subfamily of coumarin derivatives selectively targeting the 5-HT1A receptor. Previously published compounds containing an acetyl group in position C-8 of the coumarin ring showed, in general, greater affinities for both 5-HT receptor types. On the other hand, some newly synthesized 6-acetyl-7-hydroxy-4-methylcoumarins showed subnanomolar 5-HT1A receptor affinity and potent antagonistic or agonistic properties. We previously showed that in a very similar subfamily of 8-acetyl-7-hydroxy-4-methylcoumarins, where most of the compounds studied were, analogs of the ligands described in this study showed antagonistic properties [9]. Moreover, very small structural changes, e.g., between compounds 1 and 2, may result in different functional properties of ligands despite similar affinities. Finally, we showed that molecular docking to the recently solved crystal structures of 5-HT receptors could be a good preliminary indicator for estimating 5-HT1A receptor affinity, but fails in the accurate estimation of 5-HT2A affinity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24032779/s1.
Author Contributions
Conceptualization, K.O.; methodology, K.O., A.L. and B.T.; computational modeling, B.T.; formal analysis, K.O. and W.G.; resources, K.O. and B.T.; data curation, K.O., B.T. and A.L.; X-ray data, Ł.D.; writing—original draft preparation, K.O., B.T. and A.L.; writing—review and editing, M.B.-Z. and K.O.; supervision, K.O. and M.B.-Z.; funding acquisition, K.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Medical University of Warsaw, Faculty of Pharmacy, project FW24/2/F/GW/N/20.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
The authors thank Jorge Esteban Fuentes from Alcala University, Spain, for his support during the synthesis of materials. X-ray structure was determined in the Advanced Crystal Engineering Laboratory (aceLAB) at the Chemistry Department of the University of Warsaw. The radioligand binding assays were performed with the use of the Center for Preclinical Research and Technology (CePT) infrastructure.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Corvino, A.; Fiorino, F.; Severino, B.; Saccone, I.; Frecentese, F.; Perissutti, E.; Di Vaio, P.; Santagada, V.; Caliendo, G.; Magli, E. The Role of 5-HT1A Receptor in Cancer as a New Opportunity in Medicinal Chemistry. Curr. Med. Chem. 2018, 25, 3214–3227. [Google Scholar] [CrossRef]
- Nichols, D.E.; Nichols, C.D. Serotonin Receptors. Chem. Rev. 2008, 108, 1614–1641. [Google Scholar] [CrossRef] [PubMed]
- Amidfar, M.; Colic, L.; Walter, M.; Kim, Y.K. Biomarkers of Major Depression Related to Serotonin Receptors. Curr. Psychiatry Rev. 2018, 14, 239–244. [Google Scholar] [CrossRef]
- Rojas, P.; Fiedler, J.L. What Do We Really Know About 5-HT1A Receptor Signaling in Neuronal Cells? Front. Cell. Neurosci. 2016, 10, 272. [Google Scholar] [CrossRef]
- Asarch, K.B.; Ransom, R.W.; Shih, J.C. 5-HT-la and 5-HT-lb selectivity of two phenylpiperazine derivatives: Evidence for 5-HT-lb heterogeneity. Life Sci. 1985, 36, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Maj, J.; Chojnacka-Wójcik, E.; Kłodzińska, A.; Dereń, A.; Moryl, E. Hypothermia induced by m-trifluoromethylphenylpiperazine or m-chlorophenylpiperazine: An effect mediated by 5-HT1B receptors? J. Neural Transm. 1988, 73, 43–55. [Google Scholar] [CrossRef]
- Sylte, I.; Chilmończyk, Z.; Dahl, S.G.; Cybulski, J.; Edvardsen, O. The Ligand-binding Site of Buspirone Analogues at the 5-HT1A Receptor. J. Pharm. Pharmacol. 1997, 49, 698–705. [Google Scholar] [CrossRef]
- Ostrowska, K. Coumarin-piperazine derivatives as biologically active compounds. Saudi Pharm. J. 2020, 28, 220–232. [Google Scholar] [CrossRef]
- Ostrowska, K.; Młodzikowska, K.; Głuch-Lutwin, M.; Gryboś, A.; Siwek, A. Synthesis of a new series of aryl/heteroarylpiperazinyl derivatives of 8-acetyl-7-hydroxy-4-methylcoumarin with low nanomolar 5-HT1A affinities. Eur. J. Med. Chem. 2017, 137, 108–116. [Google Scholar] [CrossRef]
- Ostrowska, K.; Grzeszczuk, D.; Głuch-Lutwin, M.; Gryboś, A.; Siwek, A.; Leśniak, A.; Sacharczuk, M.; Trzaskowski, B. 5-HT1A and 5-HT2A receptors affinity, docking studies and pharmacological evaluation of a series of 8-acetyl-7-hydroxy-4-methylcoumarin derivatives. Bioorg. Med. Chem. 2018, 26, 527–535. [Google Scholar] [CrossRef]
- Ostrowska, K.; Grzeszczuk, D.; Głuch-Lutwin, M.; Gryboś, A.; Siwek, A.; Dobrzycki, Ł.; Trzaskowski, B. Development of selective agents targeting serotonin 5-HT1A receptors with subnanomolar activities based on a coumarin core. MedChemComm 2017, 8, 1690–1696. [Google Scholar] [CrossRef] [PubMed]
- Żołek, T.; Enyedy, E.A.; Ostrowska, K.; Posa, V.; Maciejewska, D. Drug likeness prediction of 5-hydroxy-substituted coumarins with high affinity for5-HT1A and 5-HT2A receptors. Eur. J. Pharm. Sci. 2018, 115, 25–36. [Google Scholar] [CrossRef]
- Żołek, T.; Domotor, O.; Ostrowska, K.; Enyedy, E.A.; Maciejewska, D. Evaluation of blood-brain barrier penetration and examination of binding to human serum albumin of 7-O-arylpiperazinylcoumarins as potential antipsychotic agents. Bioorg. Chem. 2019, 84, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Ostrowska, K.; Leśniak, A.; Karczyńska, U.; Jeleniewicz, P.; Głuch-Lutwin, M.; Mordyl, B.; Siwek, A.; Trzaskowski, B.; Sacharczuk, M.; Bujalska-Zadrozny, M. 6-Acetyl-5-hydroxy-4,7-dimethylcoumarin derivatives: Design, synthesis, modeling studies, 5-HT1A, 5-HT2A and D2 receptors affinity. Bioorg. Chem. 2020, 100, 103912. [Google Scholar] [CrossRef] [PubMed]
- Ostrowska, K.; Grzeszczuk, D.; Maciejewska, D.; Młynarczuk-Biały, I.; Czajkowska, A.; Sztokfisz, A.; Dobrzycki, L.; Kruszewska, H. Synthesis and biological screening of a new series of 5-[4-(4-aryl-1-piperazinyl)butoxy]coumarins. Monats. Chem. 2016, 147, 1615–1627. [Google Scholar] [CrossRef]
- Ostrowska, K.; Leśniak, A.; Czarnocka, Z.; Chmiel, J.; Bujalska-Zadrożny, M.; Trzaskowski, B. Design, Synthesis and Biological Evaluation of a Series of 5- and 7-Hydroxycoumarin Derivatives as 5-HT1A Serotonin Receptor Antagonists. Pharmaceuticals 2021, 14, 179. [Google Scholar] [CrossRef] [PubMed]
- Hejchman, E.; Kruszewska, H.; Maciejwska, D.; Sowirka-Taciak, B.; Tomczyk, M.; Sztokfisz-Ignasiak, A.; Jankowski, J.; Mlynarczuk-Biały, I. Design, synthesis, and biological activity of Schiff bases bearing salicyl and 7-hydroxycoumarinyl moieties. Monats. Chem. 2019, 150, 255–266. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Matsson, P.; Doak, B.C.; Over, B.; Kihlber, J. Cell permeability beyond the rule of 5. Adv. Drug. Deliver. Rev. 2016, 101, 42–61. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, H.; Yao, X.; Li, D.; Xu, L.; Li, Y.; Tian, S.; Hou, T. Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: Prediction accuracy of sampling power and scoring power. Phys. Chem. Chem. Phys. 2016, 18, 12964–12975. [Google Scholar] [CrossRef]
- Shapiro, D.A.; Renock, S.; Arrington, E.; Chiodo, L.A.; Liu, L.-X.; Sibley, D.R.; Roth, B.L.; Mailman, R. Aripiprazole, A Novel Atypical Antipsychotic Drug with a Unique and Robust Pharmacology. Neuropsychopharmacology 2003, 28, 1400–1411. [Google Scholar] [CrossRef] [PubMed]
- Keck, P.E., Jr.; McElroy, S.L. Aripiprazole: A partial dopamine D2 receptor agonist antipsychotic. Expert Opin. Investig. Drug. 2003, 12, 655–662. [Google Scholar]
- Kroeze, W.K.; Hufeisen, S.J.; Popadak, B.A.; Renock, S.M.; Steinberg, S.; Ernsberger, P.; Jayathilake, K.; Meltzer, H.Y.; Roth, B.L. H1-Histamine Receptor Affinity Predicts Short-Term Weight Gain for Typical and Atypical Antipsychotic Drugs. Neuropsychopharmacology 2003, 28, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Cross, J.B.; Thompson, D.C.; Rai, B.K.; Baber, J.C.; Fan, K.Y.; Hu, Y.; Humbler, C. Comparison of Several Molecular Docking Programs: Pose Prediction and Virtual Screening Accuracy. J. Chem. Inf. Model. 2009, 49, 1455–1474. [Google Scholar] [CrossRef] [PubMed]
- Pantsar, T.; Poso, A. Binding Affinity via Docking: Fact and Fiction. Molecules 2018, 23, 1899. [Google Scholar] [CrossRef]
- Bruker Nano Analitycs. APEX3 V2019; Bruker Nano, Inc.: Billerica, MA, USA, 2019. [Google Scholar]
- Bruker Nano Analitycs. SAINT V8.40A; Bruker Nano, Inc.: Billerica, MA, USA, 2019. [Google Scholar]
- Bruker Nano Analitycs. SADABS V2016/2; Bruker Nano, Inc.: Billerica, MA, USA, 2019. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Cryst. 2015, A71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Cryst. 2015, C71, 3–8. [Google Scholar]
- Wilson, A.J.C. (Ed.) International Tables for Crystallography; Kluwer: Dordrecht, The Netherlands, 1992; Volume C. [Google Scholar]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Cryst. 2020, 53, 226–235. [Google Scholar]
- Xu, P.; Huang, S.; Zhang, H.; Mao, C.; Zhou, X.E.; Cheng, X.; Simon, I.A.; Shen, D.-D.; Yen, H.-Y.; Robinson, C.V.; et al. Structural insights into the lipid and ligand regulation of serotonin receptors. Nature 2021, 592, 469–486. [Google Scholar] [CrossRef]
- Cao, D.; Yu, J.; Wang, H.; Luo, Z.; Liu, X.; He, L.; Qi, J.; Fan, L.; Tang, L.; Chen, Z.; et al. Structure-based discovery of nonhallucinogenic psychedelic analogs. Science 2022, 375, 403–411. [Google Scholar] [CrossRef]
- Chen, Z.; Fan, L.; Wang, H.; Yu, J.; Lu, D.; Qi, J.; Nie, F.; Luo, Z.; Liu, Z.; Cheng, J.; et al. Structure-based design of a novel third-generation antipsychotic drug lead with potential antidepressant properties. Nat. Neurosci. 2022, 25, 39–49. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pKa prediction and protonation state generation for drug-like molecules. J. Comput. Aided. Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).