Brain N-Glycosylation and Lipidomic Profile Changes Induced by a High-Fat Diet in Dyslipidemic Hamsters
Abstract
:1. Introduction
2. Results and Discussion
2.1. Body Composition, Serum Analyses, and Histological Evaluation
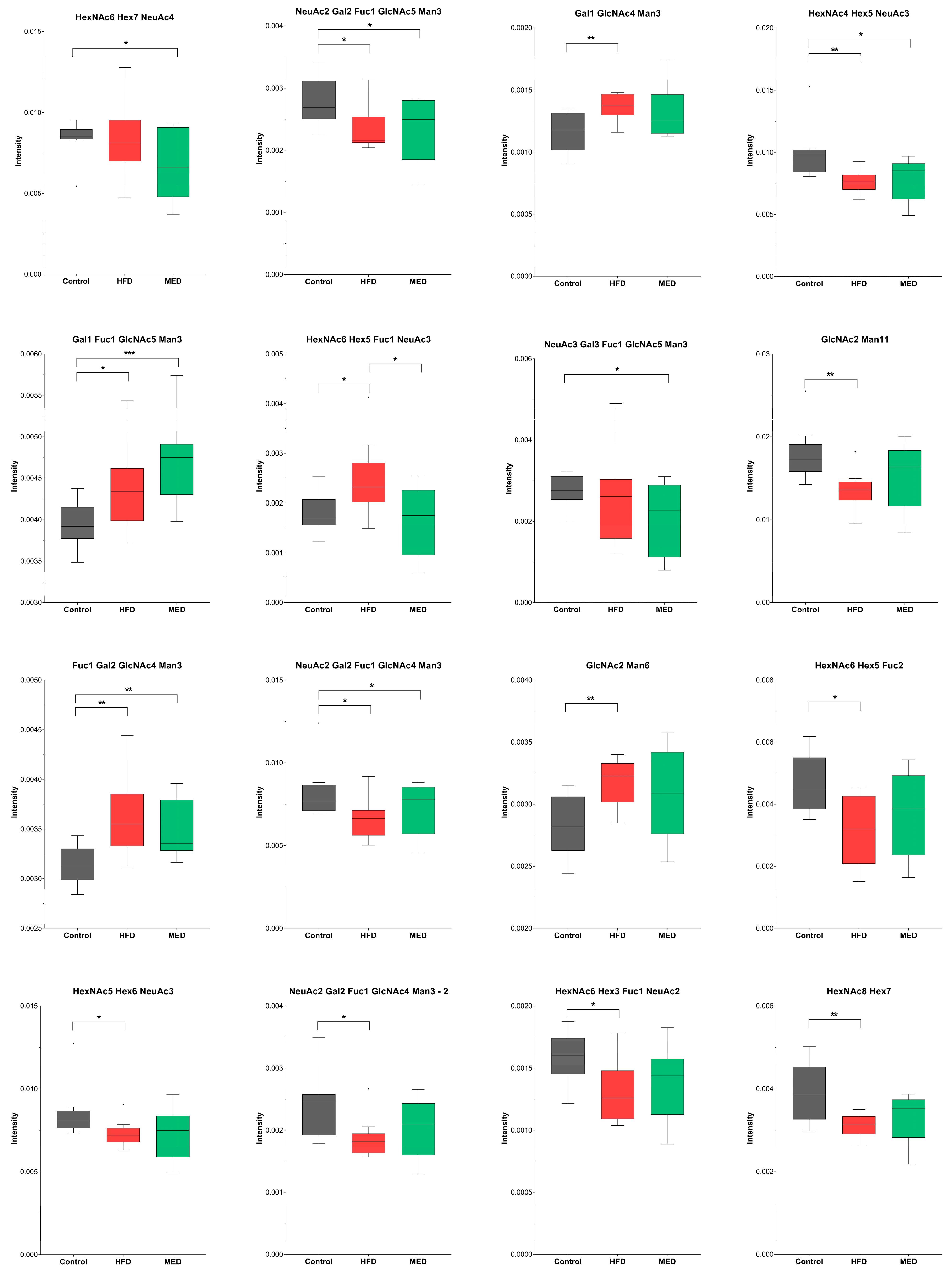
2.2. N-Glycosylation
2.3. Lipidomics
3. Materials and Methods
3.1. Reagents
3.2. Diets
3.3. Animals and Experimental Design
3.4. Histological Evaluation
3.5. Hepatic Lipid Extraction and Quantification
3.6. Body Composition Analyses
3.7. Serum Analysis
3.8. N-Glycan Analysis
3.8.1. Protein Extraction and Quantification
3.8.2. De-N-Glycosylation and Labelling of N-Glycans
3.8.3. LC-MS/MS
3.8.4. Data Processing of LC-MS Data
3.9. Lipidomics Analysis
3.9.1. Sample Preparation
3.9.2. LC-MS
3.9.3. Data Processing of LC-MS Data
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- D’innocenzo, S.; Biagi, C.; Lanari, M. Obesity and the Mediterranean Diet: A Review of Evidence of the Role and Sustainability of the Mediterranean Diet. Nutrients 2019, 11, 1306. [Google Scholar] [CrossRef] [PubMed]
- Afshin, A.; Sur, P.J.; Fay, K.A.; Cornaby, L.; Ferrara, G.; Salama, J.S.; Mullany, E.C.; Abate, K.H.; Abbafati, C.; Abebe, Z.; et al. Health effects of dietary risks in 195 countries, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2019, 393, 1958–1972. [Google Scholar] [CrossRef] [PubMed]
- Mu, M.; Xu, L.F.; Hu, D.; Wu, J.; Bai, M.J. Dietary Patterns and Overweight/Obesity: A Review Article. Iran. J. Public Health 2017, 46, 869. [Google Scholar]
- Thaler, J.P.; Yi, C.X.; Schur, E.A.; Guyenet, S.J.; Hwang, B.H.; Dietrich, M.O.; Zhao, X.; Sarruf, D.A.; Izgur, V.; Maravilla, K.R.; et al. Obesity is associated with hypothalamic injury in rodents and humans. J. Clin. Investig. 2012, 122, 153–162. [Google Scholar] [CrossRef]
- Karlsson, H.K.; Tuominen, L.; Tuulari, J.J.; Hirvonen, J.; Parkkola, R.; Helin, S.; Salminen, P.; Nuutila, P.; Nummenmaa, L. Obesity is associated with decreased µ-opioid but unaltered dopamine D2 receptor availability in the brain. J. Neurosci. 2015, 35, 3959–3965. [Google Scholar] [CrossRef] [PubMed]
- McLean, F.H.; Campbell, F.M.; Langston, R.F.; Sergi, D.; Resch, C.; Grant, C.; Morris, A.C.; Mayer, C.D.; Williams, L.M. A high-fat diet induces rapid changes in the mouse hypothalamic proteome. Nutr. Metab. 2019, 16, 26. [Google Scholar] [CrossRef]
- Freeman, L.R.; Haley-Zitlin, V.; Rosenberger, D.S.; Granholm, A.-C. Damaging effects of a high-fat diet to the brain and cognition: A review of proposed mechanisms. Nutr. Neurosci. 2014, 17, 241. [Google Scholar] [CrossRef]
- Sikalidis, A.K.; Maykish, A. The Gut Microbiome and Type 2 Diabetes Mellitus: Discussing a Complex Relationship. Biomedicines 2020, 8, 8. [Google Scholar] [CrossRef]
- Rutsch, A.; Kantsjö, J.B.; Ronchi, F. The Gut-Brain Axis: How Microbiota and Host Inflammasome Influence Brain Physiology and Pathology. Front. Immunol. 2020, 11, 3237. [Google Scholar] [CrossRef]
- Antoniazzi, L.; Arroyo-Olivares, R.; Bittencourt, M.S.; Tada, M.T.; Lima, I.; Jannes, C.E.; Krieger, J.E.; Pereira, A.C.; Quintana-Navarro, G.; Muñiz-Grijalvo, O.; et al. Adherence to a Mediterranean diet, dyslipidemia and inflammation in familial hypercholesterolemia. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 2014–2022. [Google Scholar] [CrossRef]
- Serra-Majem, L.; Roman, B.; Estruch, R. Scientific evidence of interventions using the Mediterranean diet: A systematic review. Nutr. Rev. 2006, 64, S27–S47. [Google Scholar] [CrossRef]
- Romagnolo, D.F.; Selmin, O.I. Mediterranean Diet and Prevention of Chronic Diseases. Nutr. Today 2017, 52, 208–222. [Google Scholar] [CrossRef] [PubMed]
- Rosato, V.; Temple, N.J.; La Vecchia, C.; Castellan, G.; Tavani, A.; Guercio, V. Mediterranean diet and cardiovascular disease: A systematic review and meta-analysis of observational studies. Eur. J. Nutr. 2019, 58, 173–191. [Google Scholar] [CrossRef]
- Huo, R.; Du, T.; Xu, Y.; Xu, W.; Chen, X.; Sun, K.; Yu, X. Effects of Mediterranean-style diet on glycemic control, weight loss and cardiovascular risk factors among type 2 diabetes individuals: A meta-analysis. Eur. J. Clin. Nutr. 2015, 69, 1200–1208. [Google Scholar] [CrossRef] [PubMed]
- Tangney, C.C.; Kwasny, M.J.; Li, H.; Wilson, R.S.; Evans, D.A.; Morris, M.C. Adherence to a Mediterranean-type dietary pattern and cognitive decline in a community population. Am. J. Clin. Nutr. 2011, 93, 601. [Google Scholar] [CrossRef]
- Gao, X.; Chen, H.; Fung, T.T.; Logroscino, G.; Schwarzschild, M.A.; Hu, F.B.; Ascherio, A. Prospective study of dietary pattern and risk of Parkinson disease. Am. J. Clin. Nutr. 2007, 86, 1486–1494. [Google Scholar] [CrossRef]
- Scarmeas, N.; Stern, Y.; Mayeux, R.; Luchsinger, J.A. Mediterranean diet, Alzheimer disease, and vascular mediation. Arch. Neurol. 2006, 63, 1709–1717. [Google Scholar] [CrossRef]
- Valls-Pedret, C.; Lamuela-Raventós, R.M.; Medina-Remón, A.; Quintana, M.; Corella, D.; Pintó, X.; Martínez-González, M.Á.; Estruch, R.; Ros, E. Polyphenol-rich foods in the Mediterranean diet are associated with better cognitive function in elderly subjects at high cardiovascular risk. J. Alzheimers Dis. 2012, 29, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.M.; Pereira, M.S.; Padrão, N.A.; Alves, I.; Marcos-Pinto, R.; Lago, P.; Pinho, S.S. Glycans as critical regulators of gut immunity in homeostasis and disease. Cell. Immunol. 2018, 333, 9–18. [Google Scholar] [CrossRef]
- Scott, H.; Panin, V.M. The role of protein N-glycosylation in neural transmission. Glycobiology 2014, 24, 407–417. [Google Scholar] [CrossRef]
- Ednie, A.R.; Bennett, E.S. Modulation of voltage-gated ion channels by sialylation. Compr. Physiol. 2012, 2, 1269–1301. [Google Scholar] [CrossRef]
- Tucholski, J.; Simmons, M.S.; Pinner, A.L.; Haroutunian, V.; McCullumsmith, R.E.; Meador-Woodruff, J.H. Abnormal N-linked glycosylation of cortical AMPA receptor subunits in schizophrenia. Schizophr. Res. 2013, 146, 177–183. [Google Scholar] [CrossRef]
- Lichnerova, K.; Kaniakova, M.; Park, S.P.; Skrenkova, K.; Wang, Y.X.; Petralia, R.S.; Suh, Y.H.; Horak, M. Two N-glycosylation sites in the GluN1 subunit are essential for releasing N-methyl-D-aspartate (NMDA) receptors from the endoplasmic reticulum. J. Biol. Chem. 2015, 290, 18379–18390. [Google Scholar] [CrossRef]
- Gu, W.; Fukuda, T.; Isaji, T.; Hang, Q.; Lee, H.H.; Sakai, S.; Morise, J.; Mitoma, J.; Higashi, H.; Taniguchi, N.; et al. Loss of α1,6-fucosyltransferase decreases hippocampal long term potentiation: Implications for core fucosylation in the regulation of AMPA receptor heteromerization and cellular signaling. J. Biol. Chem. 2015, 290, 17566–17575. [Google Scholar] [CrossRef] [PubMed]
- Inaba, H.; Kai, D.; Kida, S. N-glycosylation in the hippocampus is required for the consolidation and reconsolidation of contextual fear memory. Neurobiol. Learn. Mem. 2016, 135, 57–65. [Google Scholar] [CrossRef]
- Barboza, M.; Krueger, M.R.; Honeycutt, M.; Lebrilla, C.B.; Raybould, H. Multi-omics Studies Reveal Altered Hippocampal N-Glycosylation in High Fat Diet-Induced Obese Mice. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Castellanos, D.B.; Martín-Jiménez, C.A.; Rojas-Rodríguez, F.; Barreto, G.E.; González, J. Brain lipidomics as a rising field in neurodegenerative contexts: Perspectives with Machine Learning approaches. Front. Neuroendocrinol. 2021, 61, 100899. [Google Scholar] [CrossRef] [PubMed]
- Shamim, A.; Mahmood, T.; Ahsan, F.; Kumar, A.; Bagga, P. Lipids: An insight into the neurodegenerative disorders. Clin. Nutr. Exp. 2018, 20, 1–19. [Google Scholar] [CrossRef]
- Borg, M.L.; Omran, S.F.; Weir, J.; Meikle, P.J.; Watt, M.J. Consumption of a high-fat diet, but not regular endurance exercise training, regulates hypothalamic lipid accumulation in mice. J. Physiol. 2012, 590, 4377–4389. [Google Scholar] [CrossRef]
- Bravo, E.; Cantafora, A.; Calcabrini, A.; Ortu, G. Why prefer the golden Syrian hamster (Mesocricetus auratus) to the Wistar rat in experimental studies on plasma lipoprotein metabolism? Comp. Biochem. Physiol. Part B Comp. Biochem. 1994, 107, 347–355. [Google Scholar] [CrossRef]
- Yang, H.; Mayneris-Perxachs, J.; Boqué, N.; del Bas, J.M.; Arola, L.; Yuan, M.; Türkez, H.; Uhlén, M.; Borén, J.; Zhang, C.; et al. Combined Metabolic Activators Decrease Liver Steatosis by Activating Mitochondrial Metabolism in Hamsters Fed with a High-Fat Diet. Biomedicines 2021, 9, 1440. [Google Scholar] [CrossRef]
- Harvey, D.J.; Merry, A.H.; Royle, L.; Campbell, M.P.; Dwek, R.A.; Rudd, P.M. Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics 2009, 9, 3796–3801. [Google Scholar] [CrossRef]
- Lee, J.; Ha, S.; Kim, M.; Kim, S.-W.; Yun, J.; Ozcan, S.; Hwang, H.; Ji, I.J.; Yin, D.; Webster, M.J.; et al. Spatial and temporal diversity of glycome expression in mammalian brain. Proc. Natl. Acad. Sci. USA 2020, 117, 28743–28753. [Google Scholar] [CrossRef]
- Albach, C.; Klein, R.A.; Schmitz, B. Do Rodent and Human Brains Have Different N-Glycosylation Patterns? Biol. Chem. 2001, 382, 187–194. [Google Scholar] [CrossRef]
- Kleene, R.; Schachner, M. Glycans and neural cell interactions. Nat. Rev. Neurosci. 2004, 5, 195–208. [Google Scholar] [CrossRef]
- Edri-Brami, M.; Rosental, B.; Hayoun, D.; Welt, M.; Rosen, H.; Wirguin, I.; Nefussy, B.; Drory, V.E.; Porgador, A.; Lichtenstein, R.G. Glycans in sera of amyotrophic lateral sclerosis patients and their role in killing neuronal cells. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Lundström, S.L.; Yang, H.; Lyutvinskiy, Y.; Rutishauser, D.; Herukka, S.K.; Soininen, H.; Zubarev, R.A. Blood plasma IgG Fc glycans are significantly altered in Alzheimer’s disease and progressive mild cognitive impairment. J. Alzheimer’s Dis. 2014, 38, 567–579. [Google Scholar] [CrossRef]
- Váradi, C.; Nehéz, K.; Hornyák, O.; Viskolcz, B.; Bones, J. Serum N-Glycosylation in Parkinson’s Disease: A Novel Approach for Potential Alterations. Molecules 2019, 24, 2220. [Google Scholar] [CrossRef] [PubMed]
- Dotz, V.; Lemmers, R.F.H.; Reiding, K.R.; Hipgrave Ederveen, A.L.; Lieverse, A.G.; Mulder, M.T.; Sijbrands, E.J.G.; Wuhrer, M.; van Hoek, M. Plasma protein N-glycan signatures of type 2 diabetes. Biochim. Biophys. Acta-Gen. Subj. 2018, 1862, 2613–2622. [Google Scholar] [CrossRef]
- Lee, J.C.; Park, S.M.; Kim, I.Y.; Sung, H.; Seong, J.K.; Moon, M.H. High-fat diet-induced lipidome perturbations in the cortex, hippocampus, hypothalamus, and olfactory bulb of mice. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2018, 1863, 980–990. [Google Scholar] [CrossRef]
- Dahdah, N.; Gonzalez-franquesa, A.; Samino, S.; Gama-perez, P.; Herrero, L.; Perales, J.C.; Yanes, O.; Malagón, M.D.M.; Garcia-roves, P.M. Effects of lifestyle intervention in tissue-specific lipidomic profile of formerly obese mice. Int. J. Mol. Sci. 2021, 22, 3694. [Google Scholar] [CrossRef]
- van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta-Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- Suárez-García, S.; Caimari, A.; del Bas, J.M.; Suárez, M.; Arola, L. Serum lysophospholipid levels are altered in dyslipidemic hamsters. Sci. Rep. 2017, 7, 10431. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, J.H.; Noh, S.; Hur, H.J.; Sung, M.J.; Hwang, J.T.; Park, J.H.; Yang, H.J.; Kim, M.S.; Kwon, D.Y.; et al. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J. Proteome Res. 2011, 10, 722–731. [Google Scholar] [CrossRef]
- Kim, H.Y.; Kim, M.; Park, H.M.; Kim, J.; Kim, E.J.; Lee, C.H.; Yoon Park, J.H. Lysophospholipid profile in serum and liver by high-fat diet and tumor induction in obesity-resistant BALB/c mice. Nutrition 2014, 30, 1433–1441. [Google Scholar] [CrossRef]
- Estruch, R.; Ros, E.; Salas-Salvadó, J.; Covas, M.-I.; Corella, D.; Arós, F.; Gómez-Gracia, E.; Ruiz-Gutiérrez, V.; Fiol, M.; Lapetra, J.; et al. Primary Prevention of Cardiovascular Disease with a Mediterranean Diet Supplemented with Extra-Virgin Olive Oil or Nuts. N. Engl. J. Med. 2018, 378, e34. [Google Scholar] [CrossRef]
- Kris-Etherton, P.M.; Dietschy, J. Design criteria for studies examining individual fatty acid effects on cardiovascular disease risk factors: Human and animal studies. Am. J. Clin. Nutr. 1997, 65, 1590S–1596S. [Google Scholar] [CrossRef]
- Mast, N.; Shafaati, M.; Zaman, W.; Zheng, W.; Prusak, D.; Wood, T.; Ansari, G.A.S.; Lövgren-Sandblom, A.; Olin, M.; Bjorkhem, I.; et al. Marked variability in hepatic expression of cytochromes CYP7A1 and CYP27A1 as compared to cerebral CYP46A1. Lessons from a dietary study with omega 3 fatty acids in hamsters. Biochim. Biophys. Acta 2010, 1801, 674–681. [Google Scholar] [CrossRef]
- Laos, S.; Caimari, A.; Crescenti, A.; Lakkis, J.; Puiggròs, F.; Arola, L.; Del Bas, J.M. Long-term intake of soyabean phytosterols lowers serum TAG and NEFA concentrations, increases bile acid synthesis and protects against fatty liver development in dyslipidaemic hamsters. Br. J. Nutr. 2014, 112, 663–673. [Google Scholar] [CrossRef]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.-C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef]
- Apte, A.; Meitei, N.S. Bioinformatics in Glycomics: Glycan Characterization with Mass Spectrometric Data Using SimGlycan. Methods Mol. Biol. 2010, 600, 269–281. [Google Scholar] [CrossRef]
- Zhao, S.; Walsh, I.; Abrahams, J.L.; Royle, L.; Nguyen-Khuong, T.; Spencer, D.; Fernandes, D.L.; Packer, N.H.; Rudd, P.M.; Campbell, M.P. GlycoStore: A database of retention properties for glycan analysis. Bioinformatics 2018, 34, 3231–3232. [Google Scholar] [CrossRef]
- Tarancon-Diez, L.; Rodríguez-Gallego, E.; Rull, A.; Peraire, J.; Viladés, C.; Portilla, I.; Jimenez-Leon, M.R.; Alba, V.; Herrero, P.; Leal, M.; et al. Immunometabolism is a key factor for the persistent spontaneous elite control of HIV-1 infection. EBioMedicine 2019, 42, 86–96. [Google Scholar] [CrossRef]
- Tarancón-Diez, L.; Rull, A.; Herrero, P.; Vazquez-Alejo, E.; Peraire, J.; Guillén, S.; Navarro-Gomez, M.L.; Viladés, C.; Muñoz-Fernandez, M.Á.; Vidal, F. Early antiretroviral therapy initiation effect on metabolic profile in vertically HIV-1-infected children. J. Antimicrob. Chemother. 2021, 76, 2993–3001. [Google Scholar] [CrossRef]
- Samarra, I.; Masdevall, C.; Foguet-Romero, E.; Guirro, M.; Riu, M.; Herrero, P.; Canela, N.; Delpino-Rius, A. Analysis of oxylipins to differentiate between organic and conventional UHT milks. Food Chem. 2021, 343, 128477. [Google Scholar] [CrossRef]
- Peng, B.; Kopczynski, D.; Pratt, B.S.; Ejsing, C.S.; Burla, B.; Hermansson, M.; Benke, P.I.; Tan, S.H.; Chan, M.Y.; Torta, F.; et al. LipidCreator workbench to probe the lipidomic landscape. Nat. Commun. 2020, 11, 2057. [Google Scholar] [CrossRef]
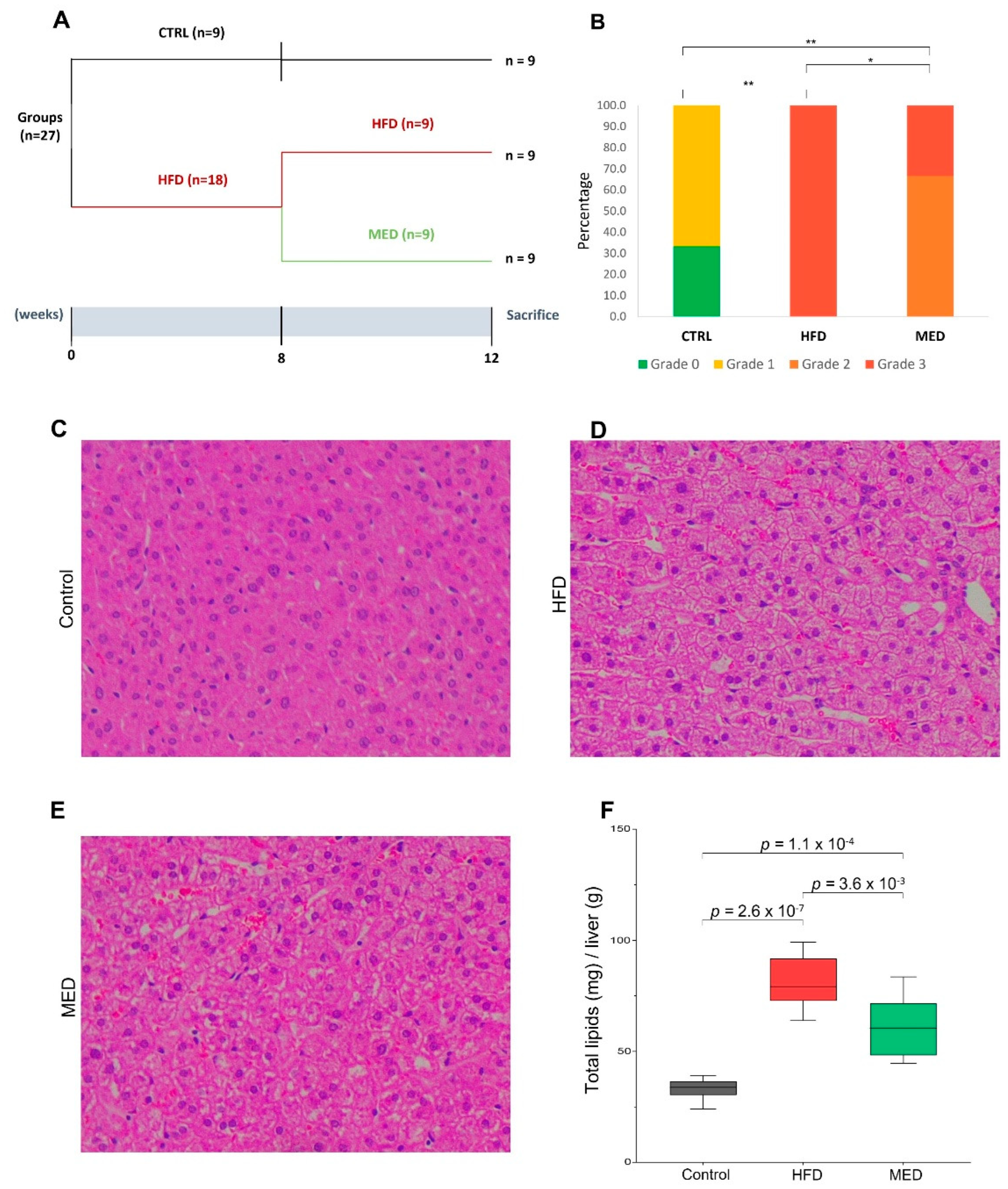


| Control (Week 8) (n = 9) | HFD (Week 8) (n = 18) | |
|---|---|---|
| Cumulative food intake (kcal) | 205.45 ± 14.08 | 198.81 ± 16.78 |
| Biometric variables | ||
| Body weight (g) | 123.05 ± 5.14 | 120.76 ± 7.08 |
| Fat mass (%) | 11.94 ± 3.20 | 12.37 ± 2.14 |
| Lean mass (%) | 84.89 ± 3.26 | 84.09 ± 2.12 |
| Lean/fat ratio | 7.83 ± 3.22 | 7.04 ± 1.51 |
| Serum parameters | ||
| CHOL (mM) * | 4.96 ± 0.66 | 8.30 ± 0.66 |
| Control (Week 12) (n = 9) | HFD (Week 12) (n = 9) | MED (Week 12) (n = 9) | |
|---|---|---|---|
| Cumulative food intake (kcal) a | 126.8 ± 14.4 | 118.7 ± 8.3 | 133.3 ± 9.9 |
| Biometric variables | |||
| Body weight (g) | 122.20 ± 5.62 | 119.60 ± 5.71 | 125.71 ± 10.71 |
| Liver weight (g) b,c | 4.23 ± 0.30 | 5.06 ± 0.43 | 5.43 ± 0.64 |
| Liver weight (%) b,c | 3.49 ± 0.25 | 4.27 ± 0.30 | 4.35 ± 0.25 |
| MWAT (%) b,c | 0.88 ± 0.24 | 1.13 ± 0.14 | 1.23 ± 0.22 |
| MUS | 0.27 ± 0.03 | 0.26 ± 0.03 | 0.26 ± 0.03 |
| Fat mass (%) | 11.28 ± 2.59 | 11.21 ± 2.12 | 13.34 ± 2.28 |
| Lean mass (%) | 85.42 ± 2.39 | 85.13 ± 2.17 | 83.32 ± 2.23 |
| Lean/fat mass ratio | 8.00 ± 2.13 | 7.88 ± 1.71 | 6.43 ± 1.21 |
| Serum variables | |||
| CHOL (mM) b,c | 3.33 ± 0.49 | 6.01 ± 0.51 | 5.47 ± 0.64 |
| HDL-C (mM) b,c | 2.51 ± 0.50 | 3.78 ± 0.38 | 3.57 ± 0.73 |
| LDL-C (mM) b,c | 0.94 ± 0.27 | 2.03 ± 0.38 | 1.97 ± 0.34 |
| TG (mM) c | 5.83 ± 1.19 | 7.10 ± 4.13 | 10.48 ± 3.09 |
| HFD vs. CTRL | |||||
|---|---|---|---|---|---|
| Glycan Name 1 | Composition 2 | Glycan Mass | m/z | p | FC |
| F(6)A2BG(4)2S(6,6)2 | NeuAc2 Gal2 Fuc1 GlcNAc5 Man3 | 2571.92 | 1442.5620 | 0.0126 | −1.20 |
| A2[6]G(4)1 | Gal1 GlcNAc4 Man3 | 1478.54 | 895.8720 | 0.0069 | 1.19 |
| - | HexNAc4 Hex5 NeuAc3 | 2513.87 | 1414.0499 | 0.0046 | −1.28 |
| F(6)A2[3]BG(4)1 | Gal1 Fuc1 GlcNAc5 Man3 | 1827.68 | 1070.4368 | 0.0426 | 1.10 |
| - | HexNAc6 Hex5 Fuc1 NeuAc3 | 3066.0951 | 1126.7695 | 0.0236 | 1.35 |
| M11 a3D1,[D2(1),D3(1)],a2D4(2) | GlcNAc2 Man11 | 2206.75 | 1259.9943 | 0.0036 | −1.32 |
| F(6)A2[6]G1Ga1 | Fuc1 Gal2 GlcNAc4 Man3 | 1786.65 | 1049.9261 | 0.0038 | 1.15 |
| F(6)A2G(4)2S(6,6)2 | NeuAc2 Gal2 Fuc1 GlcNAc4 Man3 | 2368.84 | 1341.0211 | 0.0266 | −1.23 |
| F(6)A2G(4)2S(3,3)2 | NeuAc2 Gal2 Fuc1 GlcNAc4 Man3 | 2368.84 | 1341.0211 | 0.0215 | −1.25 |
| M6 D1 | GlcNAc2 Man6 | 1396.49 | 854.8422 | 0.0050 | 1.12 |
| - | HexNAc6 Hex5 Fuc2 | 2338.8667 | 1325.9973 | 0.0103 | −1.54 |
| - | HexNAc5 Hex6 NeuAc3 | 2879.01 | 1064.7489 | 0.0366 | −1.16 |
| - | HexNAc6 Hex3 Fuc1 NeuAc2 | 2450.8940 | 1382.0477 | 0.0146 | −1.23 |
| - | HexNAc8 Hex7 | 2777.0153 | 1029.7350 | 0.0059 | −1.24 |
| MED vs. CTRL | |||||
| Glycan Name | Composition | Glycan Mass | m/z | p | FC |
| F(6)A2BG(4)2S(6,6)2 | NeuAc2 Gal2 Fuc1 GlcNAc5 Man3 | 2571.92 | 1442.5621 | 0.0478 | −1.22 |
| M7 | GlcNAc2 Man7 | 1558.54 | 935.8692 | 0.0310 | 1.15 |
| - | HexNAc4 Hex5 NeuAc3 | 2513.87 | 1414.0499 | 0.0233 | −1.29 |
| F(6)A2[3]BG(4)1 | Gal1 Fuc1 GlcNAc5 Man3 | 1827.68 | 1070.4368 | 0.0007 | 1.19 |
| F(6)A2G(4)2S(6,6)2 | NeuAc2 Gal2 Fuc1 GlcNAc4 Man3 | 2368.84 | 1341.0211 | 0.0300 | −1.26 |
| - | HexNAc6 Hex6 Fuc1 NeuAc3 | 3228.15 | 1180.7875 | 0.0226 | −1.41 |
| F(6)A3G(4)3S(3,3,3)3 | NeuAc3 Gal3 Fuc1 GlcNAc5 Man3 | 1915.69 | 1113.0953 | 0.0328 | −1.50 |
| - | HexNAc6 Hex7 NeuAc4 | 3535.24 | 1283.491 | 0.0495 | −1.30 |
| F(6)A2[6]G1Ga1 | Fuc1 Gal2 GlcNAc4 Man3 | 1786.65 | 1049.9261 | 0.0073 | 1.11 |
| MED vs. HFD | |||||
| Glycan Name | Composition | Glycan Mass | m/z | p | FC |
| - | HexNAc6 Hex5 Fuc1 NeuAc3 | 3066.0951 | 1126.7695 | 0.0285 | −1.59 |
| HFD vs. CTRL | |||
|---|---|---|---|
| Lipid Species | Compound | p | FC |
| Lysophosphatidylcholines | LPC 14:0 | 0.0476 | 1.34 |
| LPC 18:1 | 0.0447 | 1.34 | |
| Lysophosphatidylethanolamines | LPE 18:0 | 0.0307 | 1.36 |
| LPE 22:6 | 0.0239 | 1.43 | |
| Phosphatidylcholines | PC 17:0 | 0.0498 | 1.27 |
| PC 17:1 | 0.0285 | 1.30 | |
| PC 30:0 | 0.0386 | 1.28 | |
| PC 31:0 | 0.0255 | 1.31 | |
| PC 31:1 | 0.0113 | 1.37 | |
| PC 32:2 | 0.0436 | 1.29 | |
| PC 32:3 | 0.0259 | 1.33 | |
| PC 33:0 | 0.0382 | 1.29 | |
| PC 34:0 | 0.0335 | 1.28 | |
| PC 34:2 | 0.0057 | 1.36 | |
| PC 34:5 | 0.0354 | 1.35 | |
| PC 35:0 | 0.0181 | 1.39 | |
| PC 36:0 | 0.0239 | 1.30 | |
| PC 36:2 | 0.0428 | 1.26 | |
| PC 36:5 | 0.0078 | 1.32 | |
| PC 38:3 | 0.0408 | 1.29 | |
| PC 38:5 | 0.0312 | 1.41 | |
| PC 39:3 | 0.0491 | 1.27 | |
| PC 40:6 | 0.0295 | 1.27 | |
| PC 42:2 | 0.0416 | 1.85 | |
| PC 42:3 | 0.0401 | 1.42 | |
| PC 44:2 | 0.0167 | 1.44 | |
| Phosphatidylethanolamines | PE 30:1 | 0.0225 | 1.47 |
| PE 36:1 | 0.0185 | 1.37 | |
| PE 38:2 | 0.0355 | 1.39 | |
| PE 38:5 | 0.0302 | 1.33 | |
| PE 38:6 | 0.0393 | 1.22 | |
| PE 40:4 | 0.0458 | 1.32 | |
| PE 40:6 | 0.0300 | 1.26 | |
| PE 42:2 | 0.0259 | 1.30 | |
| PE 44:5 | 0.0265 | 1.23 | |
| Sterol esters | SE 27:1/18:1 | 0.0196 | 1.37 |
| SE 27:1/18:2 | 0.0013 | 1.65 | |
| SE 27:1/20:1 | 0.0008 | 1.81 | |
| SE 27:1/20:4 | 0.0027 | 1.60 | |
| SE 27:1/22:4 | 0.0339 | 1.57 | |
| SE 27:1/22:6 | 0.0401 | 1.49 | |
| Sphingomyelins | SM 32:2;2 | 0.0434 | 1.46 |
| SM 35:1;2 | 0.0418 | 1.33 | |
| SM 35:2;2 | 0.0282 | 1.31 | |
| SM 40:2;2 | 0.0283 | 1.43 | |
| Triglycerides | TAG 50:1 | 0.0497 | 1.32 |
| TAG 52:1 | 0.0091 | 1.38 | |
| TAG 56:5 | 0.0163 | 1.46 | |
| MED vs. CTRL | |||
| Lipid species | Compound | p | FC |
| Lysophosphatidylcholines | LPC 20:4 | 0.0195 | −1.53 |
| Lysophosphatidylethanolamines | LPE 20:4 | 0.0145 | −1.58 |
| Phosphatidylcholines | PC 34:2 | 0.0375 | 1.28 |
| PC 36:5 | 0.0351 | 1.27 | |
| Sterol esters | SE 27:1/16:1 | 0.0169 | −1.34 |
| SE 27:1/18:2 | 0.0399 | 1.39 | |
| SE 27:1/22:6 | 0.0010 | 2.03 | |
| MED vs. HFD | |||
| Lipid species | Compound | p | FC |
| Diacylglycerides | DAG 36:4 | 0.0494 | −1.37 |
| DAG 38:4 | 0.0396 | −1.40 | |
| Phosphatidylethanolamines | PE 30:1 | 0.0318 | −1.36 |
| PE 38:2 | 0.0186 | −1.38 | |
| Sterol esters | SE 27:1/18:3 | 0.0416 | −1.42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paton, B.; Foguet-Romero, E.; Suarez, M.; Mayneris-Perxachs, J.; Boqué, N.; Caimari, A.; Canela, N.; Herrero, P. Brain N-Glycosylation and Lipidomic Profile Changes Induced by a High-Fat Diet in Dyslipidemic Hamsters. Int. J. Mol. Sci. 2023, 24, 2883. https://doi.org/10.3390/ijms24032883
Paton B, Foguet-Romero E, Suarez M, Mayneris-Perxachs J, Boqué N, Caimari A, Canela N, Herrero P. Brain N-Glycosylation and Lipidomic Profile Changes Induced by a High-Fat Diet in Dyslipidemic Hamsters. International Journal of Molecular Sciences. 2023; 24(3):2883. https://doi.org/10.3390/ijms24032883
Chicago/Turabian StylePaton, Beatrix, Elisabet Foguet-Romero, Manuel Suarez, Jordi Mayneris-Perxachs, Noemí Boqué, Antoni Caimari, Núria Canela, and Pol Herrero. 2023. "Brain N-Glycosylation and Lipidomic Profile Changes Induced by a High-Fat Diet in Dyslipidemic Hamsters" International Journal of Molecular Sciences 24, no. 3: 2883. https://doi.org/10.3390/ijms24032883
APA StylePaton, B., Foguet-Romero, E., Suarez, M., Mayneris-Perxachs, J., Boqué, N., Caimari, A., Canela, N., & Herrero, P. (2023). Brain N-Glycosylation and Lipidomic Profile Changes Induced by a High-Fat Diet in Dyslipidemic Hamsters. International Journal of Molecular Sciences, 24(3), 2883. https://doi.org/10.3390/ijms24032883








