Molecular Analysis of MgO Nanoparticle-Induced Immunity against Fusarium Wilt in Tomato
Abstract
:1. Introduction
2. Results
2.1. RNA-seq Data Analysis
2.2. Identification of DEGs
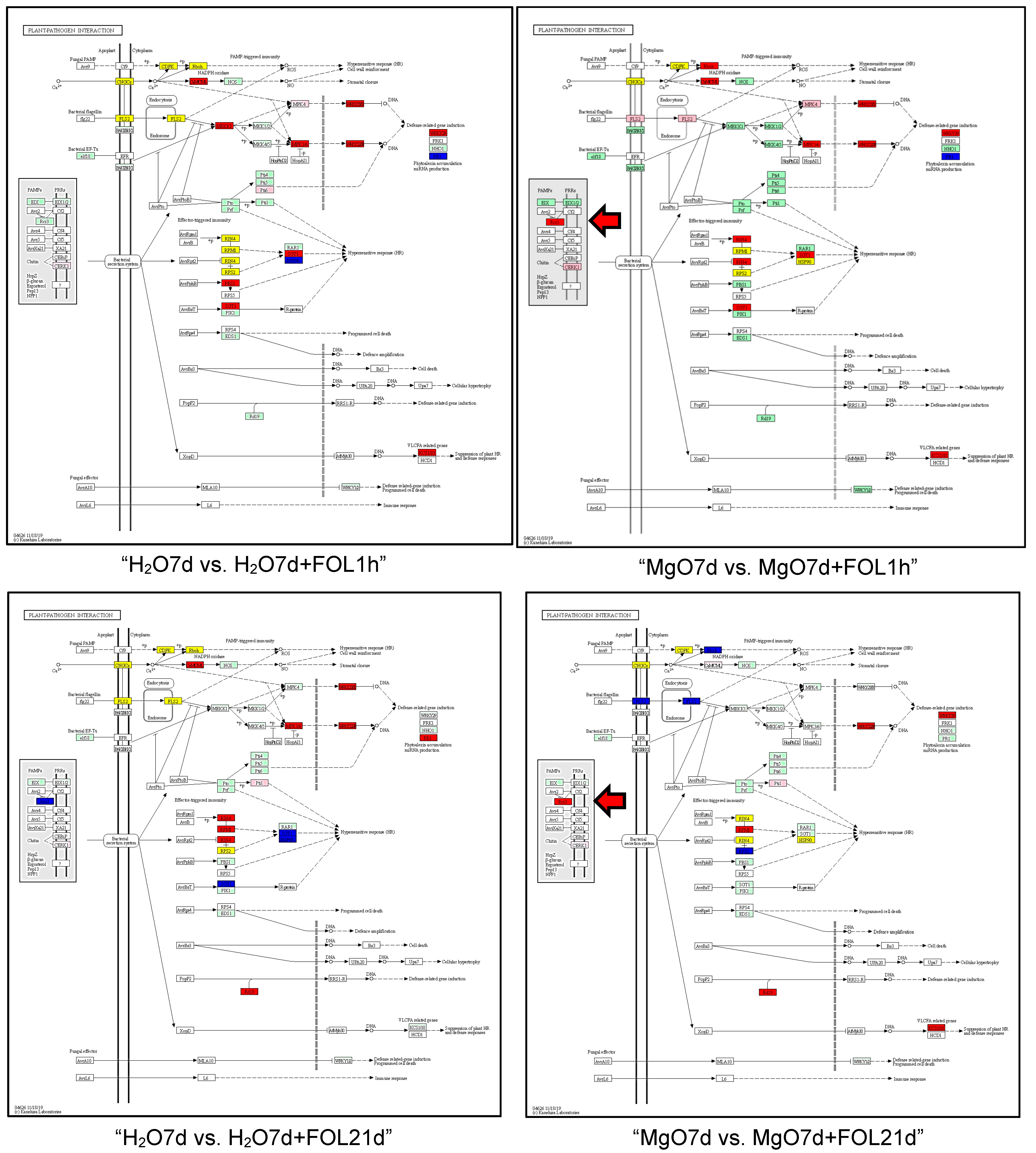
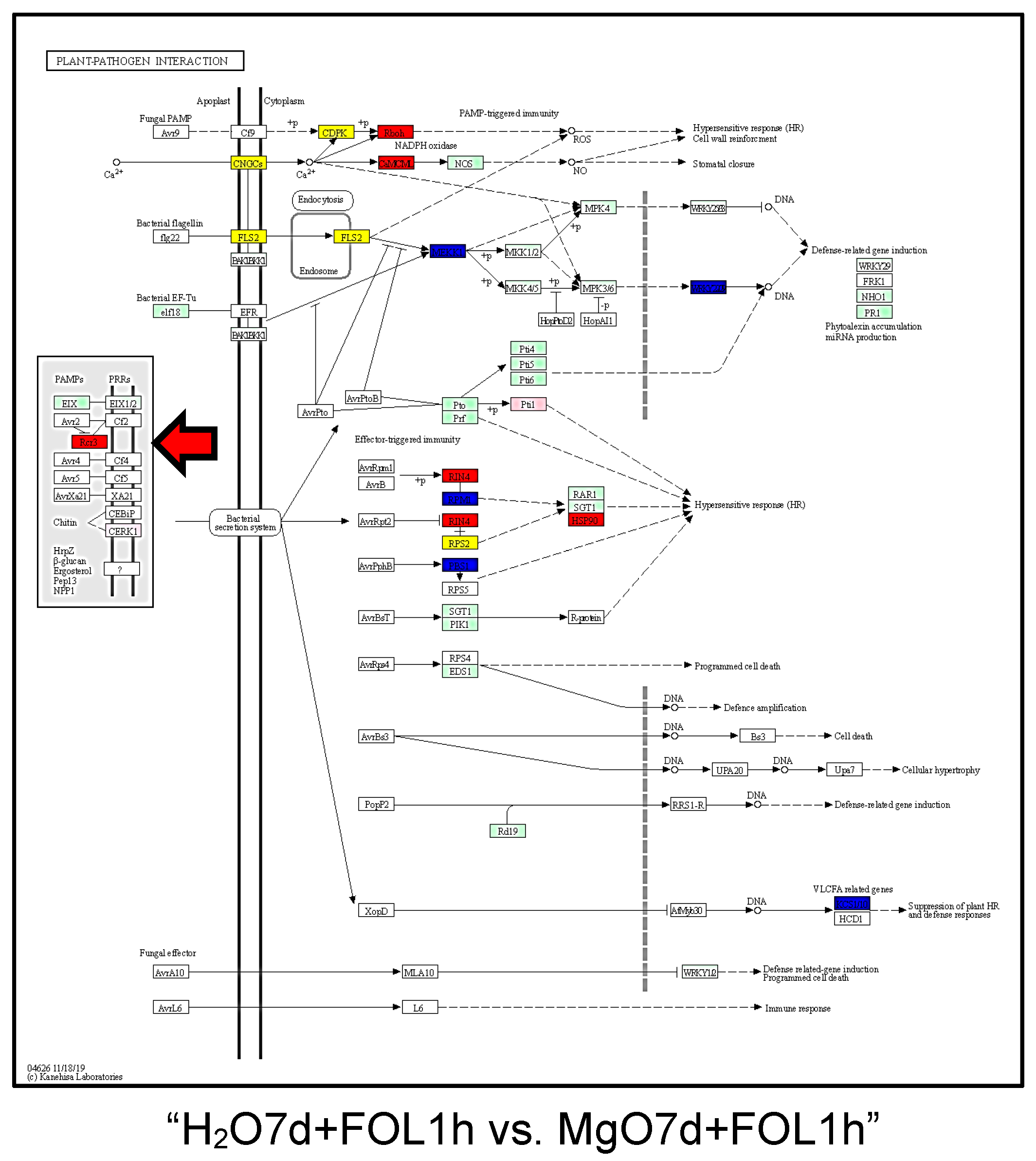
2.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analyses of DEGs
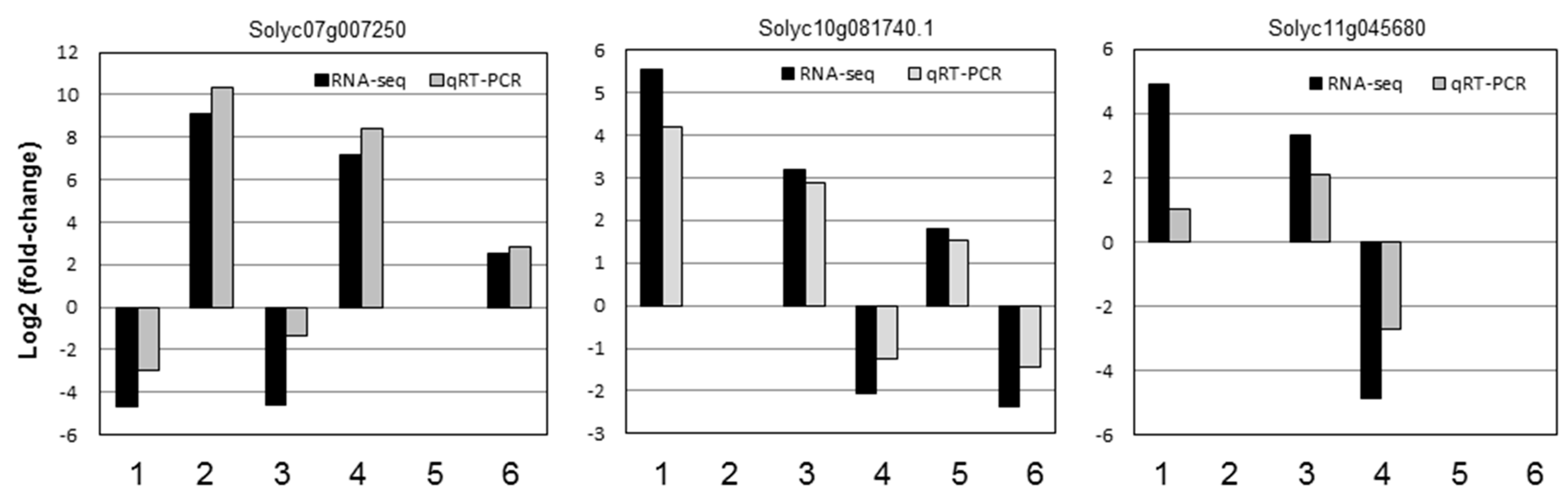
2.4. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.5. Analysis of DEGs
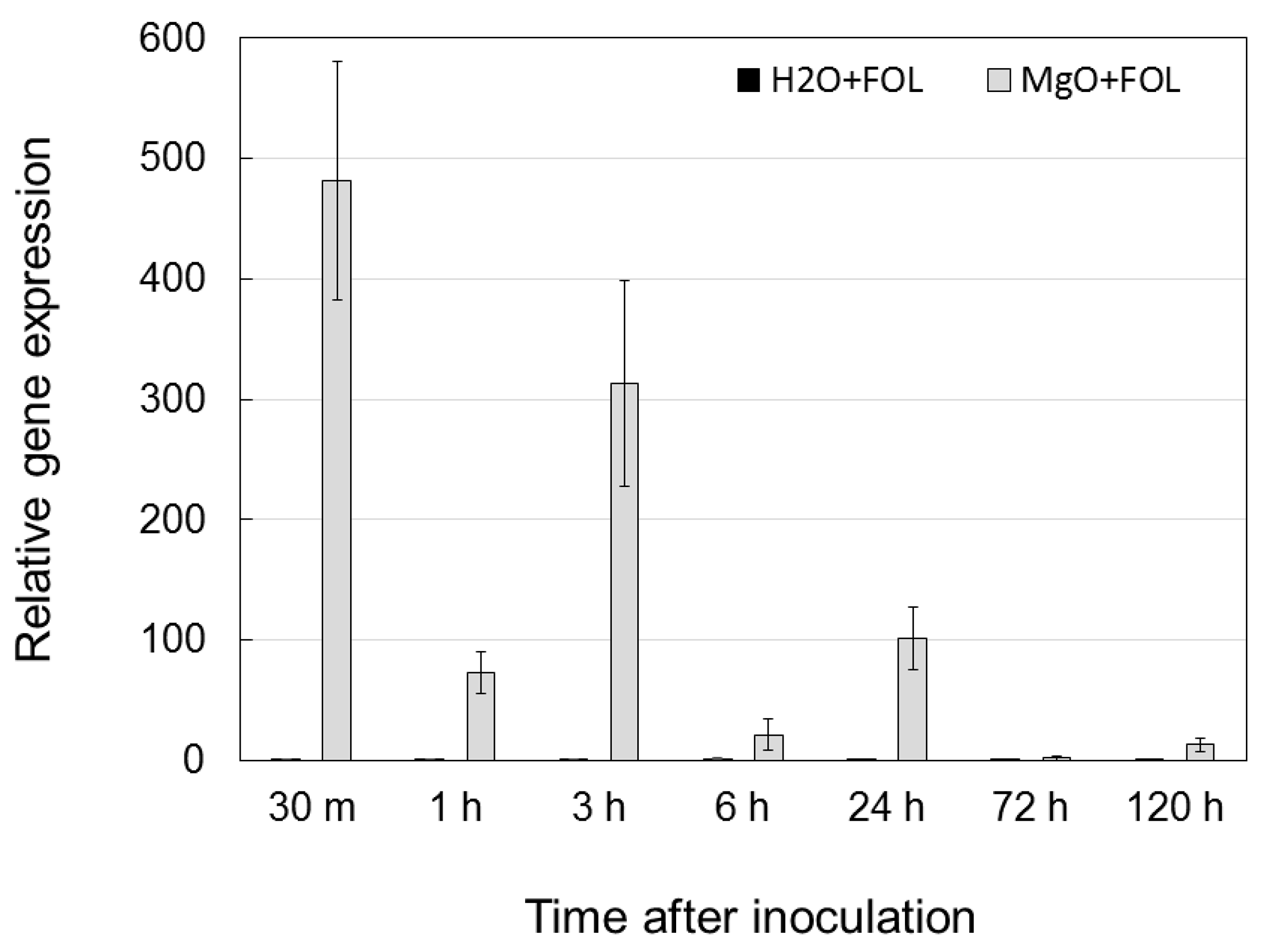
2.6. Expression Analysis of the SlGRP4 Gene
2.7. Prediction of Cis-Acting Elements in the SlGRP4 Gene
2.8. Localization of SlGRP4 in Tomato Roots
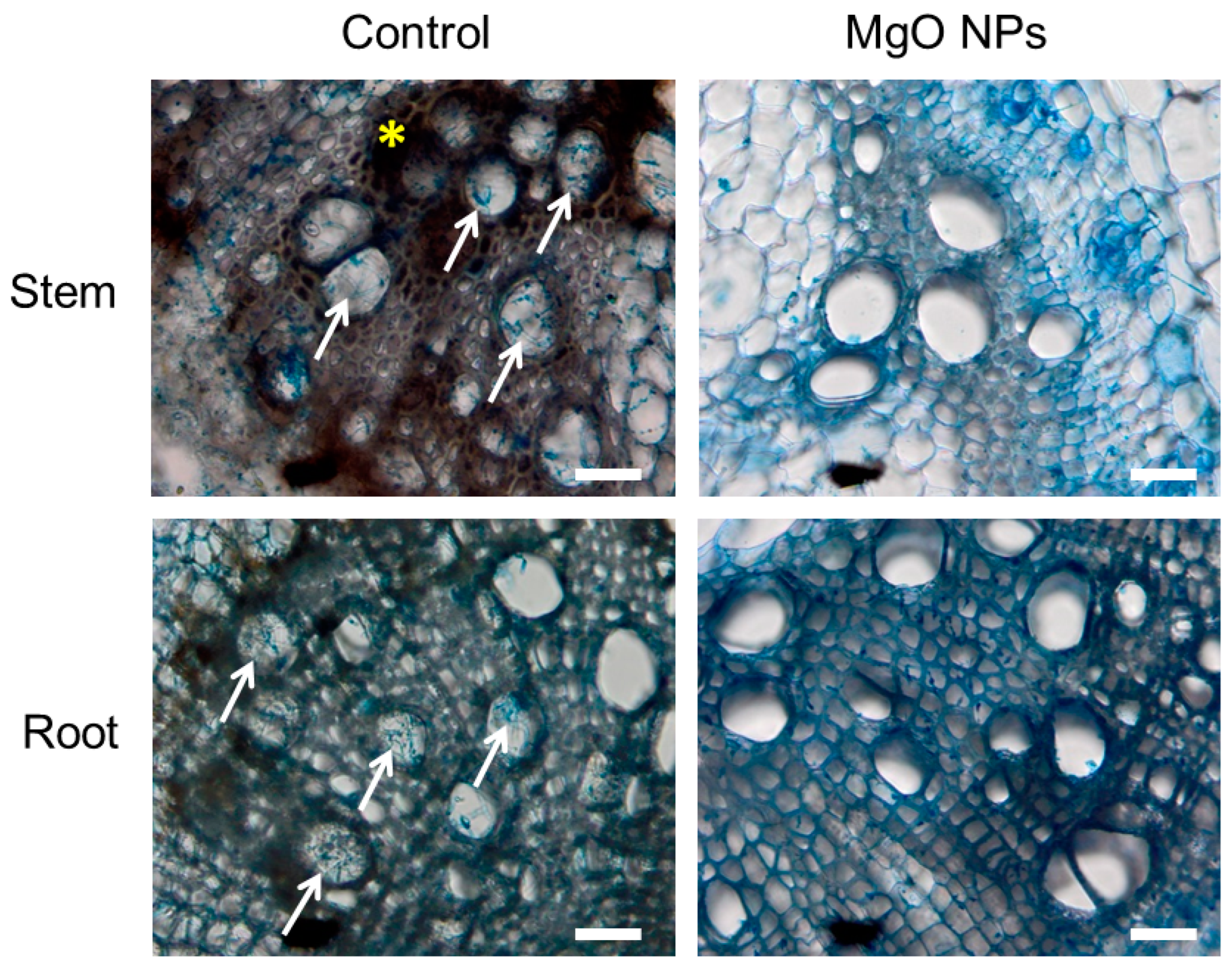
2.9. Microscopic Observation of Tomato Root Tissue Pretreated with MgO NPs and Challenge-Inoculated with FOL
3. Discussion
3.1. Response of Tomato Roots to MgO NPs
3.2. Response of Tomato Roots to Fusarium oxysporum
3.3. SlGRP4
3.4. Nanoparticles and Fusarium Wilt
4. Materials and Methods
4.1. MgO NPs
4.2. Plant Material
4.3. Fungal Strains and Growth Conditions
4.4. Inoculation Experiments
4.5. RNA Extraction
4.6. Preparation of cDNA Library and Sequencing
4.7. Screening of DEGs, Annotation, and GO and KEGG Analyses
4.8. Gene Expression Analysis
4.9. Cis-Element Prediction
4.10. Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Hirooka, T.; Ishii, H. Chemical control of plant diseases. J. Gen. Plant Pathol. 2013, 79, 390–401. [Google Scholar] [CrossRef]
- Nicolopoulou-Stamati, P.; Maipas, S.; Kotampasi, C.; Stamatis, P.; Hens, L. Chemical pesticides and human health: The urgent need for a new concept in agriculture. Front. Public Health 2016, 4, 148. [Google Scholar] [CrossRef] [PubMed]
- OECD. Report of the OECD Workshop on Integrated Pest Management (IPM) Strategies for the Adaptation and Implementation of IPM in Agriculture, Contributing to the Sustainable Use of Pesticides and to Pesticide Risk Reduction; Organisation for Economic Co-operation and Development: Paris, France, 2014; Berlin, Germany, 16–19 October 2012; Available online: https://www.oecd-ilibrary.org/docserver/9789264221635-en.pdf?expires=1669627447&id=id&accname=guest&checksum=EC1A398AF69E11603EF149104CF5C319 (accessed on 28 November 2022).
- Yang, B.; Yang, S.; Zheng, W.; Wang, Y. Plant immunity inducers: From discovery to agricultural application. Stress Biol. 2022, 2, 5. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, W. Recent advance in synthetic chemical inducers of plant immunity. Front. Plant Sci. 2018, 9, 1613. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.P.; Hartmann, A.; Gao, X.; Borriss, R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—A review. Front. Microbiol. 2015, 6, 780. [Google Scholar] [CrossRef] [PubMed]
- Dewen, Q.; Yijie, D.; Yi, Z.; Shupeng, L.; Fachao, S. Plant immunity inducer development and application. Mol. Plant Microbe Interact. 2017, 30, 355–360. [Google Scholar] [CrossRef]
- Wang, M.; Gao, L.; Dong, S.; Sun, Y.; Shen, Q.; Guo, S. Role of silicon on plant-pathogen interactions. Front. Plant Sci. 2017, 8, 701. [Google Scholar] [CrossRef]
- Suarez-Fernandez, M.; Marhuenda-Egea, F.C.; Lopez-Moya, F.; Arnao, M.B.; Cabrera-Escribano, F.; Nueda, M.J.; Gunsé, B.; Lopez-Llorca, L.V. Chitosan induces plant hormones and defenses in tomato root exudates. Front. Plant Sci. 2020, 11, 572087. [Google Scholar] [CrossRef]
- Imada, K.; Sakai, S.; Kajihara, H.; Tanaka, S.; Ito, S. Magnesium oxide nanoparticles induce systemic resistance in tomato against bacterial wilt disease. Plant Pathol. 2016, 65, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Fujikawa, I.; Takehara, Y.; Ota, M.; Imada, K.; Sasaki, K.; Kajihara, H.; Sakai, S.; Jogaiah, S.; Ito, S. Magnesium oxide induces immunity against Fusarium wilt by triggering the jasmonic acid signaling pathway in tomato. J. Biotechnol. 2021, 325, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Kessler, A.; Hedberg, J.; Blomberg, E.; Odnevall, I. Reactive oxygen species formed by metal and metal oxide nanoparticles in physiological media-A review of reactions of importance to nanotoxicity and proposal for categorization. Nanomaterials 2022, 12, 1922. [Google Scholar] [CrossRef] [PubMed]
- Ota, M.; Imada, K.; Sasaki, K.; Kajihara, H.; Sakai, S.; Ito, S. MgO-Induced defence against bacterial wilt disease in Arabidopsis thaliana. Plant Pathol. 2019, 68, 323–333. [Google Scholar] [CrossRef]
- Klabunde, K.J.; Stark, J.; Koper, O.; Mohs, C.; Park, D.G.; Decker, S.; Jiang, Y.; Lagadic, I.; Zhang, D. Nanocrystals as stoichiometric reagents with unique surface chemistry. J. Phys. Chem. 1996, 100, 12142–12153. [Google Scholar] [CrossRef]
- Zang, X.; Zheng, Y.; Feng, X.; Han, X.; Bai, Z.; Zhang, Z. Calcination temperature-dependent surface structure and physicochemical properties of magnesium oxide. RSC Adv. 2015, 5, 86102–86112. [Google Scholar] [CrossRef]
- Haneda, M.; Kato, K.; Sakai, S. Evaluation of physico-chemical properties of magnesium oxide. Annu. Rep. Adv. Ceram. Res. Cent. Nagoya Inst. Technol. 2017, 5, 44–51. [Google Scholar]
- Harikrushana, P.; Ramchandra, S.; Shah, K.R. Study of wilt producing Fusarium sp. from tomato (Lycopersicon esculentum Mill). Int. J. Curr. Microbiol. App. Sci. 2014, 3, 854–858. [Google Scholar]
- Di Pietro, A.; Madrid, M.P.; Caracuel, Z.; Delgado-Jarana, J.; Roncero, M.I.G. Fusarium oxysporum: Exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 2003, 4, 315–325. [Google Scholar] [CrossRef]
- de Lamo, F.J.; Takken, F.L.W. Biocontrol by Fusarium oxysporum using endophyte-mediated resistance. Front. Plant Sci. 2020, 11, 37. [Google Scholar] [CrossRef]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Tenhaken, R.; Dixon, R.; Lamb, C.J. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 1994, 79, 583–593. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, C.; Yang, R.; Liu, H.; Suo, Y.; Gao, G. GRP genes in potato genome and their expression response to phytohormone and Ralstonia solanacearum. J. Phytopathol. 2022, 170, 724–737. [Google Scholar] [CrossRef]
- Silva, T.D.; Almeida, C.M.A.; Malafaia, C.B.; Oliveira, L.M.S.; Silva, M.V.; Correia, M.T.S. Analysis of protein profile of tomato root infected with Fusarium oxysporum f. sp. lycopersici. Genet. Mol. Res. 2017, 16, gmr16027209. [Google Scholar] [CrossRef]
- Rentel, M.C.; Lecourieux, D.; Ouaked, F.; Usher, S.L.; Petersen, L.; Okamoto, H.; Knight, H.; Peck, S.C.; Grierson, C.S.; Hirt, H.; et al. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 2004, 427, 858–861. [Google Scholar] [CrossRef]
- Sagi, M.; Fluhr, R. Superoxide production by plant homologues of the gp91phox NADPH oxidase. Modulation of activity by calcium and by tobacco mosaic virus infection. Plant Physiol. 2001, 126, 1281–1290. [Google Scholar] [CrossRef] [PubMed]
- Saand, M.A.; Xu, Y.P.; Li, W.; Wang, J.P.; Cai, X.Z. Cyclic nucleotide gated channel gene family in tomato: Genome-wide identification and functional analyses in disease resistance. Front. Plant Sci. 2015, 6, 303. [Google Scholar] [CrossRef]
- Rutschmann, F.; Stalder, U.; Piotrowski, M.; Oecking, C.; Schaller, A. LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol. 2002, 129, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tsuda, K.; Sato, M.; Cohen, J.D.; Katagiri, F.; Glazebrook, J. Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog. 2009, 5, e1000301. [Google Scholar] [CrossRef]
- Shivnauth, V.; Pretheepkumar, S.; Marchetta, E.; Amani, K.; Castroverde, C.D.M. Structural diversity and stress regulation of the plant immunity-associated calmodulin-binding protein 60 (CBP60) family of transcription factors in Solanum lycopersicum (tomato). bioRxiv 2022. [Google Scholar] [CrossRef]
- Boncan, D.A.T.; Tsang, S.S.K.; Li, C.; Lee, I.H.T.; Lam, H.M.; Chan, T.F.; Hui, J.H.L. Terpenes and terpenoids in plants: Interactions with environment and insects. Int. J. Mol. Sci. 2020, 21, 7382. [Google Scholar] [CrossRef]
- Paulus, J.K.; Kourelis, J.; Ramasubramanian, S.; Homma, F.; Godson, A.; Horger, A.C.; Hong, T.N.; Krahn, D.; Ossorio Carballo, L.; Wang, S.; et al. Extracellular proteolytic cascade in tomato activates immune protease Rcr3. Proc. Natl. Acad. Sci. USA 2020, 117, 17409–17417. [Google Scholar] [CrossRef] [PubMed]
- Shabab, M.; Shindo, T.; Gu, C.; Kaschani, F.; Pansuriya, T.; Chintha, R.; Harzen, A.; Colby, T.; Kamoun, S.; van der Hoorn, R.A. Fungal effector protein AVR2 targets diversifying defense-related cys proteases of tomato. Plant Cell 2008, 20, 1169–1183. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Wang, Y. Apoplastic proteases: Powerful weapons against pathogen infection in plants. Plant Commun. 2020, 1, 100085. [Google Scholar] [CrossRef]
- Hernández-Aparicio, F.; Lisón, P.; Rodrigo, I.; Bellés, J.M.; López-Gresa, M.P. Signaling in the tomato immunity against Fusarium oxysporum. Molecules 2021, 26, 1818. [Google Scholar] [CrossRef] [PubMed]
- Kwak, K.J.; Kim, Y.O.; Kang, H. Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J. Exp. Bot. 2005, 56, 3007–3016. [Google Scholar] [CrossRef]
- Czolpinska, M.; Rurek, M. Plant glycine-rich proteins in stress response: An emerging, still prospective story. Front. Plant Sci. 2018, 9, 302. [Google Scholar] [CrossRef]
- Mangeon, A.; Junqueira, R.M.; Sachetto-Martins, G. Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal. Behav. 2010, 5, 99–104. [Google Scholar] [CrossRef]
- Olivain, C.; Alabouvette, C. Process of tomato root colonization by a pathogenic strain of Fusarium oxysporum f. sp. lycopersici in comparison with non-pathogenic strain. New Phytol. 1999, 141, 497–510. [Google Scholar]
- Elmer, W.; De La Torre-Roche, R.; Pagano, L.; Majumdar, S.; Zuverza-Mena, N.; Dimkpa, C.; Gardea-Torresdey, J.; White, J.C. Effect of metalloid and metal oxide nanoparticles on Fusarium wilt of watermelon. Plant Dis. 2018, 102, 1394–1401. [Google Scholar] [CrossRef]
- Farhana; Munis, M.F.H.; Alamer, K.H.; Althobaiti, A.T.; Kamal, A.; Liaquat, F.; Haroon, U.; Ahmed, J.; Chaudhary, H.J.; Attia, H. ZnO nanoparticle-mediated seed priming induces biochemical and antioxidant changes in chickpea to alleviate Fusarium wilt. J. Fungi 2022, 8, 753. [Google Scholar] [CrossRef] [PubMed]
- Turgut, B.A.; Bezirganoğlu, İ. Foliar application of CaO nanoparticles and salicylic acid on Medicago sativa seedlings enhances tolerance against Fusarium oxysporum. Physiol. Mol. Plant Pathol. 2022, 122, 101926. [Google Scholar] [CrossRef]
- Elbasuney, S.; El-Sayyad, G.S.; Attia, M.S.; Abdelaziz, A.M. Ferric oxide colloid: Towards green nano-fertilizer for tomato plant with enhanced vegetative growth and immune response against Fusarium wilt disease. J. Inorg. Organomet. Polym. Mater. 2022, 32, 4270–4283. [Google Scholar] [CrossRef]
- Ahmad, M.; Ali, A.; Ullah, Z.; Sher, H.; Dai, D.Q.; Ali, M.; Iqbal, J.; Zahoor, M.; Ali, I. Biosynthesized silver nanoparticles using Polygonatum geminiflorum efficiently control fusarium wilt disease of tomato. Front. Bioeng. Biotechnol. 2022, 10, 988607. [Google Scholar] [CrossRef]
- Noman, M.; Ahmed, T.; Ijaz, U.; Shahid, M.; Nazir, M.M.; Azizullah; White, J.C.; Li, D.; Song, F. Bio-functionalized manganese nanoparticles suppress Fusarium wilt in watermelon (Citrullus lanatus L.) by infection disruption, host defense response potentiation, and soil microbial community modulation. Small 2023, 19, 2205687. [Google Scholar] [CrossRef]
- Marslin, G.; Sheeba, C.J.; Franklin, G. Nanoparticles alter secondary metabolism in plants via ROS burst. Front. Plant Sci. 2017, 8, 832. [Google Scholar] [CrossRef]
- Namiki, F.; Shiomi, T.; Kayamura, T.; Tsuge, T. Characterization of the formae speciales of Fusarium oxysporum causing wilt of cucurbits by DNA fingerprinting with nuclear repetitive DNA sequences. Appl. Environ. Microbiol. 1994, 60, 2684–2691. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology. R Package Version 2.50.0. Available online: https://rdrr.io/bioc/topGO/ (accessed on 25 October 2022).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]







| Pathway | Up | Down | Total |
|---|---|---|---|
| Metabolic pathways | 107 | 102 | 209 |
| Biosynthesis of secondary metabolites | 65 | 59 | 124 |
| Carbon metabolism | 15 | 11 | 26 |
| Plant hormone signal transduction | 12 | 9 | 21 |
| Plant-pathogen interaction | 10 | 10 | 20 |
| MAPK signaling pathway-plant | 12 | 7 | 19 |
| Biosynthesis of amino acids | 8 | 8 | 16 |
| Biosynthesis of cofactors | 11 | 4 | 15 |
| Amino sugar and nucleotide sugar metabolism | 8 | 3 | 11 |
| Protein processing in endoplasmic reticulum | 6 | 5 | 11 |
| Pathway | Up | Down | Total |
|---|---|---|---|
| Metabolic pathways | 160 | 213 | 373 |
| Biosynthesis of secondary metabolites | 90 | 139 | 229 |
| Carbon metabolism | 17 | 25 | 42 |
| Plant hormone signal transduction | 12 | 21 | 33 |
| Biosynthesis of cofactors | 13 | 20 | 33 |
| Biosynthesis of amino acids | 12 | 20 | 32 |
| Plant-pathogen interaction | 11 | 16 | 27 |
| MAPK signaling pathway-plant | 10 | 16 | 26 |
| Amino sugar and nucleotide sugar metabolism | 8 | 18 | 26 |
| Starch and sucrose metabolism | 13 | 10 | 23 |
| Pathway | Up | Down | Total |
|---|---|---|---|
| Metabolic pathways | 195 | 121 | 316 |
| Biosynthesis of secondary metabolites | 122 | 64 | 186 |
| Plant hormone signal transduction | 20 | 20 | 40 |
| Carbon metabolism | 21 | 16 | 37 |
| MAPK signaling pathway-plant | 21 | 11 | 32 |
| Plant-pathogen interaction | 19 | 10 | 29 |
| Biosynthesis of amino acids | 16 | 11 | 27 |
| Biosynthesis of cofactors | 16 | 9 | 25 |
| Cysteine and methionine metabolism | 12 | 9 | 21 |
| Amino sugar and nucleotide sugar metabolism | 14 | 6 | 20 |
| Pathway | Up | Down | Total |
|---|---|---|---|
| Metabolic pathways | 217 | 124 | 341 |
| Biosynthesis of secondary metabolites | 128 | 72 | 200 |
| Carbon metabolism | 33 | 18 | 51 |
| Biosynthesis of amino acids | 22 | 15 | 37 |
| Plant hormone signal transduction | 15 | 21 | 36 |
| MAPK signaling pathway-plant | 20 | 10 | 30 |
| Plant-pathogen interaction | 19 | 8 | 27 |
| Biosynthesis of cofactors | 17 | 9 | 26 |
| Glycolysis/Gluconeogenesis | 13 | 8 | 21 |
| Amino sugar and nucleotide sugar metabolism | 10 | 9 | 19 |
| Pathway | Up | Down | Total |
|---|---|---|---|
| Metabolic pathways | 256 | 182 | 438 |
| Biosynthesis of secondary metabolites | 160 | 105 | 265 |
| Carbon metabolism | 34 | 32 | 66 |
| Biosynthesis of cofactors | 32 | 19 | 51 |
| Biosynthesis of amino acids | 27 | 18 | 45 |
| Ribosome | 26 | 8 | 34 |
| Glycolysis/Gluconeogenesis | 18 | 14 | 32 |
| Plant hormone signal transduction | 16 | 15 | 31 |
| Amino sugar and nucleotide sugar metabolism | 22 | 8 | 30 |
| Pyruvate metabolism | 17 | 12 | 29 |
| Pathway | Up | Down | Total |
|---|---|---|---|
| Metabolic pathways | 163 | 159 | 322 |
| Biosynthesis of secondary metabolites | 88 | 108 | 196 |
| Carbon metabolism | 20 | 20 | 40 |
| Plant hormone signal transduction | 14 | 19 | 33 |
| Biosynthesis of cofactors | 14 | 15 | 29 |
| Biosynthesis of amino acids | 9 | 16 | 25 |
| Starch and sucrose metabolism | 11 | 13 | 24 |
| MAPK signaling pathway-plant | 13 | 10 | 23 |
| Plant-pathogen interaction | 13 | 10 | 23 |
| Amino sugar and nucleotide sugar metabolism | 8 | 14 | 22 |
| Pathway | Up | Down | Total |
|---|---|---|---|
| Metabolic pathways | 147 | 115 | 262 |
| Biosynthesis of secondary metabolites | 86 | 74 | 160 |
| Carbon metabolism | 27 | 12 | 39 |
| Plant hormone signal transduction | 14 | 18 | 32 |
| Biosynthesis of amino acids | 13 | 7 | 20 |
| Biosynthesis of cofactors | 11 | 7 | 18 |
| Plant-pathogen interaction | 10 | 11 | 21 |
| MAPK signaling pathway-plant | 11 | 11 | 22 |
| Glycolysis/Gluconeogenesis | 9 | 10 | 19 |
| Starch and sucrose metabolism | 7 | 9 | 16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takehara, Y.; Fijikawa, I.; Watanabe, A.; Yonemura, A.; Kosaka, T.; Sakane, K.; Imada, K.; Sasaki, K.; Kajihara, H.; Sakai, S.; et al. Molecular Analysis of MgO Nanoparticle-Induced Immunity against Fusarium Wilt in Tomato. Int. J. Mol. Sci. 2023, 24, 2941. https://doi.org/10.3390/ijms24032941
Takehara Y, Fijikawa I, Watanabe A, Yonemura A, Kosaka T, Sakane K, Imada K, Sasaki K, Kajihara H, Sakai S, et al. Molecular Analysis of MgO Nanoparticle-Induced Immunity against Fusarium Wilt in Tomato. International Journal of Molecular Sciences. 2023; 24(3):2941. https://doi.org/10.3390/ijms24032941
Chicago/Turabian StyleTakehara, Yushi, Isamu Fijikawa, Akihiro Watanabe, Ayumi Yonemura, Tomoyuki Kosaka, Kosei Sakane, Kiyoshi Imada, Kazunori Sasaki, Hiroshi Kajihara, Shoji Sakai, and et al. 2023. "Molecular Analysis of MgO Nanoparticle-Induced Immunity against Fusarium Wilt in Tomato" International Journal of Molecular Sciences 24, no. 3: 2941. https://doi.org/10.3390/ijms24032941





