Ubiquitin-Related Modifier 1 (URM-1) Modulates Cx43 in Breast Cancer Cell Lines
Abstract
:1. Introduction
2. Results
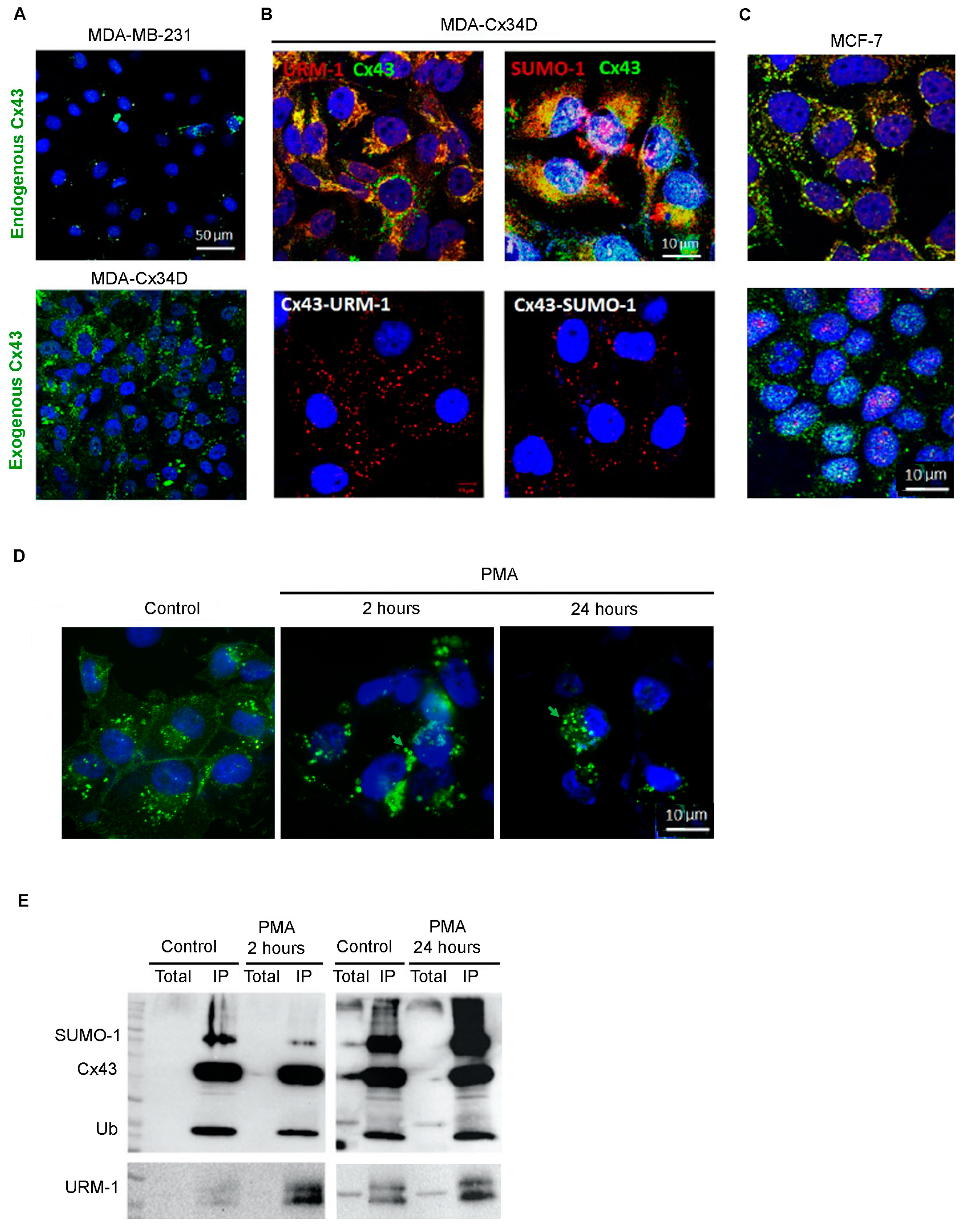
2.1. Cx43 Interacts with Ubiquitin, URM-1, and SUMO-1
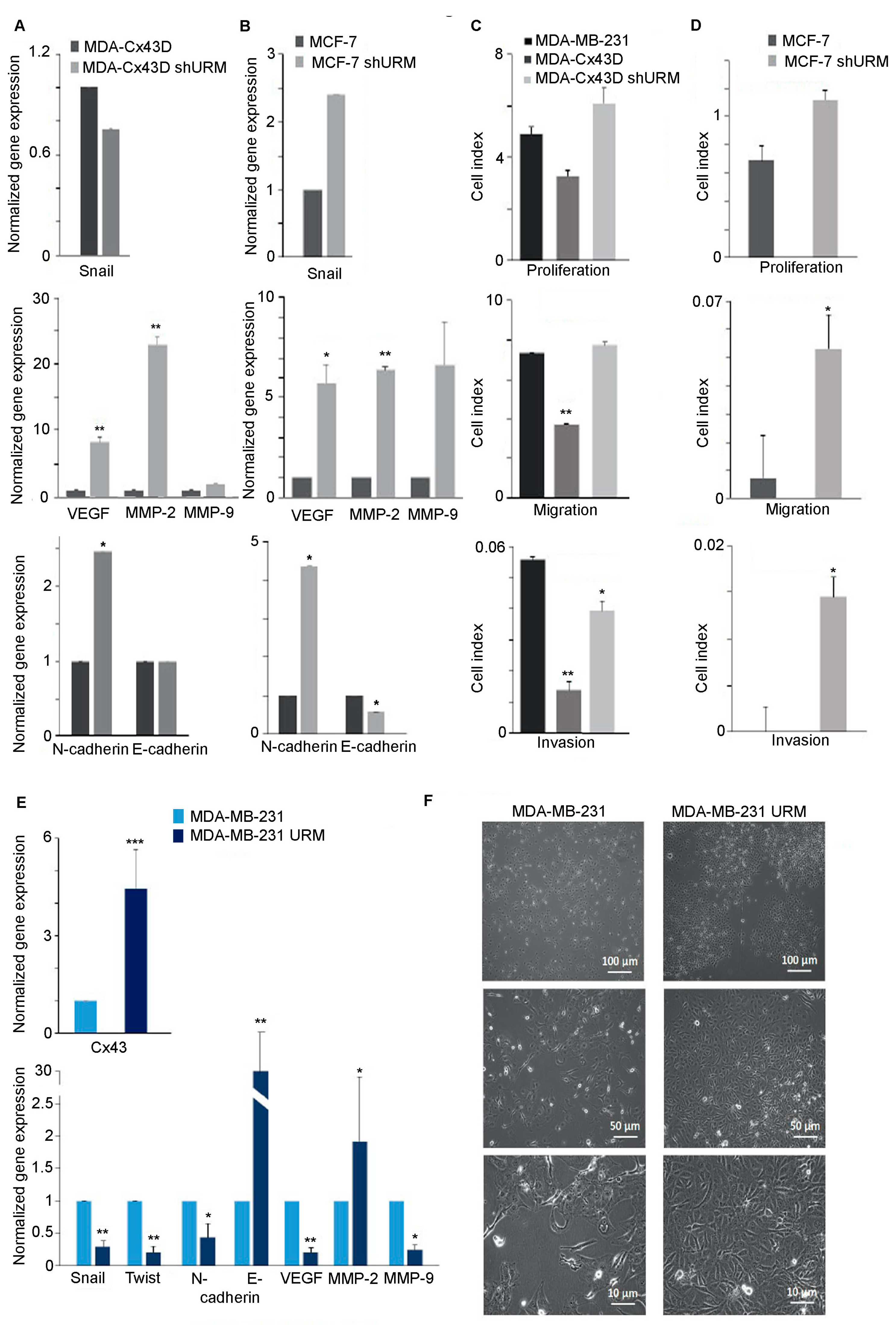
2.2. Downregulation of URM-1 Modulates Cx43 Expression
2.3. Loss of URM-1 Promotes EMT in Breast Cancer Cell Lines
3. Discussion
4. Materials and Methods
4.1. Cells and Cell Cultures
4.2. Analysis of Gene Expression
4.3. Analysis of Protein Expression and Localization
4.4. Imaging
4.5. Real-Time Cell Analyzer (RTCA) Assay for Migration, Invasion, and Proliferation
4.6. In vitro Wound Healing Assay
4.7. Dye Transfer Assays
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sorgen, P.L.; Trease, A.J.; Spagnol, G.; Delmar, M.; Nielsen, M.S. Protein–protein interactions with connexin 43: Regulation and function. Int. J. Mol. Sci. 2018, 19, 1428. [Google Scholar] [CrossRef] [PubMed]
- Trosko, J.E.; Ruch, R.J. Cell-cell communication in carcinogenesis. Front. Biosci. 1998, 3, d208–d236. [Google Scholar] [CrossRef] [PubMed]
- Mesnil, M.; Crespin, S.; Avanzo, J.-L.; Zaidan-Dagli, M.-L. Defective gap junctional intercellular communication in the carcinogenic process. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2005, 1719, 125–145. [Google Scholar] [CrossRef]
- Lin, J.H.-C.; Yang, J.; Liu, S.; Takano, T.; Wang, X.; Gao, Q.; Willecke, K.; Nedergaard, M. Connexin mediates gap junction-independent resistance to cellular injury. J. Neurosci. 2003, 23, 430–441. [Google Scholar] [CrossRef]
- Crow, D.; Beyer, E.; Paul, D.; Kobe, S.; Lau, A. Phosphorylation of connexin43 gap junction protein in uninfected and Rous sarcoma virus-transformed mammalian fibroblasts. Mol. Cell. Biol. 1990, 10, 1754–1763. [Google Scholar]
- El-Hajjar, L.; Jalaleddine, N.; Shaito, A.; Zibara, K.; Kazan, J.M.; El-Saghir, J.; El-Sabban, M. Bevacizumab induces inflammation in MDA-MB-231 breast cancer cell line and in a mouse model. Cell Signal. 2019, 53, 400–412. [Google Scholar] [CrossRef]
- Kazan, J.M.; El-Saghir, J.; Saliba, J.; Shaito, A.; Jalaleddine, N.; El-Hajjar, L.; Al-Ghadban, S.; Yehia, L.; Zibara, K.; El-Sabban, M. Cx43 Expression Correlates with Breast Cancer Metastasis in MDA-MB-231 Cells In Vitro, In a Mouse Xenograft Model and in Human Breast Cancer Tissues. Cancers 2019, 11, 460. [Google Scholar] [CrossRef]
- El-Sabban, M.E.; Pauli, B.U. Adhesion-mediated gap junctional communication between lung-metastatatic cancer cells and endothelium. Invasion Metastasis 1994, 14, 164–176. [Google Scholar]
- Talhouk, R.S.; Fares, M.B.; Rahme, G.J.; Hariri, H.H.; Rayess, T.; Dbouk, H.A.; Bazzoun, D.; Al-Labban, D.; El-Sabban, M.E. Context dependent reversion of tumor phenotype by connexin-43 expression in MDA-MB231 cells and MCF-7 cells: Role of beta-catenin/connexin43 association. Exp. Cell Res. 2013, 319, 3065–3080. [Google Scholar] [CrossRef]
- Zibara, K.; Awada, Z.; Dib, L.; El-Saghir, J.; Al-Ghadban, S.; Ibrik, A.; El-Zein, N.; El-Sabban, M. Anti-angiogenesis therapy and gap junction inhibition reduce MDA-MB-231 breast cancer cell invasion and metastasis in vitro and in vivo. Sci. Rep. 2015, 5, 12598. [Google Scholar] [CrossRef]
- Al-Ghadban, S.; Kaissi, S.; Homaidan, F.R.; Naim, H.Y.; El-Sabban, M.E. Cross-talk between intestinal epithelial cells and immune cells in inflammatory bowel disease. Sci. Rep. 2016, 6, 29783. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, K.; Mizushima, N.; Noda, T.; Ohsumi, Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 2000, 275, 7462–7465. [Google Scholar] [CrossRef] [Green Version]
- Pedrioli, P.G.; Leidel, S.; Hofmann, K. Urm1 at the crossroad of modifications. ‘Protein Modifications: Beyond the Usual Suspects’ Review Series. EMBO Rep. 2008, 9, 1196–1202. [Google Scholar] [CrossRef]
- Wang, F.; Liu, M.; Qiu, R.; Ji, C. The dual role of ubiquitin-like protein Urm1 as a protein modifier and sulfur carrier. Protein Cell 2011, 2, 612–619. [Google Scholar] [CrossRef]
- Judes, A.; Ebert, F.; Bar, C.; Thuring, K.L.; Harrer, A.; Klassen, R.; Helm, M.; Stark, M.J.; Schaffrath, R. Urmylation and tRNA thiolation functions of ubiquitin-like Uba4.Urm1 systems are conserved from yeast to man. FEBS Lett. 2015, 589, 904–909. [Google Scholar] [CrossRef]
- Alaga, K.C.; Crawford, M.; Dagnino, L.; Laird, D.W. Aberrant Cx43 Expression and Mislocalization in Metastatic Human Melanomas. J. Cancer 2017, 8, 1123–1128. [Google Scholar] [CrossRef]
- Busby, M.; Hallett, M.T.; Plante, I. The Complex Subtype-Dependent Role of Connexin 43 (GJA1) in Breast Cancer. Int. J. Mol. Sci. 2018, 19, 693. [Google Scholar] [CrossRef]
- Chasampalioti, M.; Green, A.R.; Ellis, I.O.; Rakha, E.A.; Jackson, A.M.; Spendlove, I.; Ramage, J.M. Connexin 43 is an independent predictor of patient outcome in breast cancer patients. Breast Cancer Res. Treat. 2018, 174, 93–102. [Google Scholar] [CrossRef]
- Laird, D.W.; Fistouris, P.; Batist, G.; Alpert, L.; Huynh, H.T.; Carystinos, G.D.; Alaoui-Jamali, M.A. Deficiency of connexin43 gap junctions is an independent marker for breast tumors. Cancer Res. 1999, 59, 4104–4110. [Google Scholar]
- Plante, I.; Stewart, M.K.; Barr, K.; Allan, A.L.; Laird, D.W. Cx43 suppresses mammary tumor metastasis to the lung in a Cx43 mutant mouse model of human disease. Oncogene 2011, 30, 1681–1692. [Google Scholar] [CrossRef]
- Tang, B.; Peng, Z.H.; Yu, P.W.; Yu, G.; Qian, F.; Zeng, D.Z.; Zhao, Y.L.; Shi, Y.; Hao, Y.X.; Luo, H.X. Aberrant expression of Cx43 is associated with the peritoneal metastasis of gastric cancer and Cx43-mediated gap junction enhances gastric cancer cell diapedesis from peritoneal mesothelium. PLoS ONE 2013, 8, e74527. [Google Scholar] [CrossRef] [PubMed]
- Teleki, I.; Szasz, A.M.; Maros, M.E.; Gyorffy, B.; Kulka, J.; Meggyeshazi, N.; Kiszner, G.; Balla, P.; Samu, A.; Krenacs, T. Correlations of differentially expressed gap junction connexins Cx26, Cx30, Cx32, Cx43 and Cx46 with breast cancer progression and prognosis. PloS ONE 2014, 9, e112541. [Google Scholar] [CrossRef]
- Aasen, T.; Leithe, E.; Graham, S.V.; Kameritsch, P.; Mayán, M.D.; Mesnil, M.; Pogoda, K.; Tabernero, A. Connexins in cancer: Bridging the gap to the clinic. Oncogene 2019, 38, 4429–4451. [Google Scholar] [CrossRef] [Green Version]
- Goehring, A.S.; Rivers, D.M.; Sprague, G.F., Jr. Urmylation: A ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol. Biol. Cell 2003, 14, 4329–4341. [Google Scholar] [CrossRef] [PubMed]
- Khoshnood, B.; Dacklin, I.; Grabbe, C. Urm1: An essential regulator of JNK signaling and oxidative stress in Drosophila melanogaster. Cell Mol. Life Sci. 2016, 73, 1939–1954. [Google Scholar] [CrossRef] [PubMed]
- Khoshnood, B.; Dacklin, I.; Grabbe, C. A proteomics approach to identify targets of the ubiquitin-like molecule Urm1 in Drosophila melanogaster. PLoS ONE 2017, 12, e0185611. [Google Scholar] [CrossRef] [PubMed]
- Van der Veen, A.G.; Schorpp, K.; Schlieker, C.; Buti, L.; Damon, J.R.; Spooner, E.; Ploegh, H.L.; Jentsch, S. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc. Natl. Acad. Sci. USA 2011, 108, 1763–1770. [Google Scholar] [CrossRef] [PubMed]
- Hleihel, R.; Khoshnood, B.; Dacklin, I.; Omran, H.; Mouawad, C.; Dassouki, Z.; El-Sabban, M.; Shirinian, M.; Grabbe, C.; Bazarbachi, A. The HTLV-1 oncoprotein Tax is modified by the ubiquitin related modifier 1 (Urm1). Retrovirology 2018, 15, 33. [Google Scholar] [CrossRef]
- Nimlamool, W.; Andrews, R.M.K.; Falk, M.M. Connexin43 phosphorylation by PKC and MAPK signals VEGF-mediated gap junction internalization. Mol. Biol. Cell 2015, 26, 2755–2768. [Google Scholar] [CrossRef]
- Tahara, E.; Kadara, H.; Lacroix, L.; Lotan, D.; Lotan, R. Activation of protein kinase C by phorbol 12-myristate 13-acetate suppresses the growth of lung cancer cells through KLF6 induction. Cancer Biol. Ther. 2009, 8, 801–807. [Google Scholar] [CrossRef]
- Ek-Vitorin, J.F.; King, T.J.; Heyman, N.S.; Lampe, P.D.; Burt, J.M. Selectivity of connexin 43 channels is regulated through protein kinase C-dependent phosphorylation. Circ. Res. 2006, 98, 1498–1505. [Google Scholar] [CrossRef] [PubMed]
- Berthoud, V.M.; Rook, M.B.; Traub, O.; Hertzberg, E.L.; Saez, J.C. On the mechanisms of cell uncoupling induced by a tumor promoter phorbol ester in clone 9 cells, a rat liver epithelial cell line. Eur. J. Cell Biol. 1993, 62, 384–396. [Google Scholar] [PubMed]
- Lampe, P.D.; TenBroek, E.M.; Burt, J.M.; Kurata, W.E.; Johnson, R.G.; Lau, A.F. Phosphorylation of connexin43 on serine368 by protein kinase C regulates gap junctional communication. J. Cell Biol. 2000, 149, 1503–1512. [Google Scholar] [CrossRef] [PubMed]
- Rivedal, E.; Yamasaki, H.; Sanner, T. Inhibition of gap junctional intercellular communication in Syrian hamster embryo cells by TPA, retinoic acid and DDT. Carcinogenesis 1994, 15, 689–694. [Google Scholar] [CrossRef]
- Rivedal, E.; Leithe, E. Connexin43 synthesis, phosphorylation, and degradation in regulation of transient inhibition of gap junction intercellular communication by the phorbol ester TPA in rat liver epithelial cells. Exp. Cell Res. 2005, 302, 143–152. [Google Scholar] [CrossRef]
- Leithe, E.; Rivedal, E. Ubiquitination and down-regulation of gap junction protein connexin-43 in response to 12-O-tetradecanoylphorbol 13-acetate treatment. J. Biol. Chem. 2004, 279, 50089–50096. [Google Scholar] [CrossRef]
- Oh, S.Y.; Grupen, C.G.; Murray, A.W. Phorbol ester induces phosphorylation and down-regulation of connexin 43 in WB cells. Biochim. Biophys Acta 1991, 1094, 243–245. [Google Scholar] [CrossRef]
- Lampe, P.D. Analyzing phorbol ester effects on gap junctional communication: A dramatic inhibition of assembly. J. Cell Biol. 1994, 127, 1895–1905. [Google Scholar] [CrossRef]
- Marks, D.S.; Hopf, T.A.; Sander, C. Protein structure prediction from sequence variation. Nat. Biotechnol. 2012, 30, 1072. [Google Scholar] [CrossRef]
- Johnstone, S.R.; Billaud, M.; Lohman, A.W.; Taddeo, E.P.; Isakson, B.E. Posttranslational modifications in connexins and pannexins. J. Membr. Biol. 2012, 245, 319–332. [Google Scholar] [CrossRef]
- Laing, J.G.; Beyer, E.C. The gap junction protein connexin43 is degraded via the ubiquitin proteasome pathway. J. Biol. Chem. 1995, 270, 26399–26403. [Google Scholar] [CrossRef] [PubMed]
- He, L.Q.; Fang, C.; Yu, L.; Liu, M.J.; Tan, Z.P.; Qian, P.; Fang, F.Y.; Wu, L.Q.; Long, Z.G.; Dai, H.P. Cx31 is assembled and trafficked to cell surface by ER-Golgi pathway and degraded by proteasomal or lysosomal pathways. Cell Res. 2005, 15, 455. [Google Scholar] [CrossRef] [PubMed]
- Kjenseth, A.; Fykerud, T.A.; Sirnes, S.; Bruun, J.; Yohannes, Z.; Kolberg, M.; Omori, Y.; Rivedal, E.; Leithe, E. The gap junction channel protein connexin 43 is covalently modified and regulated by SUMOylation. J. Biol. Chem. 2012, 287, 15851–15861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitoh, H.; Hinchey, J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000, 275, 6252–6258. [Google Scholar] [CrossRef] [PubMed]
- Kfoury, Y.; Setterblad, N.; El-Sabban, M.; Zamborlini, A.; Dassouki, Z.; El Hajj, H.; Hermine, O.; Pique, C.; Saïb, A.; Bazarbachi, A. Tax ubiquitylation and SUMOylation control the dynamic shuttling of Tax and NEMO between Ubc9 nuclear bodies and the centrosome. Blood 2011, 117, 190–199. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Teng, Z.; Kuang, X.; Wang, J.; Zhang, X. Real-time cell analysis–a new method for dynamic, quantitative measurement of infectious viruses and antiserum neutralizing activity. J. Virol. Methods 2013, 193, 364–370. [Google Scholar] [CrossRef] [Green Version]


| Gene | Primer Sequences | Annealing Temperature |
|---|---|---|
| Cx43 | F: CTTCACTACTTTTAAGCAAAAGAG R: TCCCTCCAGCAGTTGAG | 52 °C |
| GAPDH | F: TGGTGCTCAGTGTAGCCCAG R: GGACCTGACCTGCCGTCTAG | 52–62 °C |
| URM-1 | F: GGGCGGAGTTACTATTTGGT R: TCATAACCGATTTCACTCAAGTTT | 57 °C |
| MMP-2 | F: TTGACGGTAAGGACGGACTC R: ACTTGCAGTACTCCCCATCG | 55 °C |
| MMP-9 | F: TTGACAGCGACAAGAAGTGG R: GCCATTCACGTCGTCCTTAT | 55 °C |
| VEGF | F: AGGCCCACAGGGATTTTCTT R: ATCAAACCTCACCAAGGCCA | 55 °C |
| N-cadherin | F: GGTGGAGGAGAAGAAGACCAG R: GGCATCAGGCTCCACAGT | 58 °C |
| E-cadherin | F: CAGAAAGTTTTCCACCAAAG R: AAATGTGAGCAATTCTGCTT | 58 °C |
| Snail | F: CTTCCAGCAGCCCTACGAC R: CGGTGGGGTTGAGGATCT | 58 °C |
| Twist | F: AGCTACGCCTTCTCGGTCT R: CCTTCTCTGGAAACAATGACATC | 58 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Hajjar, L.; Saliba, J.; Karam, M.; Shaito, A.; El Hajj, H.; El-Sabban, M. Ubiquitin-Related Modifier 1 (URM-1) Modulates Cx43 in Breast Cancer Cell Lines. Int. J. Mol. Sci. 2023, 24, 2958. https://doi.org/10.3390/ijms24032958
El-Hajjar L, Saliba J, Karam M, Shaito A, El Hajj H, El-Sabban M. Ubiquitin-Related Modifier 1 (URM-1) Modulates Cx43 in Breast Cancer Cell Lines. International Journal of Molecular Sciences. 2023; 24(3):2958. https://doi.org/10.3390/ijms24032958
Chicago/Turabian StyleEl-Hajjar, Layal, Jessica Saliba, Mario Karam, Abdullah Shaito, Hiba El Hajj, and Marwan El-Sabban. 2023. "Ubiquitin-Related Modifier 1 (URM-1) Modulates Cx43 in Breast Cancer Cell Lines" International Journal of Molecular Sciences 24, no. 3: 2958. https://doi.org/10.3390/ijms24032958






