1. Introduction
Laccases are multi-copper oxidases that catalyze the oxidation of different substrates, such as
p-diphenols, while simultaneously reducing molecular oxygen to water. Laccases have been found in bacteria, fungi, plants, and insects. Laccases were studied by spectroscopic methods [
1], and then their structures were determined by X-ray crystallography [
2] (review of results up to 2015).
Laccases are globular proteins comprised of two or three domains of cupredoxin fold [
2]. Their active site includes four copper ions arranged in two separated centers: the T1 copper site and the trinuclear copper cluster (TNC). Three types of copper ions [
3] in laccases were well-established in earlier studies [
1,
4]. The TNC contains a triangle of closely located copper ions, including a pair of T3 copper ions coordinated by three histidine residues each, and one T2 copper ion coordinated by two histidine residues. Electrons are accepted from substrate at the T1 copper site. Via an electron-transfer chain electrons are transferred to the TNC where the molecular oxygen reduction reaction O
2 + 4e
− + 4H
+ → 2H
2O occurs [
1,
5]. The TNC is connected to the surrounding solvent via the T3 and T2 channels clearly observed in structures as chains of water molecules [
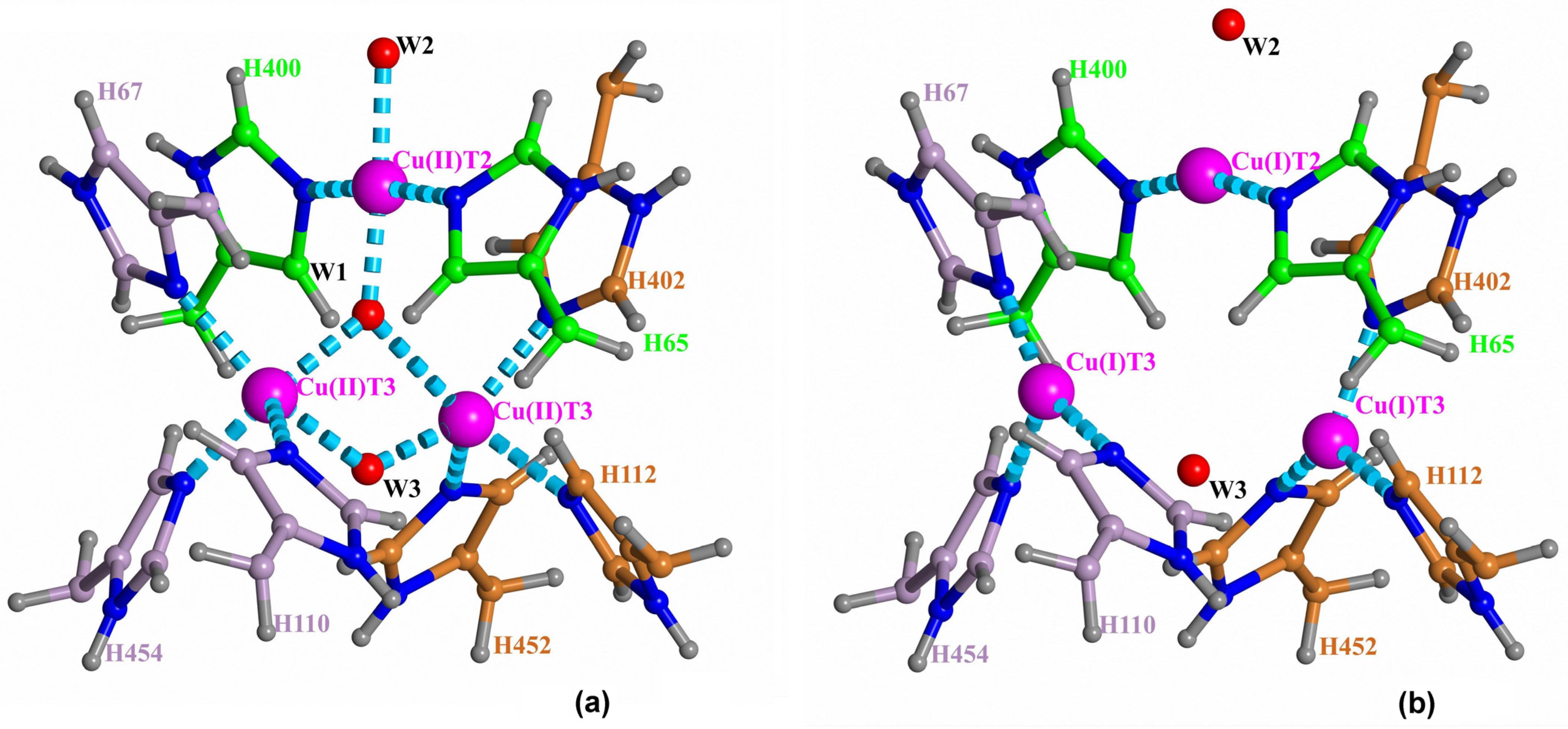
6] (
Figure 1). Molecular oxygen gets to the TNC through the T3 channel.
The molecular oxygen reduction at the TNC was considered in works by Solomon and coworkers [
4,
5] based on spectroscopy and early X-ray studies of ascorbate oxidase [
7]. It includes a couple of two-electron reduction stages. Among the TNC states at the oxygen reduction, the following ones can be distinguished. In the fully reduced state of the TNC, all three copper ions are reduced to charge +1e and two oxygen ligands are involved (one between two T3 copper ions outside TNC, and the other one near the T2 copper ion in channel T2). The peroxy intermediate (PI) state includes deprotonated hydrogen peroxide located inside the copper ion triangle. The T2 and one of the T3 copper ions are oxidized to Cu(II), and there is an oxygen ligand near the T2 copper ion. In the native intermediate (NI) state, all three copper ions are oxidized to charge +2e, and there are three oxygen ligands involved. One is the oxygen ligand near ion T2 in the channel, one is in the center of the copper ions triangle, and one is between two copper ions T3. The last two appear from peroxide when the O–O bond is broken. Each of the T3 copper ions is five-coordinated by three nitrogen atoms of histidine residues and by two oxygen ligands between the T3 copper ions in the TNC.
The fully reduced and NI states are clearly observed from X-ray data [
6], whereas the PI state has been deduced entirely from spectroscopic data. The features of this oxygen reduction scheme are the asymmetric location of the two oxygen ligands in respect to copper ions T3 in the PI state, and the involvement of proton transfers between an aspartic acid residue in the T2 channel and the oxygen ligand near the T2 copper ion. This leads to the appearance of a hydroxyl ion instead of a water molecule as the T2 copper coordinating oxygen ligand in channel T2 [
8]. The fully reduced state is stable, whereas the NI decays, but this process is very slow [
9].
Based on the above-mentioned scheme of molecular oxygen reduction in laccases [
5], studies were conducted using quantum mechanical (QM) and molecular mechanical methods [
10,
11,
12,
13]. Those QM calculations based on the density functional theory (DFT) agreed with the suggested reaction mechanism and demonstrated a strong dependence of obtained energies on the protonation state of the oxygen ligands coordinating the TNC copper ions [
12,
13]. However, the oxygen ligand transfer from the center of the trinuclear cluster to the copper coordinating position in the T2 channel [
12] is questionable. According to high-resolution X-ray data structures of fungal laccase [
6], surrounding residues involved in the coordination of the TNC copper ions leave no space for such a move because of steric restrictions in the trinuclear center.
Recently, there have been reported serial structures of laccase with different degrees of TNC copper ions reduction solved at high resolution for sets of X-ray data collected from one crystal [
6,
14]. These studies, as well as other reported results for serial crystallography [
15,
16], revealed a picture that contradicted, to some extent, the scheme of oxygen reduction suggested earlier. Due to the high quality of subatomic resolution data for sets of structures with different degrees of oxidation, both oxidized and reduced states of the TNC were resolved and clearly interpreted. As a result, it was possible to determine the coordination of the TNC copper ions in the oxidized (a) and reduced (b) states of the TNC (
Figure 1). There were two detected positions of each copper T3 ion corresponding to its Cu(I) or Cu(II) state. In state Cu(I), the copper ion is three-coordinated by three histidine residues. In the oxidized state, Cu(II) is five-coordinated by those histidine residues and two oxygen ligands. In the reduced state, the distance between the T3 ions is about 5.2 Å, and it is about 3.3 Å in the oxidized state. For the oxygen ligand coordinating the T2 copper ion in channel T2, there was detected switching between two ligand positions corresponding to the two states, which clearly indicates a change of the T2 copper ion coordination at its oxidation. In the reduced TNC state, Cu(I) ion T2 is linearly coordinated to nitrogen atoms of two surrounding histidine residues. The neighboring oxygen ligand in the T2 channel interacts with copper ion T2 via electrostatic forces. When an oxygen ligand is found at the center of the trinuclear cluster, the T2 Cu(II) ion has square-planar four-coordination with the addition of this oxygen ligand and the oxygen ligand in the T2 channel. As a result, the distance between the T2 copper ion and the oxygen ligand in the T2 channel diminishes.
Based on those serial X-ray data, some corrections to the oxygen reduction scheme were discussed [
6]. In this reaction model, an oxygen molecule, being an induced dipole attracted by the trinuclear cluster of ions Cu(I), penetrates between two copper ions T3, oxidizing both of them. The appearing PI state is symmetrical in respect to the five-coordinated T3 ions. It will wait for the next coming electron that reduces one of the T3 copper ions. The following cleavage of the peroxide O–O bond is caused by the oxidation of this copper ion and copper ion T2 by the two oxygen atoms and the change in coordinating the T3 and T2 copper ions. The final structure corresponds to the NI state of the TNC. The next reductions of TNC copper ions due to the electron supply release two oxygen ligands from the copper ion cluster, restoring the reduced state of the TNC. According to this scheme, the change of T2 copper ion coordination is accompanied by the cleavage of the peroxide O–O bond. In this scheme, all of the stable TNC states are quasi-symmetrical structures in respect to the pair of copper ions T3. Copper ion T2 is oxidized last, and there is no need in appearance of a hydroxyl ion in the T2 channel near the T2 copper ion.
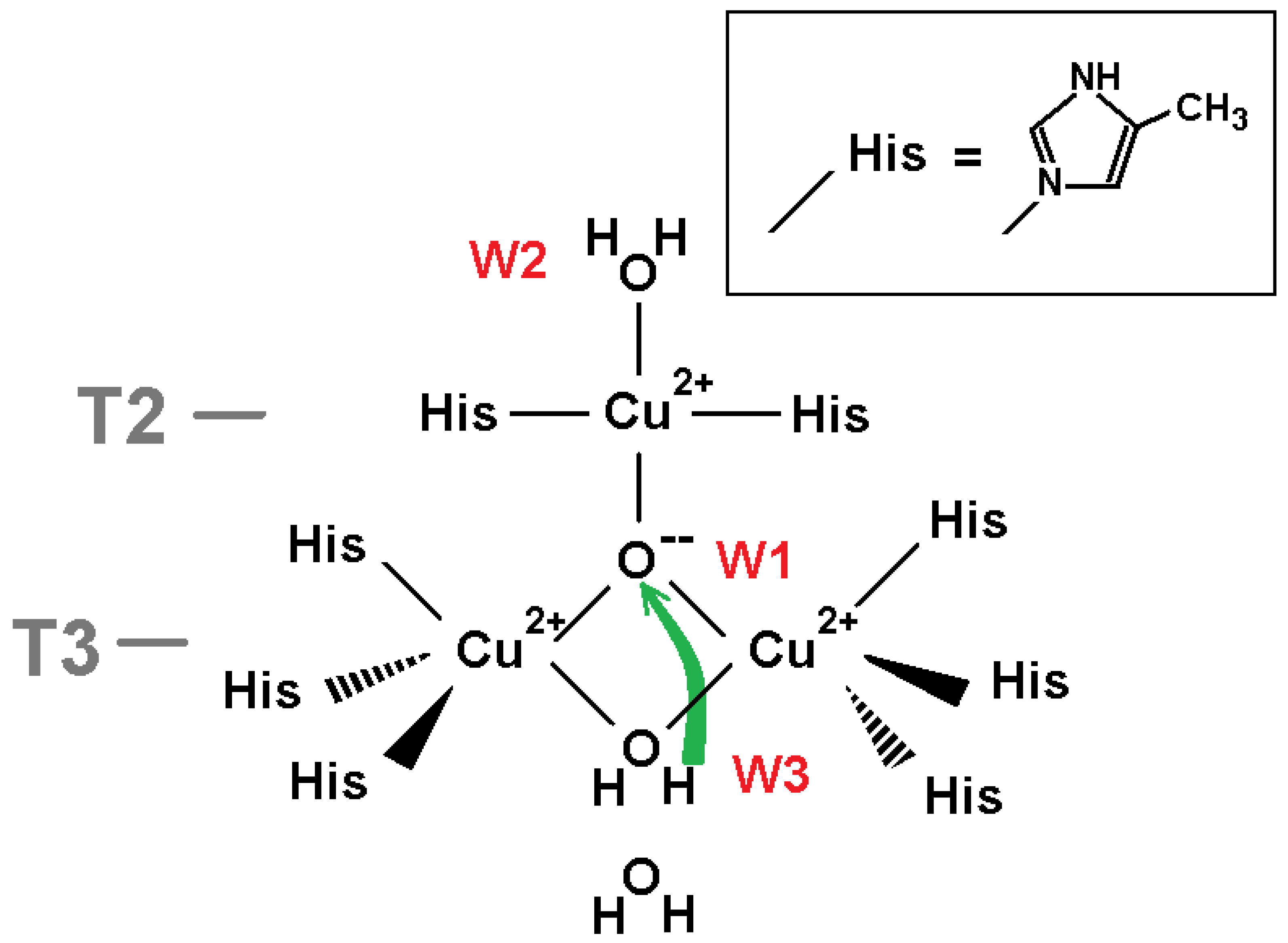
The oxidized (NI) and reduced states of the TNC were well-interpreted from the serial X-ray data. Positions of the TNC atoms and ligands in the oxidized state of the TNC (the native intermediate) are shown in
Figure 1a. The oxygen ligand in the center of the copper triangle is denoted as W1, the oxygen ligand in channel T2 as W2, and the oxygen ligand in channel T3 as W3. In the reduced state, ion Cu(I) T2 is linearly coordinated by two nitrogen atoms of the histidine residues, and oxygen ligand W2 is connected to the ion Cu(I) by only ion-dipole electrostatic interaction. In the fully oxidized state, position W2 is closer to the ion due to involvement of the oxygen ligand W2 into the T2 Cu(II) coordination. Positions Cu(I) T2 and Cu(II) T2 almost coincide.
In the previous quantum mechanical calculations for TNC models [
13], there was shown a strong dependence of the QM-calculated energies of states at cleavage of the O–O bond upon protonation of the oxygen ion in the center of the trinuclear cluster. It indicates an importance of true protonation states of the oxygen ligands involved. Protons cannot be resolved from a solution of X-ray data structures where the protonation of the oxygen ligands remains undetermined even at atomic resolution.
In the present work, DFT calculations have been applied for a study of the protonation of the TNC oxygen ligands (
Figure 1) for the observed stable oxidized (NI) and fully reduced states of TNC based on serial one-crystal X-ray data at subatomic resolution [
6,
14]. The QM calculations have also been supplemented by continuum-electrostatics calculations of energies of the proton migration in the outer parts of the laccase’s channels. Based on the determined protonation of the TNC oxygen ligands, the scheme of the oxygen reduction mechanism has been improved.
3. Discussion
The present work is a development of the study of molecular oxygen reduction in laccases [
6,
14]. The previous exploration was entirely based on X-ray mono-crystal serial data with an increasing absorbed radiation dose at subatomic resolution. Those studies allowed the process of molecular oxygen reduction in TNC to be traced, and the TNC structures in their two stable states were determined (NI and reduced states,
Figure 1). In the present work, the protonation of the laccase’s oxygen ligands in these two stable states is estimated via DFT calculations.
For QM calculations, the coordinates of non-hydrogen atoms were taken from X-ray data solved at subatomic resolution. Those X-ray data allowed us to assign the degree of oxidation to each copper ion position. It reduced QM optimization due to the fixation of copper ions and restrained the number of models studied compared to the previous research [
10].
The present QM and PB calculations together allow us to draw some conclusions about the protonation of the oxygen ligands involved into the oxygen reduction cycle in laccases. The pKa of dissociation of ligand will be pKa = −log(e)ΔG/RT, where ΔG (in kJ/mol) is a sum of the QM and estimated electrostatic energy changes at the proton transfer from infinity to ligand. According to the Henderson-Hasselbalch relation, for the dissociation of ligand in the active site pKa = pH − log([OH−]/[H2O]), where [OH−] and [H2O] are concentrations of the bound ligand in its deprotonated and protonated states, respectively. Then the protonated state occupancy will be η = [H2O]/([H2O] + [OH−]) = 1/(1 + 10pH-pKa). Using the definition of pH = −log(Np0/Nw0) and the relation between pKa and ΔG, the expression of η can be rewritten as η = α/(1 + α) where α = (Np0/Nw0)exp(−ΔG/RT). Here, Np0 is the hydroxonium ion number density in the bulk solution, Nw0 is the water molecule number density in the bulk solution, and ΔG is the free energy difference for the proton transfer from bulk to ligand. Assuming that laccase works in the acidic environment (pH = 5), the ratio Np0/Nw0 is 10−5. One can rewrite the expression of α as exp(−(ΔG0 + ΔG)/RT), where ΔG0 = −RT ln(Np0/Nw0). This value is +6.8 kcal/mol at temperature 298K and pH = 5. In other words, for the protonated state occupancy higher than 0.5, the energy of proton transfer from the bulk to this site must be lower than −6.8 kcal/mol at pH = 5 and even less at higher pH.
For the reduced state of TNC, both W2 and W3 oxygen ligands should be water molecules. Indeed, as follows from the QM and PB calculations, the rest of the transfer of a proton from position W3+2 to the hydroxyl ion at position W3 leads to the energy decrease of 31 kcal/mol (
Table 2), whereas a proton transfer from the bulk solution to position W3+2 rather decreases the system energy more (the maximal estimate of its possible increase does not exceed 4 kcal/mol) (PB calculations,
Table S1). Thus, the calculated total decrease of the energy at protonation of a hydroxyl ion at position W3 in the reduced state of TNC exceeds 27 kcal/mol. This value cannot be compensated neither by the external field neglected in the QM calculations (a few kcal/mol) nor by the Boltzmann factor for hydroxonium ions in the bulk (6.8 kcal/mol). A similar picture is observed in channel T2: the QM calculations give the energy decrease of 36 kcal/mol (
Table 3) and the preceding proton transfer from the bulk to position W2+2 (PB calculations) can diminish this value by no more than 4 kcal/mol (
Table S1). In other words, in the case of the reduced state of TNC, the entire proton migration from the bulk solution to the hydroxyl ion at position W2 in channel T2 is energetically quite favorable as well.
These results are in accordance with expectations. For oxygen ligand W3 as a hydroxyl ion, the induced polarization of an oxygen molecule would hardly be competitive with doubled energy of ionic pair OH−–Cu+ to approach the pair of copper ions T3. On the other hand, the competition between an induced dipole of molecular oxygen and a permanent dipole of a water molecule looks plausible enough.
For the oxidized state of TNC, the picture is more complicated. The oxygen ligand W3 in the NI state should be a hydroxyl ion. The proton transfer from the hydroxonium at position W3+1 to the hydroxyl ion at W3 increases energy by 6 kcal/mol as follows from the QM calculations (
Section 2.1.2,
Figure 3). The migration of a positive charge from bulk to position W3+2 in the T3 channel significantly increases this energy irrespective of oxygen ligand W2. Taking into account the Boltzmann factor for hydroxonium ions, one has to conclude that a hydroxyl ion at W3 cannot be protonated in the oxidized state of the TNC.
After the cleavage of the peroxide bond, a proton migration from the bulk to water at position W3+2 is quite favorable, according to the PB calculations (
Section 4). Finally, it leads to the further jump of a proton from hydroxonium ion at W2+2 to hydroxyl ion at W2+1. Thus, we conclude that there can easily be achieved a state of oxidized TNC when there are ion O
2− at position W1, OH
− at position W3, and water molecules as other oxygen ligands in channel T3.
The next step of oxygen ligands’ protonation in the oxidized state of the TNC is the second migration of a proton in channel T3 to get a water molecule instead a hydroxyl ion at position W3 when ion O
2− remains at position W1 (this is the starting configuration of the QM calculations described in
Section 2.1.1). I.e., there are ion O
2− at position W1, ion OH
− at position W3, and molecules H
2O at positions W3+1 and W3+2. The PB calculations indicate that a proton migration to position W3+2 meets a potential barrier in the channel despite the negatively charged aspartic acid Asp 456 on the way of the transfer. Even when we choose a hydroxyl ion at position W2 instead of the water molecule, the migration of a proton to position W3+2 can meet a barrier estimated from 1 to 9 kcal/mol (a more precise value is beyond the accuracy of our model). When we assume a more polarizable protein interior, this barrier is much lower (up to 2 kcal/mol,
Table S2). On the other hand, the rest of the proton transfer from position W3+2 to hydroxyl ion at position W3 decreases energy by 10 kcal/mol, according to the QM calculations (
Section 2.1.6,
Table 5). Thus, one may admit that there could appear the water molecule at position W3 needed for the proton transfer to the central oxygen ligand (
Figure 2), but it seems questionable at some possible values of the unknown model parameters.
Finally, we will consider proton transfers in channel T2 when the TNC is in its oxidized state. The results of the PB calculations for the proton transfer in channel T2 were obtained for a model of the oxidized TNC with a hydroxyl ion at the centre of the copper ions triangle. As follows from these data, the proton transfer to position W2+2 in the oxidized state of TNC increases the energy of the system by several kcal/mol. Again, the accuracy of the model is low, and the true value cannot be calculated. As follows from the QM calculations, a further transfer of a proton from a hydroxonium ion at position W2+2 to a hydroxyl ion at position W2 also slightly increases the energy (about 4 kcal/mol,
Table 4). Although the sum gives a growth of energy, the definite conclusion cannot be drawn. First, the lower limit of this sum is about 6 kcal/mol (
Table 4 and the second line of
Table S2). Second, at such low energies, the mean field of the protein environment (neglected in the QM calculations) could affect the result by values of comparable magnitude, decreasing the proton transfer energy up to negative values. One could only say that the water molecule at position W2 can lose its proton in the oxidized state of the TNC. In such a case, this proton will migrate in the channel T2 towards exit from the channel. In addition, we have only considered a case of hydroxyl ion at position W1. If ion O
2− is present there, such a loss of a proton looks much less probable due to additional attraction of the proton by this ion.
Thus, a determination of the degree of protonation of the oxygen ligand at position W2 remains questionable for the oxidized state of TNC. Experimental data rather suggest in favor of a water molecule there. It follows from the fact that halogen ions are bound at this position [
14], and they inhibit the molecular oxygen reduction. The plausible explanation is that their negative charge impedes the reduction of the T2 copper ion, whereas a water molecule does not [
14].
4. Materials and Methods
All initial coordinates of non-hydrogen atoms were taken from the 6RGP record of the Protein Data Bank [
14]. The 6RGP structure was solved at a subatomic resolution (0.97 Å) and presents clearly distinguishable oxidized and reduced enzyme forms (
Figure 1). To model the proton transfers along the distal parts of the water wires in both channels, the QM calculations included the TNC’s adjacent environment in a gas phase. Such an approach is possible due to the fact that the TNC is immersed into the hydrophobic core of the laccase macromolecule. In all models, there were included three TNC copper ions along with side chains of eight histidine residues and oxygen ligands coordinating them.
The chosen molecular model around the active site was a compromise between delivering a minimal adequate chemical environment and computational cost. The input structures were prepared in the following way. Initially, the 6RGP structure was imported into Maestro 11.8 graphical interface within Schrodinger 2018-4 software [
24]. For oxidized and reduced laccase forms, the corresponding copper and oxygen atom coordinates were chosen from X-ray data according to the analysis performed previously [
14]. Then, all hydrogens of the model were added, and bond orders were assigned employing the Protein Preparation Wizard and manually edited as necessary for the model chosen. In most cases, histidine residues were truncated to their side chains with beta carbons substituted by methyl groups. When proton transfers in the T2 or T3 channels were considered, the water-coordinating functional groups (C=O, NH, COO-) were also preserved, as well as other oxygen ligands. The side chains of amino acids, other than histidine, were removed and backbone fragments were capped (N-methylated C-termini and acetylated N-termini). Aspartatic side chains were truncated to acetates. To fill up the free valences where the backbone and side chain fragments were cut off, the hydrogens were added.
All the DFT calculations were performed with Gaussian 16 Revision B 01 [
25] software using TPSSh density functional (meta-hybrid density functional [
26,
27]). It performs well in describing reactions of transition metal systems [
28,
29]. The basis sets were composite: the Pople’s split-valence 3-21G* and 6-31G** for
p-elements and effective core potentials (ECP) for copper atoms [
30,
31]. The pseudopotentials and basis sets themselves were taken for 10 core electrons, multi-electron approximation, and completely relativistic (ECP10MDF) [
25,
32].
The positions of copper ions and most of the non-hydrogen atoms of residues were taken from the X-ray data structure and kept frozen, whereas all hydrogen atoms, oxygen ligands chosen for a particular model, and carbon atoms of capping methyl groups were free. The initial geometry optimization was carried out using smaller 3-21G* basis sets for s- and p-elements. There were calculated energy second derivatives for a final optimization that was done with a larger 6-31G** basis set. Geometry optimizations followed by frequency calculations to confirm the correctness of stationary points were found, as well as thermochemistry calculations, including zero-point energy.
Oxygen ligand protonation was evaluated via a comparison of the energies of a system with different protonation of oxygen ligands. The energy barrier of a proton transfer from the W3 to W1 oxygen ligands was also estimated. The potential barrier at the proton transfer was studied by means of determining the transition state using the quadratic synchronous transit 3 (QST3) algorithm [
17]. The transition state finding was verified by the intrinsic reaction coordinate (IRC) approach [
33].
The continuum-electrostatics calculations were applied to estimate the electrostatic energies of the proton transfer from infinity (~17 Å apart from the protein surface) to oxygen ligands surrounding the copper ions T2 and T3. The calculations were based on solutions of the Poisson-Boltzmann (PB) equation for an atom model (PDB entry 6RGP without hydrogen atoms) of the protein immersed into an electrolyte solution. The energy difference at the proton transfer was described as a difference of electrostatic energies for different positions of the hydroxonium ion, which was described as a spherical cation at positions of crystallization water inside the protein molecule or arbitrary located far away from the macromolecule. The PB finite-difference grid spacing was 0.75 Å. The 0.12 M monovalent electrolyte solution was used. Its dielectric constant was 78.5. The thickness of ion exclusion layer around the macromolecule atoms (and the hydroxonium cation) was 2.5 Å. The dielectric interface with solution was shifted 0.8 Å away from the VdW radius of atoms. The VdW radius of the hydroxonium cation was set to 1 Å, and also to 1.5 Å for comparison. The partial charges of the protein atoms were described as only charged groups, and also as partial charges from the Amber ff99SB force field [
34]. The PB calculations were done at a protein dielectric constant of 2, 4, and 10, as its true value is unknown (the value of 4 is regarded as the most plausible one). All PB calculations were carried out using the software developed by one of the authors of this article [
35].
5. Conclusions
The reported results are in accordance with the scheme of the molecular oxygen reduction discussed [
6]. That scheme was entirely deduced from interpretation of serial X-ray data. In the present work, we have confirmed/corrected what is suggested in the scheme protonation of oxygen ligands W1, W2, and W3 in the NI and reduced states of the TNC determined from X-ray data.
One definite correction is a hydroxyl ion instead of a water molecule at position W3 in the oxidized NI state. It just means that this ion will be protonated later when released from the T3 channel at the reduction of the TNC copper ions. It is shown that the central oxygen ligand W1 becomes a hydroxyl ion in the TNC oxidized state if oxygen ligand at position W3 has become a water molecule. However, the lack of accuracy for the estimation of W3 ligand protonation does not allow us to draw a definite conclusion (oxygen ligand W1 may remain ion O
2− in the NI state). What we can conclude is that after cleavage of the peroxide bond, one migration of a proton to oxygen ligand W3 takes place as was suggested in the scheme of the reaction [
6]. One may admit the second migration of a proton in the fully oxidized state of TNC. The third proton migration described in the published scheme [
6] is impossible.
Oxygen ligand W2 plays crucial role in the previously [
5] and recently [
6] suggested schemes of molecular oxygen reduction, but in the two schemes, this role is quite different in respect to the peroxide bond cleavage. It has been confirmed that this ligand is a water molecule in the fully reduced state of the TNC. For the oxidized TNC, the reported calculations do not allow us to determine the W2 protonation precisely. At least these results do not reject its form as a water molecule. Even admitting a loss of a proton by this molecule when the TNC copper ions get oxidized, the suggested reaction scheme will not be changed. The proton can just migrate along the T2 channel. When the copper ions get reduced, migration in the opposite direction must take place and the W2 ligand becomes a water molecule.
Anyhow, the intermediate and short-living PI state and the cleavage of the peroxide covalent bond require further QM studies.