PDIA3 Expression Is Altered in the Limbic Brain Regions of Triple-Transgenic Mouse Model of Alzheimer’s Disease
Abstract
1. Introduction
2. Results
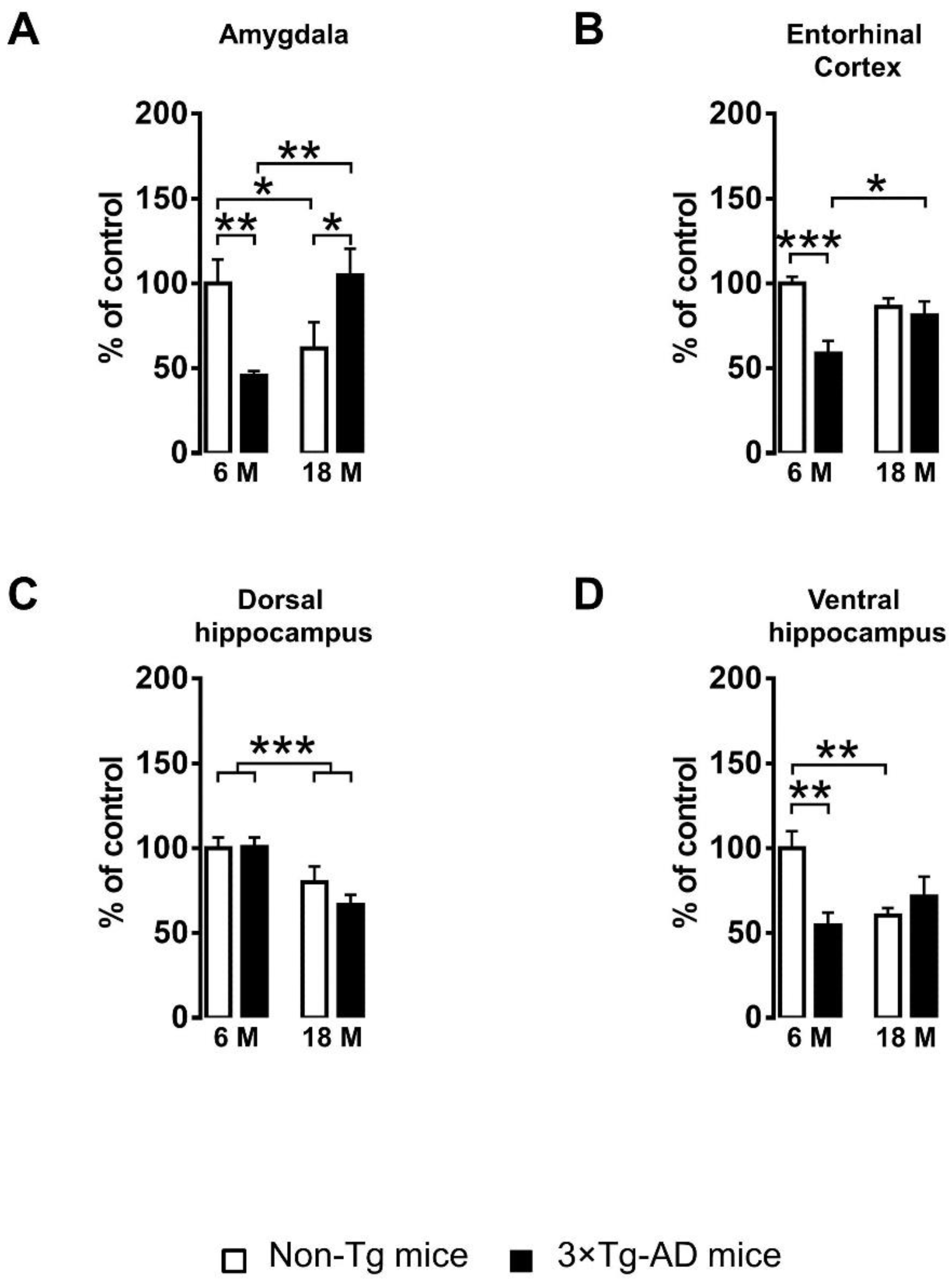
2.1. Alteration of PDIA3 Protein Expression in the Limbic Brain Regions
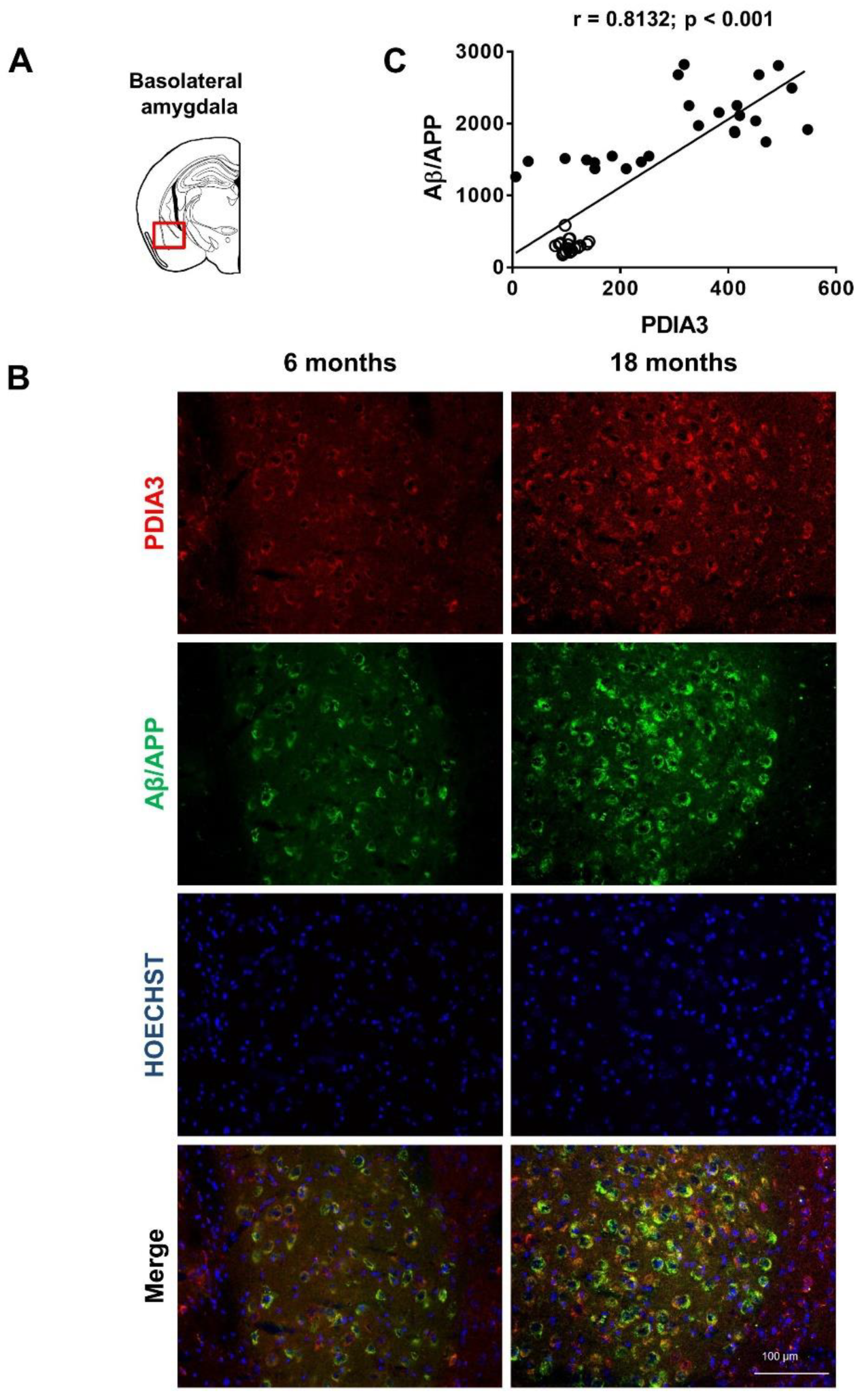
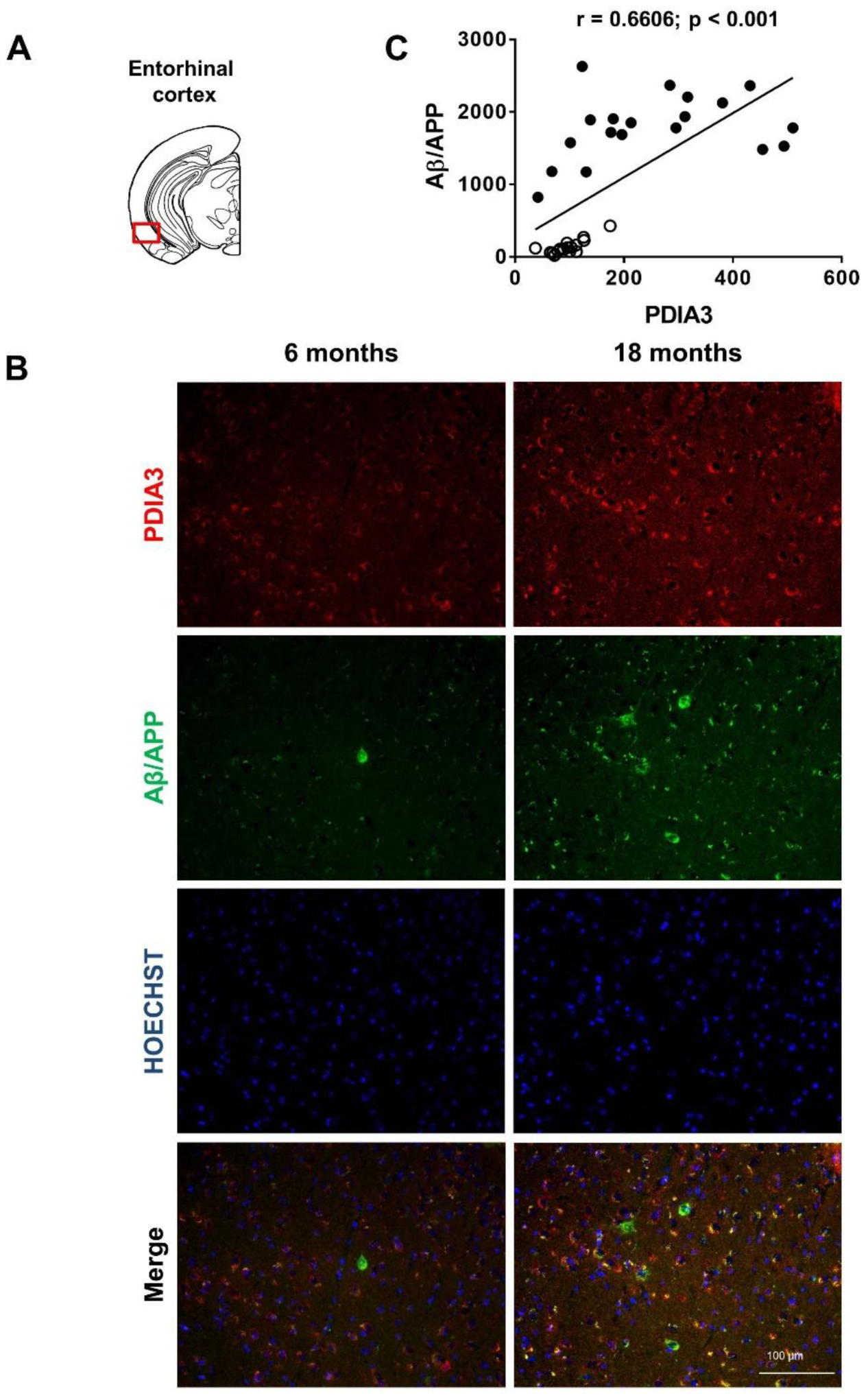
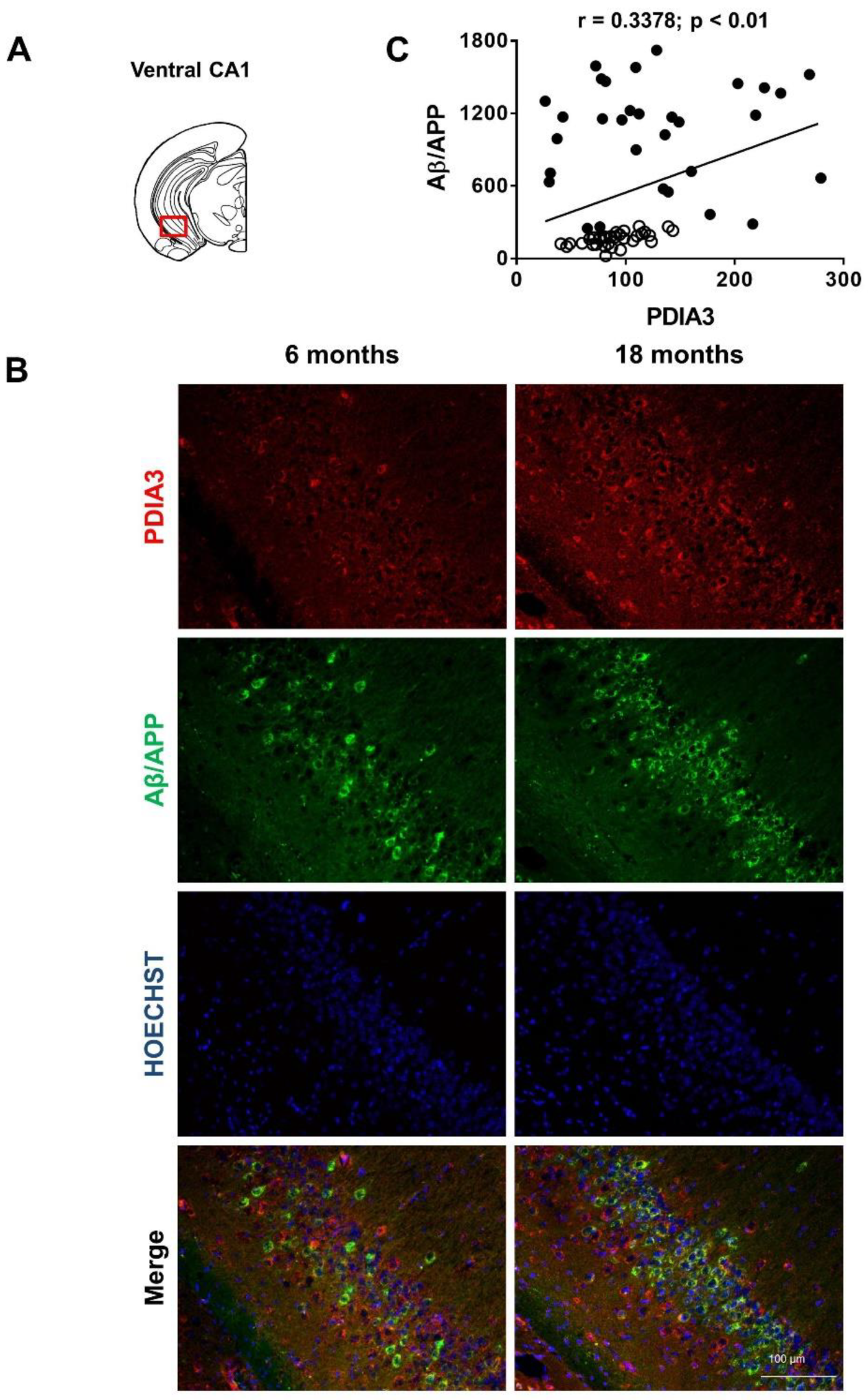
2.2. Aβ/APP-PDIA3 Double-Fluorescent Immunostaining
2.3. PDIA3-NeuN-GFAP Triple-Fluorescent Immunostaining
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Protein Isolation and Blotting Analysis
4.3. Double-Immunofluorescence
4.4. Triple-Immunofluorescence
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cummings, J.L. Alzheimer’s Disease. N. Engl. J. Med. 2004, 351, 56–67. [Google Scholar] [CrossRef]
- Hashimoto, S.; Saido, T.C. Critical review: Involvement of endoplasmic reticulum stress in the aetiology of Alzheimer’s disease. Open Biol. 2018, 8, 180024. [Google Scholar] [CrossRef] [PubMed]
- Alberdi, E.; Wyssenbach, A.; Alberdi, M.; Sánchez-Gómez, M.V.; Cavaliere, F.; Rodríguez, J.J.; Verkhratsky, A.; Matute, C. Ca2+-dependent endoplasmic reticulum stress correlates with astrogliosis in oligomeric amyloid β-treated astrocytes and in a model of Alzheimer’s disease. Aging Cell 2013, 12, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Nishitsuji, K.; Tomiyama, T.; Ishibashi, K.; Ito, K.; Teraoka, R.; Lambert, M.P.; Klein, W.L.; Mori, H. The E693Δ Mutation in Amyloid Precursor Protein Increases Intracellular Accumulation of Amyloid β Oligomers and Causes Endoplasmic Reticulum Stress-Induced Apoptosis in Cultured Cells. Am. J. Pathol. 2009, 174, 957–969. [Google Scholar] [CrossRef]
- Seyb, K.I.; Ansar, S.; Bean, J.; Michaelis, M.L. β-Amyloid and Endoplasmic Reticulum Stress Reponses in Primary Neurons: Effects of Drugs That Interact With the Cytoskeleton. J. Mol. Neurosci. 2006, 28, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.O.; Lacor, P.N.; Ferreira, I.L.; Resende, R.; Auberson, Y.P.; Klein, W.L.; Oliveira, C.R.; Rego, A.C.; Pereira, C.M.F. Endoplasmic reticulum stress occurs downstream of GluN2B subunit ofN-methyl-D-aspartate receptor in mature hippocampal cultures treated with amyloid-β oligomers. Aging Cell 2012, 11, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Hettinghouse, A.; Liu, R.; Liu, C.-J. Multifunctional molecule ERp57: From cancer to neurodegenerative diseases. Pharmacol. Ther. 2018, 181, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, M.; Rozas, P.; Hetz, C.; Medinas, D.B. ERp57 as a novel cellular factor controlling prion protein biosynthesis: Therapeutic potential of protein disulfide isomerases. Prion 2016, 10, 50–56. [Google Scholar] [CrossRef]
- Kim-Han, J.S.; O’Malley, K.L. Cell Stress Induced by the Parkinsonian Mimetic, 6-Hydroxydopamine, is Concurrent with Oxidation of the Chaperone, ERp57, and Aggresome Formation. Antioxid. Redox Signal. 2007, 9, 2255–2264. [Google Scholar] [CrossRef]
- Hurben, A.K.; Erber, L.N.; Tretyakova, N.Y.; Doran, T.M. Proteome-Wide Profiling of Cellular Targets Modified by Dopamine Metabolites Using a Bio-Orthogonally Functionalized Catecholamine. ACS Chem. Biol. 2021, 16, 2581–2594. [Google Scholar] [CrossRef]
- Erickson, R.R.; Dunning, L.M.; Olson, D.A.; Cohen, S.J.; Davis, A.T.; Wood, W.G.; Kratzke, R.A.; Holtzman, J.L. In cerebrospinal fluid ER chaperones ERp57 and calreticulin bind β-amyloid. Biochem. Biophys. Res. Commun. 2005, 332, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Chichiarelli, S.; Altieri, F.; Paglia, G.; Rubini, E.; Minacori, M.; Eufemi, M. ERp57/PDIA3: New insight. Cell. Mol. Biol. Lett. 2022, 27, 12. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, G.; Maattanen, P.; Schrag, J.D.; Pollock, S.; Cygler, M.; Nagar, B.; Thomas, D.; Gehring, K. Crystal Structure of the bb′ Domains of the Protein Disulfide Isomerase ERp57. Structure 2006, 14, 1331–1339. [Google Scholar] [CrossRef]
- Okuda, A.; Shimizu, M.; Morishima, K.; Inoue, R.; Sato, N.; Urade, R.; Sugiyama, M. Solution structure of multi-domain protein ER-60 studied by aggregation-free SAXS and coarse-grained-MD simulation. Sci. Rep. 2021, 11, 5655. [Google Scholar] [CrossRef]
- High, S.; LeComte, F.J.; Russell, S.J.; Abell, B.M.; Oliver, J.D. Glycoprotein folding in the endoplasmic reticulum: A tale of three chaperones? FEBS Lett. 2000, 476, 38–41. [Google Scholar] [CrossRef]
- Matsusaki, M.; Kanemura, S.; Kinoshita, M.; Lee, Y.-H.; Inaba, K.; Okumura, M. The Protein Disulfide Isomerase Family: From proteostasis to pathogenesis. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2019, 1864, 129338. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Wearsch, P.A.; Peaper, D.R.; Cresswell, P.; Reinisch, K.M. Insights into MHC Class I Peptide Loading from the Structure of the Tapasin-ERp57 Thiol Oxidoreductase Heterodimer. Immunity 2009, 30, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Fisette, O.; Schröder, G.F.; Schäfer, L.V. Atomistic structure and dynamics of the human MHC-I peptide-loading complex. Proc. Natl. Acad. Sci. USA 2020, 117, 20597–20606. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Sandoval, W.; Lam, C.; Haley, B.; Liu, P.; Xue, D.; Roy, D.; Patapoff, T.; Louie, S.; Snedecor, B.; et al. UBR E3 ligases and the PDIA3 protease control degradation of unfolded antibody heavy chain by ERAD. J. Cell Biol. 2020, 219, e201908087. [Google Scholar] [CrossRef]
- Jessop, C.E.; Chakravarthi, S.; Garbi, N.; Hämmerling, G.J.; Lovell, S.; Bulleid, N. ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. EMBO J. 2007, 26, 28–40. [Google Scholar] [CrossRef]
- Tang, Z.; Bereczki, E.; Zhang, H.; Wang, S.; Li, C.; Ji, X.; Branca, R.; Lehtiö, J.; Guan, Z.; Filipcik, P.; et al. Mammalian Target of Rapamycin (mTor) Mediates Tau Protein Dyshomeostasis. J. Biol. Chem. 2013, 288, 15556–15570. [Google Scholar] [CrossRef]
- Coppari, S.; Altieri, F.; Ferraro, A.; Chichiarelli, S.; Eufemi, M.; Turano, C. Nuclear localization and DNA interaction of protein disulfide isomerase ERp57 in mammalian cells. J. Cell. Biochem. 2002, 85, 325–333. [Google Scholar] [CrossRef]
- Nemere, I.; Garbi, N.; Hammerling, G.; Hintze, K.J. Role of the 1,25D3-MARRS receptor in the 1,25(OH)2D3-stimulated uptake of calcium and phosphate in intestinal cells. Steroids 2012, 77, 897–902. [Google Scholar] [CrossRef] [PubMed]
- Yavuz, U.; Alaylıoğlu, M.; Şengül, B.; Karras, S.N.; Gezen-Ak, D.; Dursun, E. Protein disulfide isomerase A3 might be involved in the regulation of 24-dehydrocholesterol reductase via vitamin D equilibrium in primary cortical neurons. In Vitro Cell. Dev. Biol.-Anim. 2021, 57, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Plácido, A.I.; Pereira, C.M.F.; Duarte, A.I.; Candeias, E.; Correia, S.C.; Santos, R.X.; Carvalho, C.; Cardoso, S.; Oliveira, C.R.; Moreira, P.I. The role of endoplasmic reticulum in amyloid precursor protein processing and trafficking: Implications for Alzheimer’s disease. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2014, 1842, 1444–1453. [Google Scholar] [CrossRef]
- Selivanova, A.; Winblad, B.; Dantuma, N.P.; Farmery, M.R. Biogenesis and processing of the amyloid precursor protein in the early secretory pathway. Biochem. Biophys. Res. Commun. 2007, 357, 1034–1039. [Google Scholar] [CrossRef]
- Medinas, D.B.; Malik, S.; Yıldız-Bölükbaşı, E.; Borgonovo, J.; Saaranen, M.J.; Urra, H.; Pulgar, E.; Afzal, M.; Contreras, D.; Wright, M.T.; et al. Mutation in protein disulfide isomerase A3 causes neurodevelopmental defects by disturbing endoplasmic reticulum proteostasis. EMBO J. 2021, 41, e105531. [Google Scholar] [CrossRef]
- Nakamura, T.; Lipton, S.A. Cell death: Protein misfolding and neurodegenerative diseases. Apoptosis 2009, 14, 455–468. [Google Scholar] [CrossRef]
- Yoo, D.Y.; Bin Cho, S.; Jung, H.Y.; Kim, W.; Lee, K.Y.; Kim, J.W.; Moon, S.M.; Won, M.-H.; Choi, J.H.; Yoon, Y.S.; et al. Protein disulfide-isomerase A3 significantly reduces ischemia-induced damage by reducing oxidative and endoplasmic reticulum stress. Neurochem. Int. 2019, 122, 19–30. [Google Scholar] [CrossRef]
- Uehara, T.; Nakamura, T.; Yao, D.; Shi, Z.-Q.; Gu, Z.; Ma, Y.; Masliah, E.; Nomura, Y.; Lipton, S.A. S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 2006, 441, 513–517. [Google Scholar] [CrossRef]
- Shen, L.; Chen, C.; Yang, A.; Chen, Y.; Liu, Q.; Ni, J. Redox proteomics identification of specifically carbonylated proteins in the hippocampi of triple transgenic Alzheimer’s disease mice at its earliest pathological stage. J. Proteom. 2015, 123, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Honjo, Y.; Ito, H.; Horibe, T.; Takahashi, R.; Kawakami, K. Protein disulfide isomerase-immunopositive inclusions in patients with Alzheimer disease. Brain Res. 2010, 1349, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Andreu, C.I.; Woehlbier, U.; Torres, M.; Hetz, C. Protein disulfide isomerases in neurodegeneration: From disease mechanisms to biomedical applications. FEBS Lett. 2012, 586, 2826–2834. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.-J.; Lee, H.-W.; Choi, J.-W.; Choi, M.-S. The Role of Protein S-Nitrosylation in Protein Misfolding-Associated Diseases. Life 2021, 11, 705. [Google Scholar] [CrossRef]
- Conway, M.E.; Eharris, M. S-nitrosylation of the thioredoxin-like domains of protein disulfide isomerase and its role in neurodegenerative conditions. Front. Chem. 2015, 3, 27. [Google Scholar] [CrossRef]
- Tohda, C.; Urano, T.; Umezaki, M.; Nemere, I.; Kuboyama, T. Diosgenin is an exogenous activator of 1,25D3-MARRS/Pdia3/ERp57 and improves Alzheimer’s disease pathologies in 5XFAD mice. Sci. Rep. 2012, 2, 535. [Google Scholar] [CrossRef]
- Lecanu, L.; Rammouz, G.; McCourty, A.; Sidahmed, E.; Greeson, J.; Papadopoulos, V. Caprospinol reduces amyloid deposits and improves cognitive function in a rat model of Alzheimer’s disease. Neuroscience 2010, 165, 427–435. [Google Scholar] [CrossRef]
- Yoo, Y.; Byun, K.; Kang, T.; Bayarsaikhan, D.; Kim, J.Y.; Oh, S.; Kim, Y.H.; Kim, S.-Y.; Chung, W.-I.; Kim, S.U.; et al. Amyloid-Beta-Activated Human Microglial Cells Through ER-Resident Proteins. J. Proteome Res. 2015, 14, 214–223. [Google Scholar] [CrossRef]
- Di Risola, D.; Ricci, D.; Marrocco, I.; Giamogante, F.; Grieco, M.; Francioso, A.; Vasco-Vidal, A.; Mancini, P.; Colotti, G.; Mosca, L.; et al. ERp57 chaperon protein protects neuronal cells from Aβ-induced toxicity. J. Neurochem. 2022, 162, 322–336. [Google Scholar] [CrossRef]
- Oddo, S.; Caccamo, A.; Shepherd, J.D.; Murphy, M.P.; Golde, T.E.; Kayed, R.; Metherate, R.; Mattson, M.P.; Akbari, Y.; LaFerla, F.M. Triple-Transgenic Model of Alzheimer’s Disease with Plaques and Tangles: Intracellular Abeta and Synaptic Dysfunction. Neuron 2003, 39, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Cassano, T.; Magini, A.; Giovagnoli, S.; Polchi, A.; Calcagnini, S.; Pace, L.; Lavecchia, M.A.; Scuderi, C.; Bronzuoli, M.R.; Ruggeri, L.; et al. Early intrathecal infusion of everolimus restores cognitive function and mood in a murine model of Alzheimer’s disease. Exp. Neurol. 2019, 311, 88–105. [Google Scholar] [CrossRef] [PubMed]
- Scuderi, C.; Bronzuoli, M.R.; Facchinetti, R.; Pace, L.; Ferraro, L.; Broad, K.D.; Serviddio, G.; Bellanti, F.; Palombelli, G.; Carpinelli, G.; et al. Ultramicronized palmitoylethanolamide rescues learning and memory impairments in a triple transgenic mouse model of Alzheimer’s disease by exerting anti-inflammatory and neuroprotective effects. Transl. Psychiatry 2018, 8, 32. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Pace, L.; Tempesta, B.; Lavecchia, A.M.; Macheda, T.; Bedse, G.; Petrella, A.; Cifani, C.; Serviddio, G.; Vendemiale, G.; et al. Depressive-Like Behavior Is Paired to Monoaminergic Alteration in a Murine Model of Alzheimer’s Disease. Int. J. Neuropsychopharmacol. 2014, 18, pyu020. [Google Scholar] [CrossRef] [PubMed]
- Cassano, T.; Serviddio, G.; Gaetani, S.; Romano, A.; Dipasquale, P.; Cianci, S.; Bellanti, F.; Laconca, L.; Romano, A.D.; Padalino, I.; et al. Glutamatergic alterations and mitochondrial impairment in a murine model of Alzheimer disease. Neurobiol. Aging 2012, 33, 1121.e1–1121.e12. [Google Scholar] [CrossRef] [PubMed]
- Cassano, T.; Romano, A.; Macheda, T.; Colangeli, R.; Cimmino, C.S.; Petrella, A.; LaFerla, F.M.; Cuomo, V.; Gaetani, S. Olfactory memory is impaired in a triple transgenic model of Alzheimer disease. Behav. Brain Res. 2011, 224, 408–412. [Google Scholar] [CrossRef]
- Bellanti, F.; Iannelli, G.; Blonda, M.; Tamborra, R.; Villani, R.; Romano, A.; Calcagnini, S.; Mazzoccoli, G.; Vinciguerra, M.; Gaetani, S.; et al. Alterations of Clock Gene RNA Expression in Brain Regions of a Triple Transgenic Model of Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 59, 615–631. [Google Scholar] [CrossRef] [PubMed]
- Landel, V.; Stephan, D.; Cui, X.; Eyles, D.; Feron, F. Differential expression of vitamin D-associated enzymes and receptors in brain cell subtypes. J. Steroid Biochem. Mol. Biol. 2018, 177, 129–134. [Google Scholar] [CrossRef]
- Lavezzi, A.M.; Corna, M.F.; Matturri, L. Neuronal nuclear antigen (NeuN): A useful marker of neuronal immaturity in sudden unexplained perinatal death. J. Neurol. Sci. 2013, 329, 45–50. [Google Scholar] [CrossRef]
- Montibeller, L.; de Belleroche, J. Amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease (AD) are characterised by differential activation of ER stress pathways: Focus on UPR target genes. Cell Stress Chaperones 2018, 23, 897–912. [Google Scholar] [CrossRef]
- Gezen-Ak, D.; Atasoy, I.L.; Candaş, E.; Alaylioglu, M.; Yılmazer, S.; Dursun, E. Vitamin D Receptor Regulates Amyloid Beta 1-42 Production with Protein Disulfide Isomerase A3. ACS Chem Neurosci. 2017, 8, 2335–2346. [Google Scholar] [CrossRef]
- Erickson, R.R.; Dunning, L.M.; Holtzman, J.L. The Effect of Aging on the Chaperone Concentrations in the Hepatic, Endoplasmic Reticulum of Male Rats: The Possible Role of Protein Misfolding Due to the Loss of Chaperones in the Decline in Physiological Function Seen With Age. J. Gerontol. Ser. A 2006, 61, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-W.; Thompson, R.; Zhang, H.; Xu, H. APP processing in Alzheimer’s disease. Mol. Brain 2011, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, N.; Suelves, N.; Perrin, F.; Vadukul, D.M.; Vrancx, C.; Constantinescu, S.N.; Kienlen-Campard, P. Structural Determinant of β-Amyloid Formation: From Transmembrane Protein Dimerization to β-Amyloid Aggregates. Biomedicines 2022, 10, 2753. [Google Scholar] [CrossRef] [PubMed]
- Fourriere, L.; Gleeson, P.A. Amyloid β production along the neuronal secretory pathway: Dangerous liaisons in the Golgi? Traffic 2021, 22, 319–327. [Google Scholar] [CrossRef]
- Nowakowska-Gołacka, J.; Czapiewska, J.; Sominka, H.; Sowa-Rogozińska, N.; Słomińska-Wojewódzka, M. EDEM1 Regulates Amyloid Precursor Protein (APP) Metabolism and Amyloid-β Production. Int. J. Mol. Sci. 2021, 23, 117. [Google Scholar] [CrossRef]
- Klaips, C.L.; Jayaraj, G.G.; Hartl, F.U. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 2017, 217, 51–63. [Google Scholar] [CrossRef]
- Samant, R.S.; Livingston, C.M.; Sontag, E.M.; Frydman, J. Distinct proteostasis circuits cooperate in nuclear and cytoplasmic protein quality control. Nature 2018, 563, 407–411. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Buchberger, A.; Bukau, B.; Sommer, T. Protein Quality Control in the Cytosol and the Endoplasmic Reticulum: Brothers in Arms. Mol. Cell 2010, 40, 238–252. [Google Scholar] [CrossRef]
- Hipp, M.S.; Park, S.-H.; Hartl, F.U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014, 24, 506–514. [Google Scholar] [CrossRef]
- Holtzman, J.L. The roles of the thiol: Protein disulfide oxidoreductases in membrane and secretory protein synthesis within the lumen of the endoplasmic reticulum. J. Investig. Med. 1997, 45, 28–34. [Google Scholar]
- Khodagholi, F.; Digaleh, H.; Motamedi, F.; Foolad, F.; Shaerzadeh, F. Nitric Oxide and Protein Disulfide Isomerase Explain the Complexities of Unfolded Protein Response Following Intra-hippocampal Aβ Injection. Cell. Mol. Neurobiol. 2016, 36, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, M.; Oddo, S.; Yamasaki, T.R.; Green, K.N.; LaFerla, F.M. Lipopolysaccharide-Induced Inflammation Exacerbates Tau Pathology by a Cyclin-Dependent Kinase 5-Mediated Pathway in a Transgenic Model of Alzheimer’s Disease. J. Neurosci. 2005, 25, 8843–8853. [Google Scholar] [CrossRef] [PubMed]
- Barone, E.; Di Domenico, F.; Cassano, T.; Arena, A.; Tramutola, A.; Lavecchia, M.A.; Coccia, R.; Butterfield, D.A.; Perluigi, M. Impairment of biliverdin reductase-A promotes brain insulin resistance in Alzheimer disease: A new paradigm. Free. Radic. Biol. Med. 2016, 91, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Cassano, T.; Calcagnini, S.; Pace, L.; De Marco, F.; Romano, A.; Gaetani, S. Cannabinoid Receptor 2 Signaling in Neurodegenerative Disorders: From Pathogenesis to a Promising Therapeutic Target. Front. Neurosci. 2017, 11, 30. [Google Scholar] [CrossRef]
- Di Domenico, F.; Barone, E.; Perluigi, M.; Butterfield, D.A.; Reeder, B.J.; Tramutola, A.; Arena, A.; Giorgi, A.; di Francesco, L.; Schininà, M.E.; et al. The Triangle of Death in Alzheimer’s Disease Brain: The Aberrant Cross-Talk Among Energy Metabolism, Mammalian Target of Rapamycin Signaling, and Protein Homeostasis Revealed by Redox Proteomics. Antioxid. Redox Signal. 2017, 26, 364–387. [Google Scholar] [CrossRef]
- Crino, P.B. The mTOR signalling cascade: Paving new roads to cure neurological disease. Nat. Rev. Neurol. 2016, 12, 379–392. [Google Scholar] [CrossRef]
- An, W.-L.; Cowburn, R.F.; Li, L.; Braak, H.; Alafuzoff, I.; Iqbal, K.; Iqbal, I.-G.; Winblad, B.; Pei, J.-J. Up-Regulation of Phosphorylated/Activated p70 S6 Kinase and Its Relationship to Neurofibrillary Pathology in Alzheimer’s Disease. Am. J. Pathol. 2003, 163, 591–607. [Google Scholar] [CrossRef] [PubMed]
- Talboom, J.S.; Velazquez, R.; Oddo, S. The mammalian target of rapamycin at the crossroad between cognitive aging and Alzheimer’s disease. NPJ Aging Mech. Dis. 2015, 1, 15008. [Google Scholar] [CrossRef]
- Caccamo, A.; Majumder, S.; Richardson, A.; Strong, R.; Oddo, S. Molecular Interplay between Mammalian Target of Rapamycin (mTOR), Amyloid-β, and Tau. J. Biol. Chem. 2010, 285, 13107–13120. [Google Scholar] [CrossRef]
- Caccamo, A.; Maldonado, M.A.; Majumder, S.; Medina, D.X.; Holbein, W.; Magrí, A.; Oddo, S. Naturally Secreted Amyloid-β Increases Mammalian Target of Rapamycin (mTOR) Activity via a PRAS40-mediated Mechanism. J. Biol. Chem. 2011, 286, 8924–8932. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.-B.; Zhang, X.; Sun, J.; Bennink, J.R.; Yewdell, J.W.; Patterson, C. mTORC1 Links Protein Quality and Quantity Control by Sensing Chaperone Availability. J. Biol. Chem. 2010, 285, 27385–27395. [Google Scholar] [CrossRef]
- Ramírez-Rangel, I.; Bracho-Valdés, I.; Vázquez-Macías, A.; Carretero-Ortega, J.; Reyes-Cruz, G.; Vázquez-Prado, J. Regulation of mTORC1 Complex Assembly and Signaling by GRp58/ERp57. Mol. Cell. Biol. 2011, 31, 1657–1671. [Google Scholar] [CrossRef] [PubMed]
- Beynon-Jones, S.M.; Antoniou, A.N.; Powis, S.J. Mutational analysis of the oxidoreductase ERp57 reveals the importance of the two central residues in the redox motif. FEBS Lett. 2006, 580, 1897–1902. [Google Scholar] [CrossRef] [PubMed]
- Onisko, B.C. The Hydroxyproline Proteome of HeLa Cells with Emphasis on the Active Sites of Protein Disulfide Isomerases. J. Proteome Res. 2020, 19, 756–768. [Google Scholar] [CrossRef]
- Satoh, M.; Shimada, A.; Kashiwai, A.; Saga, S.; Hosokawa, M. Differential cooperative enzymatic activities of protein disulfide isomerase family in protein folding. Cell Stress Chaperones 2005, 10, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Sabatini, D.M. Redox Regulation of the Nutrient-sensitive Raptor-mTOR Pathway and Complex. J. Biol. Chem. 2005, 280, 39505–39509. [Google Scholar] [CrossRef]
- Neklesa, T.K.; Davis, R.W. Superoxide anions regulate TORC1 and its ability to bind Fpr1:rapamycin complex. Proc. Natl. Acad. Sci. USA 2008, 105, 15166–15171. [Google Scholar] [CrossRef]
- Hetz, C.; Saxena, S. ER stress and the unfolded protein response in neurodegeneration. Nat. Rev. Neurol. 2017, 13, 477–491. [Google Scholar] [CrossRef]
- Shin, Y.; Cho, H.; Choi, B.Y.; Kim, J.; Ha, J.; Suh, S.W.; Park, S.B. Phenotypic Discovery of Neuroprotective Agents by Regulation of Tau Proteostasis via Stress-Responsive Activation of PERK Signaling. Angew. Chem. Int. Ed. 2021, 60, 1831–1838. [Google Scholar] [CrossRef]
- Morales, I.; Guzmã¡n-Martã nez, L.; Cerda-Troncoso, C.; Farías, G.A.; Maccioni, R.B. Neuroinflammation in the pathogenesis of Alzheimer’s disease. A rational framework for the search of novel therapeutic approaches. Front. Cell. Neurosci. 2014, 8, 112. [Google Scholar] [CrossRef] [PubMed]
- Cassano, T.; Gaetani, S.; Morgese, M.G.; Macheda, T.; Laconca, L.; DiPasquale, P.; Taltavull, J.; Shippenberg, T.S.; Cuomo, V.; Gobbi, G. Monoaminergic Changes in Locus Coeruleus and Dorsal Raphe Nucleus Following Noradrenaline Depletion. Neurochem. Res. 2009, 34, 1417–1426. [Google Scholar] [CrossRef]
- Höhn, A.; König, J.; Grune, T. Protein oxidation in aging and the removal of oxidized proteins. J. Proteom. 2013, 92, 132–159. [Google Scholar] [CrossRef] [PubMed]
- Gruneab, T.; Reinheckelac, T.; Lia, R.; North, J.A.; Davies, K.J. Proteasome-Dependent Turnover of Protein Disulfide Isomerase in Oxidatively Stressed Cells. Arch. Biochem. Biophys. 2002, 397, 407–413. [Google Scholar] [CrossRef]
- Conn, C.S.; Qian, S.-B. mTOR signaling in protein homeostasis. Cell Cycle 2011, 10, 1940–1947. [Google Scholar] [CrossRef] [PubMed]
- Kranz, P.; Neumann, F.; Wolf, A.; Classen, F.; Pompsch, M.; Ocklenburg, T.; Baumann, J.; Janke, K.; Baumann, M.; Goepelt, K.; et al. PDI is an essential redox-sensitive activator of PERK during the unfolded protein response (UPR). Cell Death Dis. 2017, 8, e2986. [Google Scholar] [CrossRef]
- Giamogante, F.; Marrocco, I.; Cervoni, L.; Eufemi, M.; Chichiarelli, S.; Altieri, F. Punicalagin, an active pomegranate component, is a new inhibitor of PDIA3 reductase activity. Biochimie 2018, 147, 122–129. [Google Scholar] [CrossRef]
- Kaneya, Y.; Takata, H.; Wada, R.; Kure, S.; Ishino, K.; Kudo, M.; Kondo, R.; Taniai, N.; Ohashi, R.; Yoshida, H.; et al. Inhibitor for protein disulfide-isomerase family A member 3 enhances the antiproliferative effect of inhibitor for mechanistic target of rapamycin in liver cancer: An in vitro study on combination treatment with everolimus and 16F16. Oncol. Lett. 2021, 21, 1. [Google Scholar] [CrossRef]

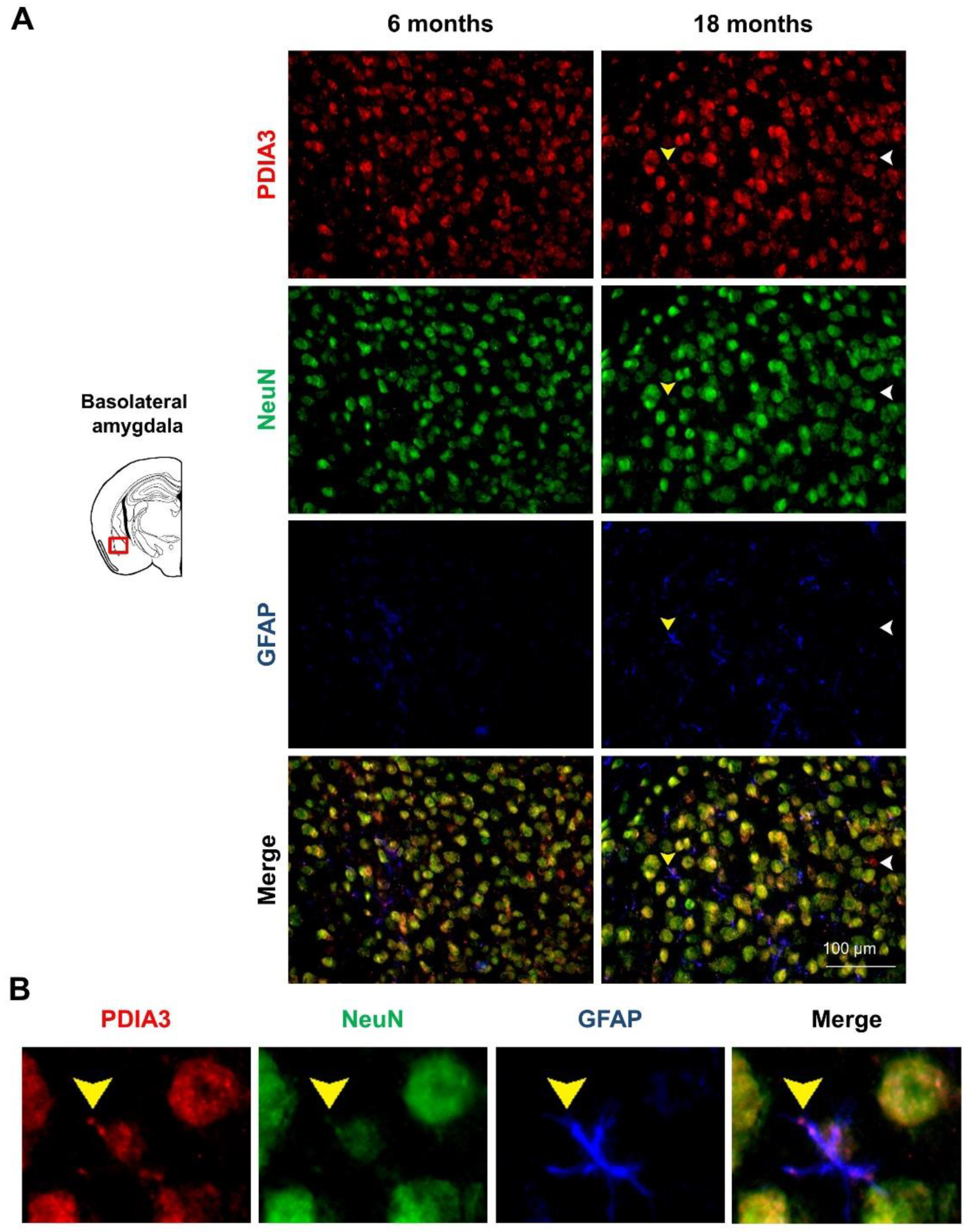
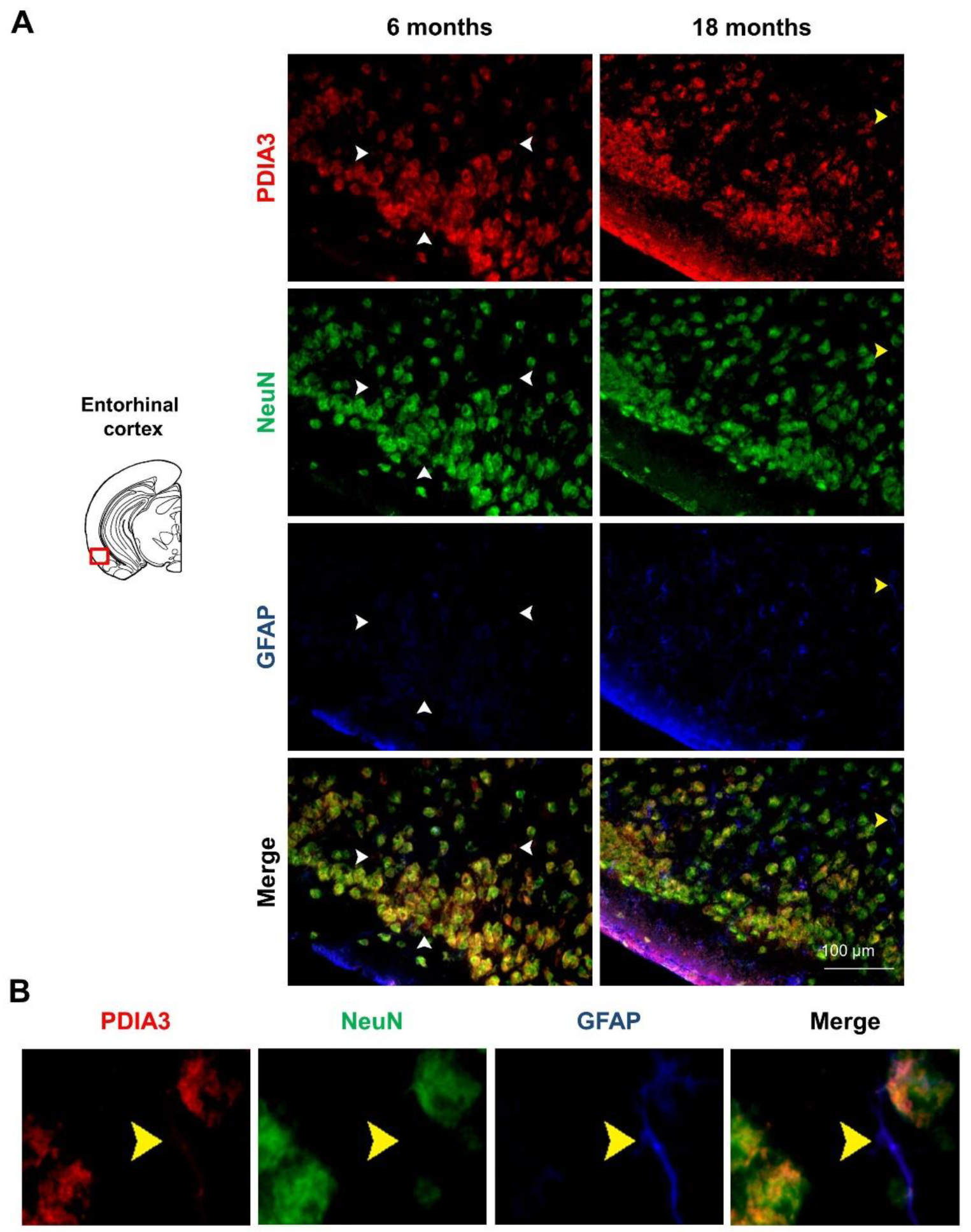

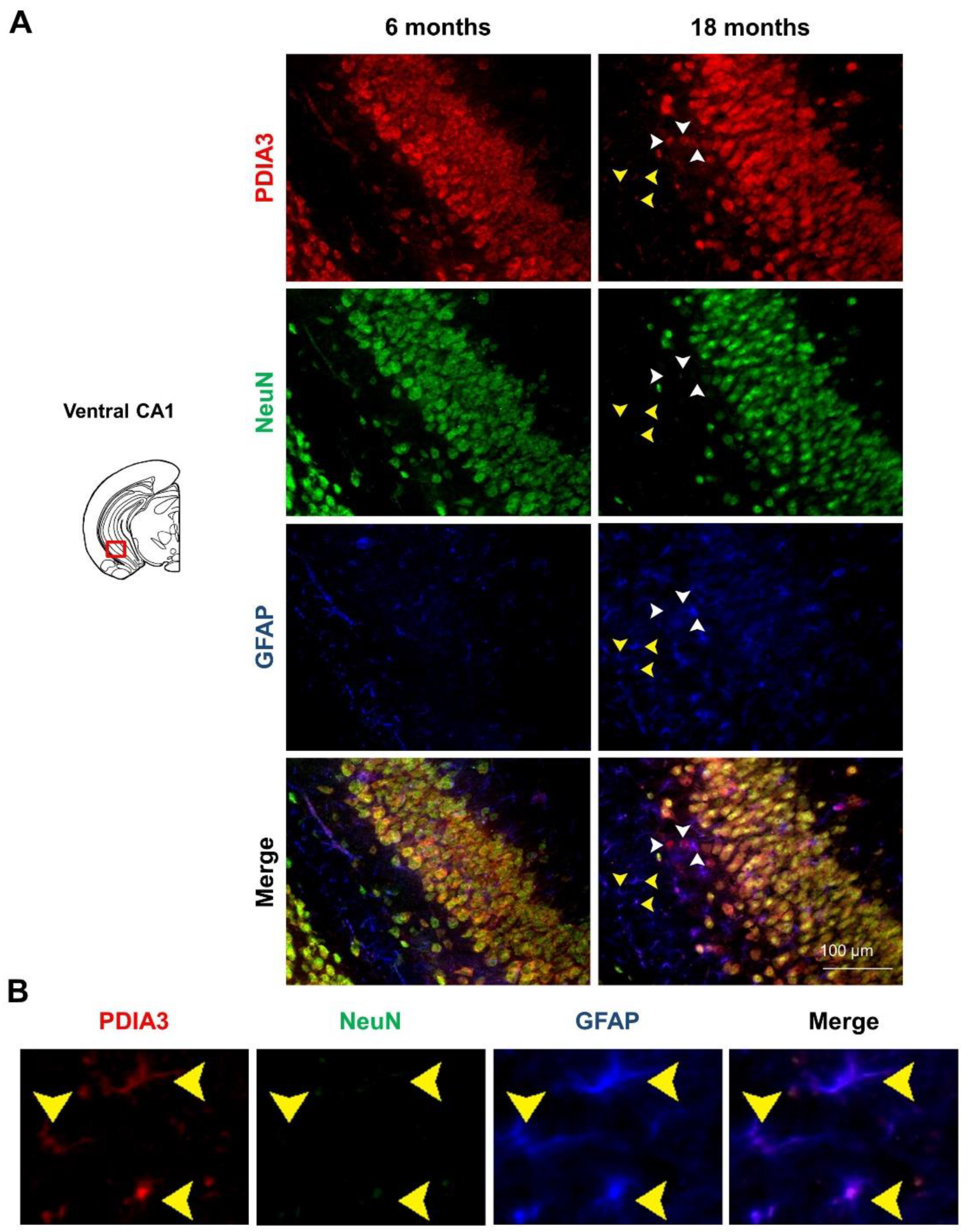
| Brain Region | Genotype (G) | Age (A) | Interaction (G × A) |
|---|---|---|---|
| Amygdala | F(1,35) = 0.192, n.s. | F(1,35) = 0.622, n.s. | F(1,35) = 13.657, p < 0.001 |
| Entorhinal cortex | F(1,35) = 12.781, p < 0.01 | F(1,35) = 0.433, n.s. | F(1,35) = 7.787, p < 0.01 |
| Dorsal hippocampus | F(1,35) = 0.826, n.s. | F(1,35) = 14.961, p < 0.001 | F(1,35) = 1.013, n.s. |
| Ventral hippocampus | F(1,35) = 3.789, n.s. | F(1,35) = 1.619, n.s. | F(1,35) = 10.287, p < 0.01 |
| Amygdala | Entorhinal Cortex | Dorsal Hippocampus | Ventral Hippocampus | |
|---|---|---|---|---|
| Pearson correlation coefficient (r) | 0.8132 | 0.6606 | 0.8457 | 0.3378 |
| p value | <0.001 | <0.001 | <0.001 | <0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cassano, T.; Giamogante, F.; Calcagnini, S.; Romano, A.; Lavecchia, A.M.; Inglese, F.; Paglia, G.; Bukke, V.N.; Romano, A.D.; Friuli, M.; et al. PDIA3 Expression Is Altered in the Limbic Brain Regions of Triple-Transgenic Mouse Model of Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 3005. https://doi.org/10.3390/ijms24033005
Cassano T, Giamogante F, Calcagnini S, Romano A, Lavecchia AM, Inglese F, Paglia G, Bukke VN, Romano AD, Friuli M, et al. PDIA3 Expression Is Altered in the Limbic Brain Regions of Triple-Transgenic Mouse Model of Alzheimer’s Disease. International Journal of Molecular Sciences. 2023; 24(3):3005. https://doi.org/10.3390/ijms24033005
Chicago/Turabian StyleCassano, Tommaso, Flavia Giamogante, Silvio Calcagnini, Adele Romano, Angelo Michele Lavecchia, Francesca Inglese, Giuliano Paglia, Vidyasagar Naik Bukke, Antonino Davide Romano, Marzia Friuli, and et al. 2023. "PDIA3 Expression Is Altered in the Limbic Brain Regions of Triple-Transgenic Mouse Model of Alzheimer’s Disease" International Journal of Molecular Sciences 24, no. 3: 3005. https://doi.org/10.3390/ijms24033005
APA StyleCassano, T., Giamogante, F., Calcagnini, S., Romano, A., Lavecchia, A. M., Inglese, F., Paglia, G., Bukke, V. N., Romano, A. D., Friuli, M., Altieri, F., & Gaetani, S. (2023). PDIA3 Expression Is Altered in the Limbic Brain Regions of Triple-Transgenic Mouse Model of Alzheimer’s Disease. International Journal of Molecular Sciences, 24(3), 3005. https://doi.org/10.3390/ijms24033005









