Prunus Knotted-like Genes: Genome-Wide Analysis, Transcriptional Response to Cytokinin in Micropropagation, and Rootstock Transformation
Abstract
1. Introduction
2. Results
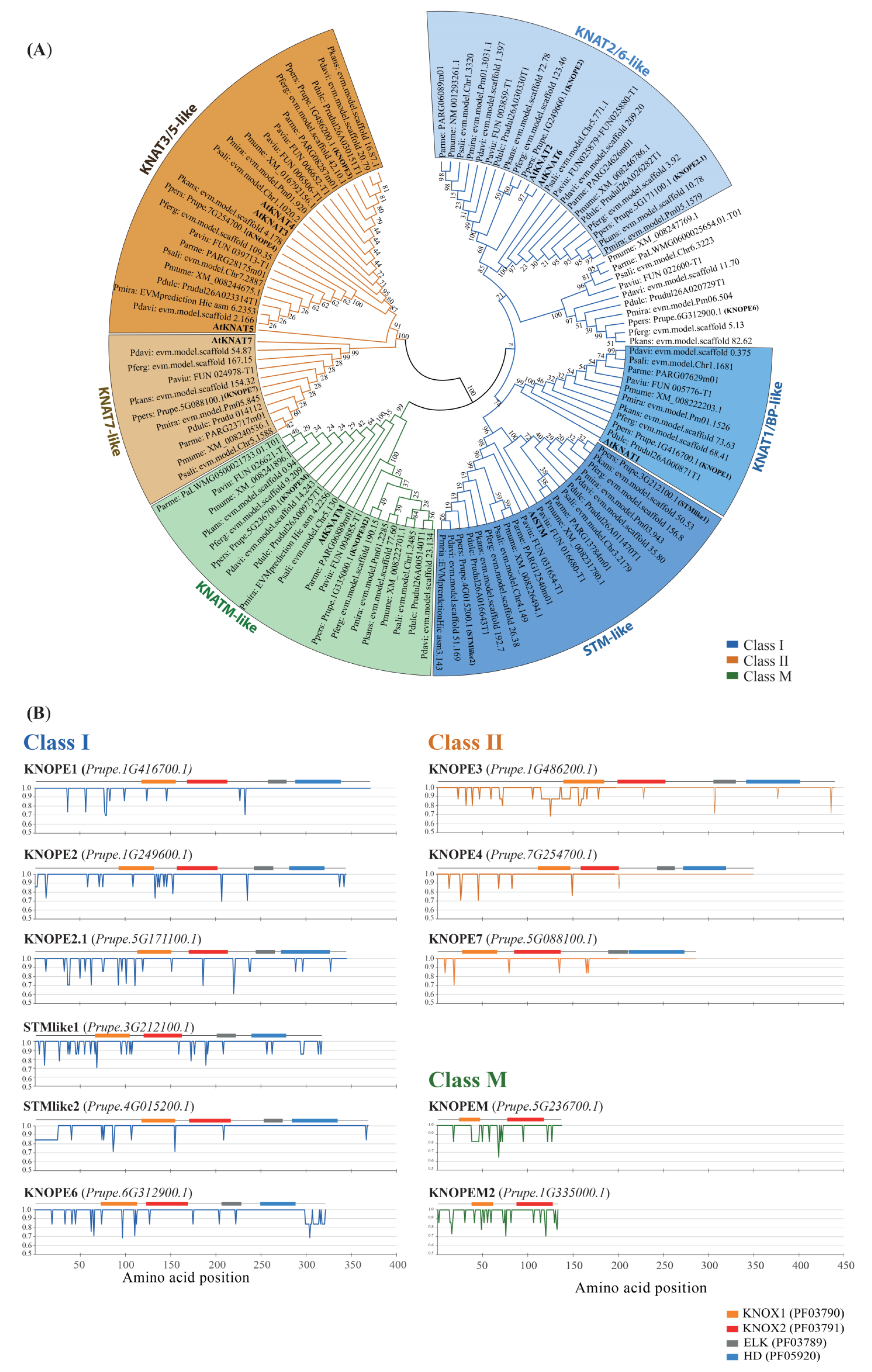
2.1. Genome-Wide Characterization of KNOX from Prunus Diploid Fruit Trees
2.1.1. Protein Features
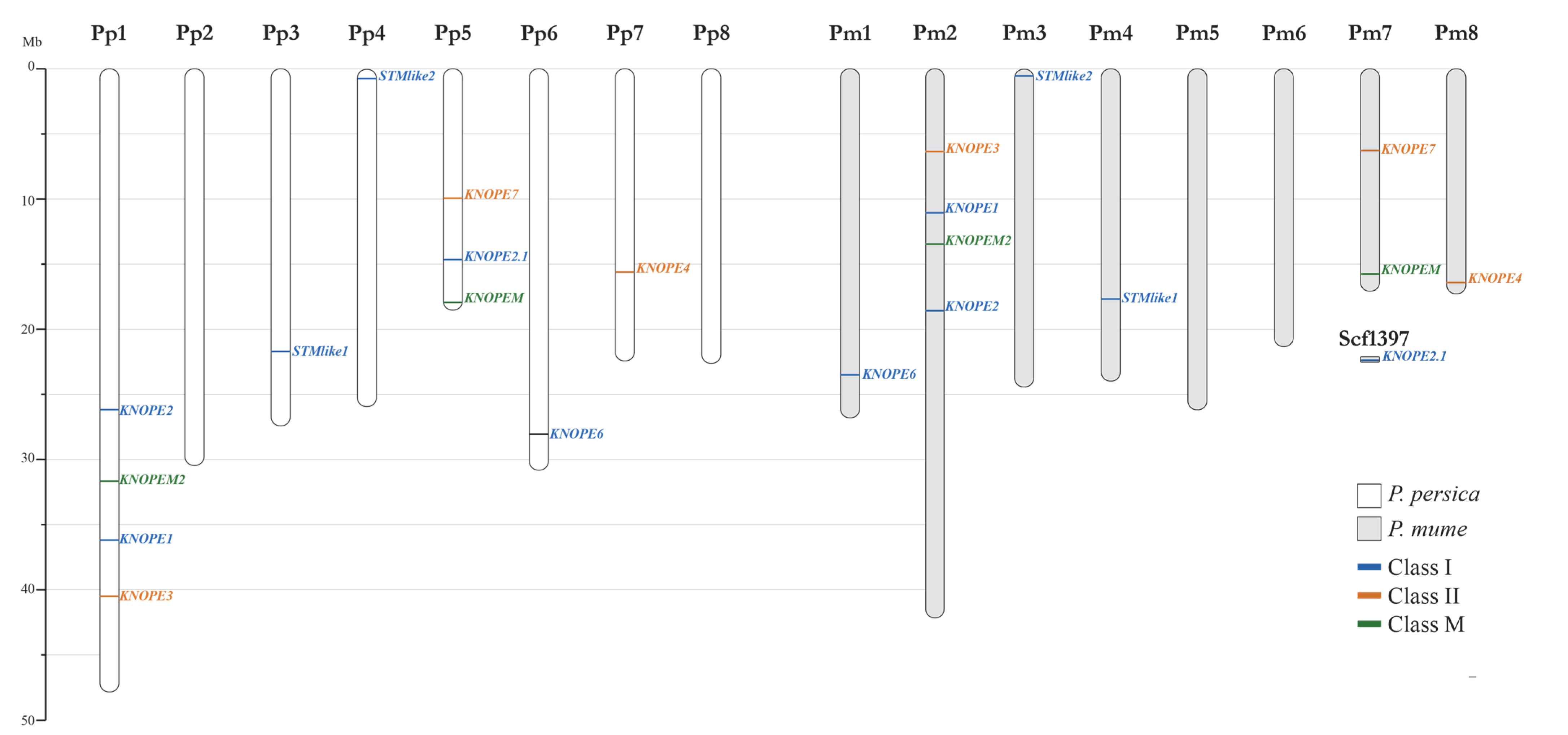
2.1.2. Genomic Features
2.1.3. Transcriptomic Features
2.2. PRUKNOX Responses to Cytokinin in GF677 Rootstock Microcuttings
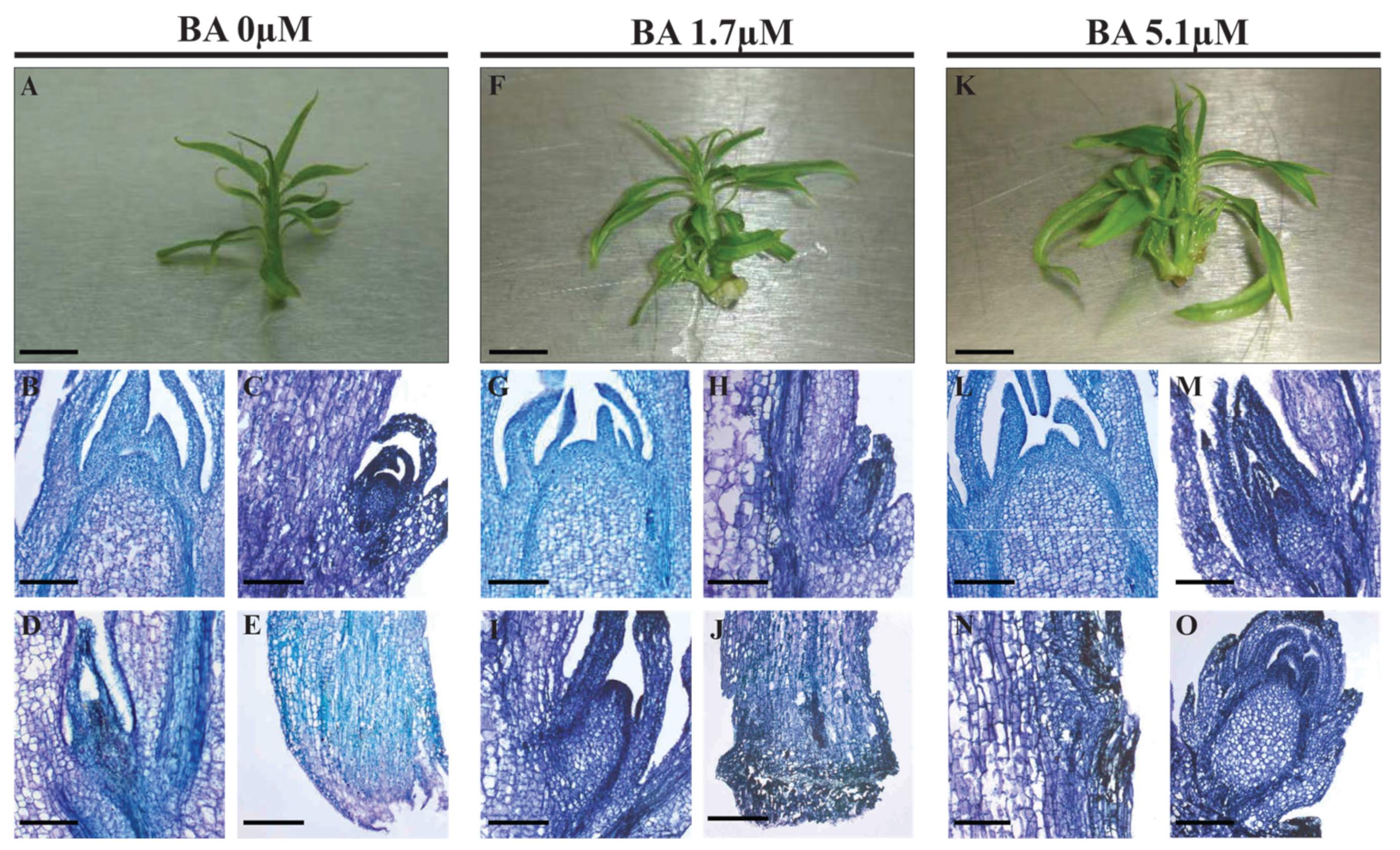
2.2.1. Pheno-Histological Features
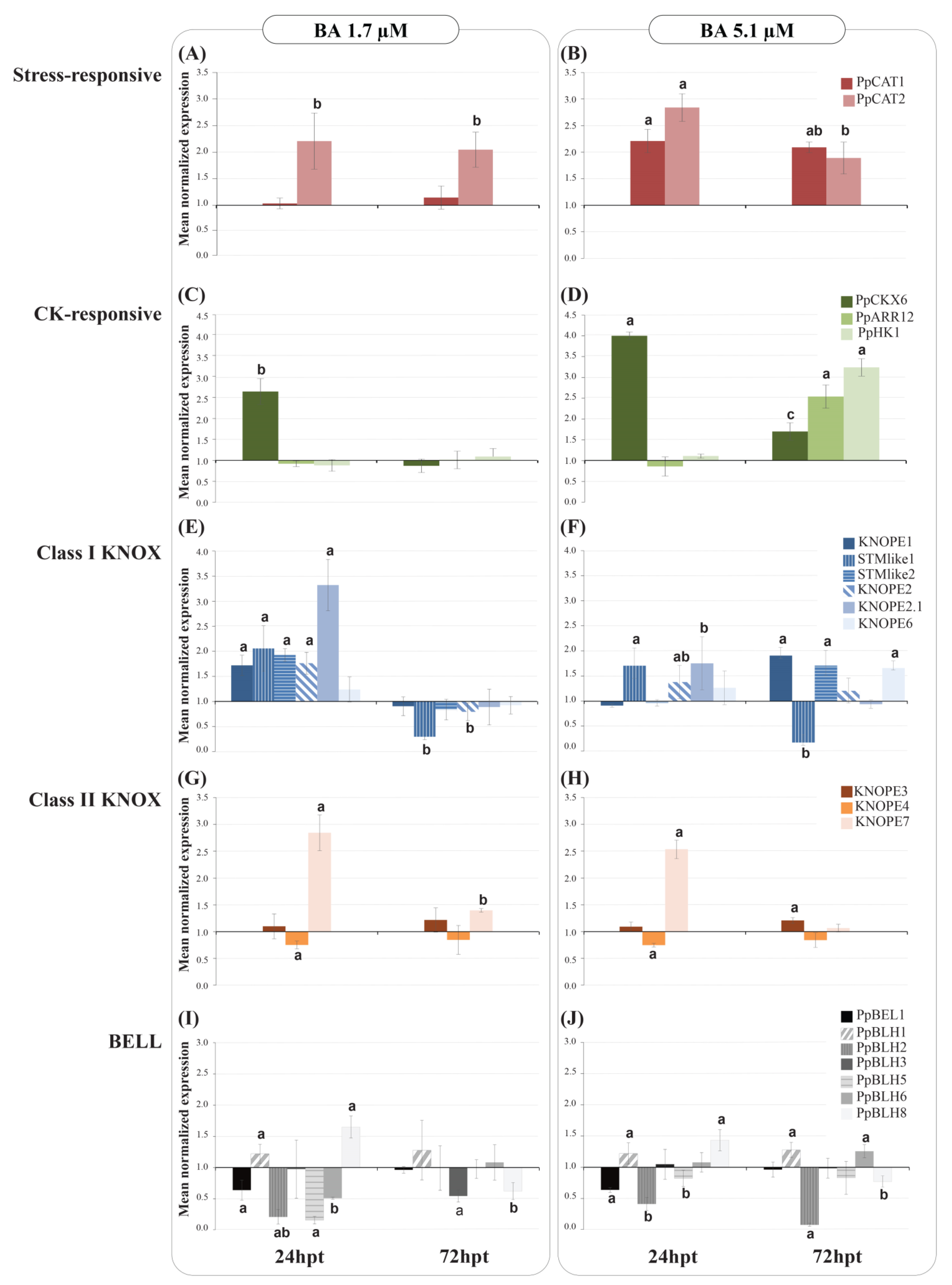
2.2.2. PRUNOX Expression Patterns
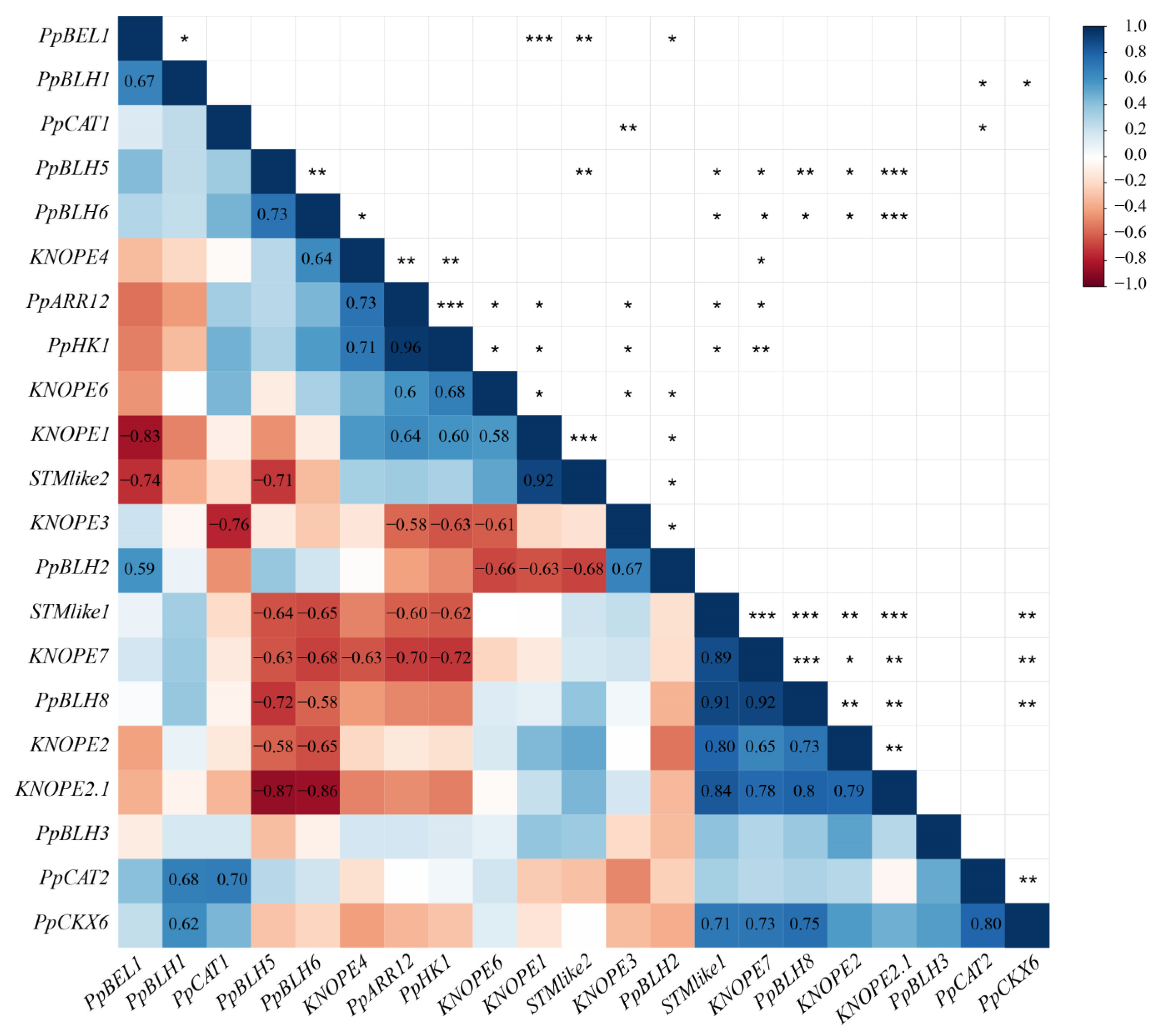
2.2.3. Gene Coexpression Analysis
2.3. KNOPE1 Overexpression in Gisela 6 Rootstock
3. Discussion
3.1. PRUNOX
3.2. PRUNOXI and -II Transcription Response to BA in Micropropagation
3.3. Transformation of Gisela 6 and KNOPE1 Phenotypes
4. Materials and Methods
4.1. Bioinformatic Surveys
4.2. Cytokinin Assays
4.2.1. Micropropagation, Treatments, Sampling, and Histological Analyses
4.2.2. Morpho-Histological Analyses
4.2.3. Gene Expression Analyses
4.3. Gene Transfer into Gisela 6
4.3.1. Plant Materials and In Vitro Culture
4.3.2. Shoot Regeneration
4.3.3. Genetic Constructs and Transformation
4.3.4. Transgene Analyses
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yang, X.; Zhao, W.; Lang, T.; Samuelsson, T. Evolution, diversification, and expression of KNOX proteins in plants. Front. Plant Sci. 2015, 6, 882. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Tsiantis, M. KNOX genes: Versatile regulators of plant development and diversity. Development 2010, 137, 3153–3165. [Google Scholar] [CrossRef] [PubMed]
- Scofield, S.; Dewitte, W.; Murray, J.A. A model for Arabidopsis class-1 KNOX gene function. Plant Signal Behav. 2008, 3, 257–259. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, S.; Chen, C.Y.; Li, C.; Shan, W.; Lu, W.; Cui, Y.; Liu, X.; Wu, K. Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture. PLoS Genet. 2015, 11, e1005125. [Google Scholar] [CrossRef]
- Wang, S.; Yamaguchi, M.; Grienenberger, E.; Martone, P.T.; Samuels, A.L.; Mansfield, S.D. The Class II KNOX genes KNAT3 and KNAT7 work cooperatively to influence deposition of secondary cell walls that provide mechanical support to Arabidopsis stems. Plant J. 2020, 101, 293–309. [Google Scholar] [CrossRef]
- Nookaraju, A.; Pandey, S.K.; Ahlawat, Y.K.; Joshi, C.P. Understanding the Modus Operandi of Class II KNOX Transcription Factors in Secondary Cell Wall Biosynthesis. Plants 2022, 11, 493. [Google Scholar] [CrossRef]
- Furumizu, C.; Alvarez, J.P.; Sakakibara, K.; Bowman, J.L. Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PLoS Genet. 2015, 11, e1004980. [Google Scholar] [CrossRef]
- Keren-Keiserman, A.; Shtern, A.; Levy, M.; Chalupowicz, D.; Furumizu, C.; Alvarez, J.P.; Amsalem, Z.; Arazi, T.; Alkalai-Tuvia, S.; Efroni, I.; et al. CLASS-II KNOX genes coordinate spatial and temporal ripening in tomato. Plant Physiol. 2022, 190, 657–668. [Google Scholar] [CrossRef]
- Hepworth, S.R.; Pautot, V.A. Beyond the Divide: Boundaries for Patterning and Stem Cell Regulation in Plants. Front. Plant Sci. 2015, 6, 1052. [Google Scholar] [CrossRef]
- Kim, D.; Cho, Y.H.; Ryu, H.; Kim, Y.; Kim, T.H.; Hwang, I. BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J. 2013, 75, 755–766. [Google Scholar] [CrossRef]
- Jia, P.; Zhang, C.; Xing, L.; Li, Y.; Shah, K.; Zuo, X.; Zhang, D.; An, N.; Han, M.; Ren, X. Genome-Wide Identification of the MdKNOX Gene Family and Characterization of Its Transcriptional Regulation in Malus domestica. Front. Plant Sci. 2020, 11, 128. [Google Scholar] [CrossRef]
- Cheng, X.; Li, M.; Abdullah, M.; Li, G.; Zhang, J.; Manzoor, M.A.; Wang, H.; Jin, Q.; Jiang, T.; Cai, Y.; et al. In Silico Genome-Wide Analysis of the Pear (Pyrus bretschneideri) KNOX Family and the Functional Characterization of PbKNOX1, an Arabidopsis BREVIPEDICELLUS Orthologue Gene, Involved in Cell Wall and Lignin Biosynthesis. Front. Genet. 2019, 10, 632. [Google Scholar] [CrossRef]
- Yang, Q.; Yuan, C.; Cong, T.; Wang, J.; Zhang, Q. Genome-wide identification of three-amino-acid-loop-extension gene family and their expression profile under hormone and abiotic stress treatments during stem development of Prunus mume. Front. Plant Sci. 2022, 13, 1006360. [Google Scholar] [CrossRef]
- Srinivasan, C.; Liu, Z.; Scorza, R. Ectopic expression of class 1 KNOX genes induce adventitious shoot regeneration and alter growth and development of tobacco (Nicotiana tabacum L.) and European plum (Prunus domestica L.). Plant Cell Rep. 2011, 30, 655–664. [Google Scholar] [CrossRef]
- Jia, P.; Xing, L.; Zhang, C.; Chen, H.; Li, Y.; Zhang, D.; Ma, J.; Zhao, C.; Han, M.; Ren, X.; et al. MdKNOX15, a class I knotted-like transcription factor of apple, controls flowering and plant height by regulating GA levels through promoting the MdGA2ox7 transcription. Environ. Exp. Bot. 2021, 185, 104411. [Google Scholar] [CrossRef]
- Testone, G.; Bruno, L.; Condello, E.; Chiappetta, A.; Bruno, A.; Mele, G.; Tartarini, A.; Spano, L.; Innocenti, A.M.; Mariotti, D.; et al. Peach [Prunus persica (L.) Batsch] KNOPE1, a class 1 KNOX orthologue to Arabidopsis BREVIPEDICELLUS/KNAT1, is misexpressed during hyperplasia of leaf curl disease. J. Exp. Bot. 2008, 59, 389–402. [Google Scholar] [CrossRef]
- Zeng, R.F.; Fu, L.M.; Deng, L.; Liu, M.F.; Gan, Z.M.; Zhou, H.; Hu, S.F.; Hu, C.G.; Zhang, J.Z. CiKN1 and CiKN6 are involved in leaf development in citrus by regulating CimiR164. Plant J. 2022, 110, 828–848. [Google Scholar] [CrossRef]
- Scofield, S.; Murray, J.A. KNOX gene function in plant stem cell niches. Plant Mol. Biol. 2006, 60, 929–946. [Google Scholar] [CrossRef]
- Tsuda, K.; Hake, S. Diverse functions of KNOX transcription factors in the diploid body plan of plants. Curr. Opin. Plant Biol. 2015, 27, 91–96. [Google Scholar] [CrossRef]
- Testone, G.; Condello, E.; Verde, I.; Nicolodi, C.; Caboni, E.; Dettori, M.T.; Vendramin, E.; Bruno, L.; Bitonti, M.B.; Mele, G.; et al. The peach (Prunus persica L. Batsch) genome harbours 10 KNOX genes, which are differentially expressed in stem development, and the class 1 KNOPE1 regulates elongation and lignification during primary growth. J. Exp. Bot. 2012, 63, 5417–5435. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.F.; Zhou, H.; Fu, L.M.; Yan, Z.; Ye, L.X.; Hu, S.F.; Gan, Z.M.; Ai, X.Y.; Hu, C.G.; Zhang, J.Z. Two citrus KNAT-like genes, CsKN1 and CsKN2, are involved in the regulation of spring shoot development in sweet orange. J. Exp. Bot. 2021, 72, 7002–7019. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Xing, L.; Zhang, C.; Zhang, D.; Ma, J.; Zhao, C.; Han, M.; Ren, X.; An, N. MdKNOX19, a class II knotted-like transcription factor of apple, plays roles in ABA signalling/sensitivity by targeting ABI5 during organ development. Plant Sci. 2021, 302, 110701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gottschalk, C.; van Nocker, S. Genetic mechanisms in the repression of flowering by gibberellins in apple (Malus × domestica Borkh.). BMC Genom. 2019, 20, 747. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, P.T.; Orłowska, R. Plant tissue culture environment as a switch-key of (epi)genetic changes. Plant Cell Tissue Organ. Cult. 2020, 140, 245–257. [Google Scholar] [CrossRef]
- Hassan, S.A.M.; Zayed, N.S. Factor Controlling Micropropagation of Fruit Trees: A Review. Sci. Int. 2018, 6, 1–10. [Google Scholar] [CrossRef]
- George, E.F.; Hall, M.A.; Klerk, G.J.D. Plant Tissue Culture Procedure—Background. In Plant Propagation by Tissue Culture: Volume 1. The Background; George, E.F., Hall, M.A., Klerk, G.-J.D., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 1–28. [Google Scholar]
- Cheong, E.J. Biotechnological approaches for improvement and conservation of Prunus species. Plant Biotechnol. Rep. 2012, 6, 17–28. [Google Scholar] [CrossRef]
- Gentile, A.; Jàquez Gutiérrez, M.; Martinez, J.; Frattarelli, A.; Nota, P.; Caboni, E. Effect of meta-Topolin on micropropagation and adventitious shoot regeneration in Prunus rootstocks. Plant Cell Tissue Org. Cult. 2014, 118, 373–381. [Google Scholar] [CrossRef]
- Van Nocker, S.; Gardiner, S.E. Breeding better cultivars, faster: Applications of new technologies for the rapid deployment of superior horticultural tree crops. Hortic. Res. 2014, 1, 14022. [Google Scholar] [CrossRef]
- Zong, X.; Denler, B.J.; Danial, G.H.; Chang, Y.; Song, G. Adventitious shoot regeneration and agrobacterium tumefaciens-mediated transient transformation of almond × peach hybrid rootstock ‘Hansen 536’. HortScience 2019, 54, 936–940. [Google Scholar] [CrossRef]
- Ricci, A.; Sabbadini, S.; Prieto, H.; Padilla, I.M.; Dardick, C.; Li, Z.; Scorza, R.; Limera, C.; Mezzetti, B.; Perez-Jimenez, M.; et al. Genetic Transformation in Peach (Prunus persica L.): Challenges and Ways Forward. Plants 2020, 9, 971. [Google Scholar] [CrossRef]
- Petri, C.; Alburquerque, N.; Faize, M.; Scorza, R.; Dardick, C. Current achievements and future directions in genetic engineering of European plum (Prunus domestica L.). Transgen. Res. 2018, 27, 225–240. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, W.; Chen, F.; Song, A. The Role of Transcription Factors in the Regulation of Plant Shoot Branching. Plants 2022, 11, 1997. [Google Scholar] [CrossRef]
- Bianchi, V.J.; Rubio, M.; Trainotti, L.; Verde, I.; Bonghi, C.; Martinez-Gomez, P. Prunus transcription factors: Breeding perspectives. Front. Plant Sci. 2015, 6, 443. [Google Scholar] [CrossRef]
- Mukherjee, K.; Brocchieri, L.; Burglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef]
- Shi, S.; Li, J.; Sun, J.; Yu, J.; Zhou, S. Phylogeny and classification of Prunus sensu lato (Rosaceae). J. Integr. Plant Biol. 2013, 55, 1069–1079. [Google Scholar] [CrossRef]
- Ramireddy, E.; Brenner, W.G.; Pfeifer, A.; Heyl, A.; Schmulling, T. In planta analysis of a cis-regulatory cytokinin response motif in Arabidopsis and identification of a novel enhancer sequence. Plant Cell Physiol. 2013, 54, 1079–1092. [Google Scholar] [CrossRef]
- Brenner, W.G.; Schmulling, T. Summarizing and exploring data of a decade of cytokinin-related transcriptomics. Front. Plant Sci. 2015, 6, 29. [Google Scholar] [CrossRef]
- Zhang, D.; Lan, S.; Yin, W.L.; Liu, Z.J. Genome-Wide Identification and Expression Pattern Analysis of KNOX Gene Family in Orchidaceae. Front. Plant Sci. 2022, 13, 901089. [Google Scholar] [CrossRef]
- Chin, S.W.; Shaw, J.; Haberle, R.; Wen, J.; Potter, D. Diversification of almonds, peaches, plums and cherries—Molecular systematics and biogeographic history of Prunus (Rosaceae). Mol. Phylogenet. Evol. 2014, 76, 34–48. [Google Scholar] [CrossRef]
- Hodel, R.G.; Zimmer, E.; Wen, J. A phylogenomic approach resolves the backbone of Prunus (Rosaceae) and identifies signals of hybridization and allopolyploidy. Mol. Phylogenet. Evol. 2021, 160, 107118. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Liu, Y.B.; Zhang, X.S. Auxin-cytokinin interaction regulates meristem development. Mol. Plant 2011, 4, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Polivanova, O.B.; Bedarev, V.A. Hyperhydricity in Plant Tissue Culture. Plants 2022, 11, 3313. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Pan, Y.; Zhi, C.; Zheng, Y.; Wang, X.; Li, X.; Cheng, Z. Genome-Wide Identification and Characterization of KNOTTED-Like Homeobox (KNOX) Homologs in Garlic (Allium sativum L.) and Their Expression Profilings Responding to Exogenous Cytokinin and Gibberellin. Int. J. Mol. Sci. 2021, 22, 9237. [Google Scholar] [CrossRef]
- Soucek, P.; Klima, P.; Rekova, A.; Brzobohaty, B. Involvement of hormones and KNOXI genes in early Arabidopsis seedling development. J. Exp. Bot. 2007, 58, 3797–3810. [Google Scholar] [CrossRef]
- Brenner, W.G.; Romanov, G.A.; Kollmer, I.; Burkle, L.; Schmulling, T. Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 2005, 44, 314–333. [Google Scholar] [CrossRef]
- Alvarez, J.M.; Bueno, N.; Cuesta, C.; Feito, I.; Ordas, R.J. Hormonal and gene dynamics in de novo shoot meristem formation during adventitious caulogenesis in cotyledons of Pinus pinea. Plant Cell Rep. 2020, 39, 527–541. [Google Scholar] [CrossRef]
- Perez-Jimenez, M.; Cantero-Navarro, E.; Perez-Alfocea, F.; Cos-Terrer, J. Endogenous hormones response to cytokinins with regard to organogenesis in explants of peach (Prunus persica L. Batsch) cultivars and rootstocks (P. persica × Prunus dulcis). Plant Physiol. Biochem. 2014, 84, 197–202. [Google Scholar] [CrossRef]
- Ziv, M. Quality of micropropagated plants—Vitrification. In Vitro Cell Dev. Biol.—Plant 1991, 27, 64–69. [Google Scholar] [CrossRef]
- Groover, A.T.; Mansfield, S.D.; DiFazio, S.P.; Dupper, G.; Fontana, J.R.; Millar, R.; Wang, Y. The Populus homeobox gene ARBORKNOX1 reveals overlapping mechanisms regulating the shoot apical meristem and the vascular cambium. Plant Mol. Biol. 2006, 61, 917–932. [Google Scholar] [CrossRef]
- Kim, M.H.; Cho, J.S.; Jeon, H.W.; Sangsawang, K.; Shim, D.; Choi, Y.I.; Park, E.J.; Lee, H.; Ko, J.H. Wood Transcriptome Profiling Identifies Critical Pathway Genes of Secondary Wall Biosynthesis and Novel Regulators for Vascular Cambium Development in Populus. Genes 2019, 10, 690. [Google Scholar] [CrossRef]
- Santos, A.M.; Oliver, M.J.; Sánchez, A.M.; Oliveira, M.M. Expression of almond Knotted1 homologue (PdKn1) anticipates adventitious shoot initiation. In Vitro Cell Dev. Biol.—Plant 2012, 48, 40–49. [Google Scholar] [CrossRef]
- Prieto, H. Genetic Transformation Strategies in Fruit Crops. In Genetic Transformation; Alvarez, M., Ed.; IntechOpen: Rijeka, Croatia, 2011; pp. 81–99. [Google Scholar]
- Jia, H.F.; Guo, J.X.; Qin, L.; Shen, Y.Y. Virus-induced PpCHLH gene silencing in peach leaves (Prunus persica). J. Hortic. Sci. Biotechnol. 2010, 85, 528–532. [Google Scholar] [CrossRef]
- Song, G.Q.; Sink, K.C. Transformation of Montmorency sour cherry (Prunus cerasus L.) and Gisela 6 (P. cerasus × P. canescens) cherry rootstock mediated by Agrobacterium tumefaciens. Plant Cell Rep. 2006, 25, 117–123. [Google Scholar] [CrossRef]
- Song, G.Q.; Sink, K.C. Transformation of cherry: Prunus cerasus l. ‘Montmorency’ and Prunus cerasus × P. Canescen ‘Gisela 6’ mediated by Agrobacterium tumefaciens and a two-step selection system. Acta Hortic. 2007, 738, 683–690. [Google Scholar] [CrossRef]
- Zong, X.; Xu, L.; Tan, Y.; Wei, H. Development of genetically modified sweet cherry rootstock ‘Gisela 6’ with overexpression of PcMPK3-HA gene by Agrobacterium-mediated genetic transformation. Plant Cell Tissue Org. Cult. 2022, 151, 375–384. [Google Scholar] [CrossRef]
- Rajeevkumar, S.; Anunanthini, P.; Sathishkumar, R. Epigenetic silencing in transgenic plants. Front. Plant Sci. 2015, 6, 693. [Google Scholar] [CrossRef]
- Lowder, L.G.; Malzahn, A.; Qi, Y. Plant Gene Regulation Using Multiplex CRISPR-dCas9 Artificial Transcription Factors. Methods Mol. Biol. 2018, 1676, 197–214. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- He, Y.; Liu, X.; Ye, L.; Pan, C.; Chen, L.; Zou, T.; Lu, G. Genome-Wide Identification and Expression Analysis of Two-Component System Genes in Tomato. Int. J. Mol. Sci. 2016, 17, 1204. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Giannino, D.; Frugis, G.; Ticconi, C.; Florio, S.; Mele, G.; Santini, L.; Cozza, R.; Bitonti, M.B.; Innocenti, A.; Mariotti, D. Isolation and molecular characterisation of the gene encoding the cytoplasmic ribosomal protein S28 in Prunus persica [L.] Batsch. Mol. Gen. Genet. 2000, 263, 201–212. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 1. [Google Scholar] [CrossRef]
- Testone, G.; Condello, E.; Di Giacomo, E.; Nicolodi, C.; Caboni, E.; Rasori, A.; Bonghi, C.; Bruno, L.; Bitonti, M.B.; Giannino, D. The KNOTTED-like genes of peach (Prunus persica L. Batsch) are differentially expressed during drupe growth and the class 1 KNOPE1 contributes to mesocarp development. Plant Sci. 2015, 237, 69–79. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Quoirin, M.; Lepoivre, P. Improved media for in vitro culture of Prunus sp. In Symposium on Tissue Culture for Horticultural Purposes 78; International Society for Horticultural Science: Leuven, Belgium, 1977; pp. 437–442. [Google Scholar]




| Genus | Prunus | Arabidopsis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subgenus | Prunus | Cerasus | - | ||||||||
| Section | Persicae | Armeniaca | Amygdalus | Prunus | - | - | |||||
| Species 2: | P. dav | P. ferg | P. kan | P. mir | P. per | P. arm | P. mum | P. dul | P. sal | P. avi | A. thaliana |
| Size (Mb): | 237 | 237 | 238 | 243 | 265 | 240 | 280 | 240 | 308 | 338 | 135 |
| Class I | |||||||||||
| STM-like | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 |
| KNAT1-like | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KNAT2/6-like | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| extra group | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| subtotal | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 4 |
| Class II | |||||||||||
| KNAT3/4/5-like | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 |
| KNAT7-like | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| subtotal | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 |
| Class M | |||||||||||
| KNATM-like | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| KNATM2-like | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 0 |
| subtotal | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 |
| Total KNOX | 11 | 11 | 11 | 11 | 11 | 11 | 11 | 11 | 11 | 12 | 9 |
| BA (µM) | Stem Height (cm) | Leaves on Main Axis | Lateral Shoots |
|---|---|---|---|
| 0.0 | 0.9 ± 0.2 | 12.8 ± 1.5 b | 0.0 ± 0.0 c |
| 1.7 | 1.2 ± 0.3 | 17.2 ± 3.4 a | 2.3 ± 0.1 b |
| 5.1 | 1.1 ± 0.2 | 20.3 ± 2.8 a | 2.9 ± 0.2 a |
| Significance | n.s. | *** | ** |
| Group | Gene | Dose | Time | DxT |
|---|---|---|---|---|
| Stress-responsive | PpCAT1 | ** | n.s. | n.s. |
| PpCAT2 | *** | n.s. | * | |
| CK-responsive | PpCKX6 | *** | *** | n.s. |
| PpARR12 | n.s. | n.s. | *** | |
| PpHK1 | n.s. | n.s. | *** | |
| Class I KNOX | STMlike1 | n.s. | ** | n.s. |
| STMlike2 | *** | *** | *** | |
| KNOPE1 | *** | *** | *** | |
| KNOPE2 | n.s. | ** | * | |
| KNOPE2.1 | ** | *** | * | |
| KNOPE6 | n.s. | n.s. | * | |
| Class II KNOX | KNOPE3 | n.s. | n.s. | n.s. |
| KNOPE4 | n.s. | n.s. | n.s. | |
| KNOPE7 | n.s. | *** | n.s. | |
| BELL | PpBEL1 | ** | ** | *** |
| PpBLH1 | ** | n.s. | * | |
| PpBLH2 | n.s. | ** | ** | |
| PpBLH3 | n.s. | n.s. | n.s. | |
| PpBLH5 | ** | *** | ** | |
| PpBLH6 | ** | ** | n.s. | |
| PpBLH8 | n.s. | *** | n.s. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Testone, G.; Caboni, E.; D’Angeli, S.; Altamura, M.M.; Giannino, D. Prunus Knotted-like Genes: Genome-Wide Analysis, Transcriptional Response to Cytokinin in Micropropagation, and Rootstock Transformation. Int. J. Mol. Sci. 2023, 24, 3046. https://doi.org/10.3390/ijms24033046
Testone G, Caboni E, D’Angeli S, Altamura MM, Giannino D. Prunus Knotted-like Genes: Genome-Wide Analysis, Transcriptional Response to Cytokinin in Micropropagation, and Rootstock Transformation. International Journal of Molecular Sciences. 2023; 24(3):3046. https://doi.org/10.3390/ijms24033046
Chicago/Turabian StyleTestone, Giulio, Emilia Caboni, Simone D’Angeli, Maria Maddalena Altamura, and Donato Giannino. 2023. "Prunus Knotted-like Genes: Genome-Wide Analysis, Transcriptional Response to Cytokinin in Micropropagation, and Rootstock Transformation" International Journal of Molecular Sciences 24, no. 3: 3046. https://doi.org/10.3390/ijms24033046
APA StyleTestone, G., Caboni, E., D’Angeli, S., Altamura, M. M., & Giannino, D. (2023). Prunus Knotted-like Genes: Genome-Wide Analysis, Transcriptional Response to Cytokinin in Micropropagation, and Rootstock Transformation. International Journal of Molecular Sciences, 24(3), 3046. https://doi.org/10.3390/ijms24033046








