Selection of Novel Reference Genes by RNA-Seq and Their Evaluation for Normalising Real-Time qPCR Expression Data of Anthocyanin-Related Genes in Lettuce and Wild Relatives
Abstract
1. Introduction
2. Results
2.1. Selection of Candidate reference genes (RGs) Based on RNA-seq Data
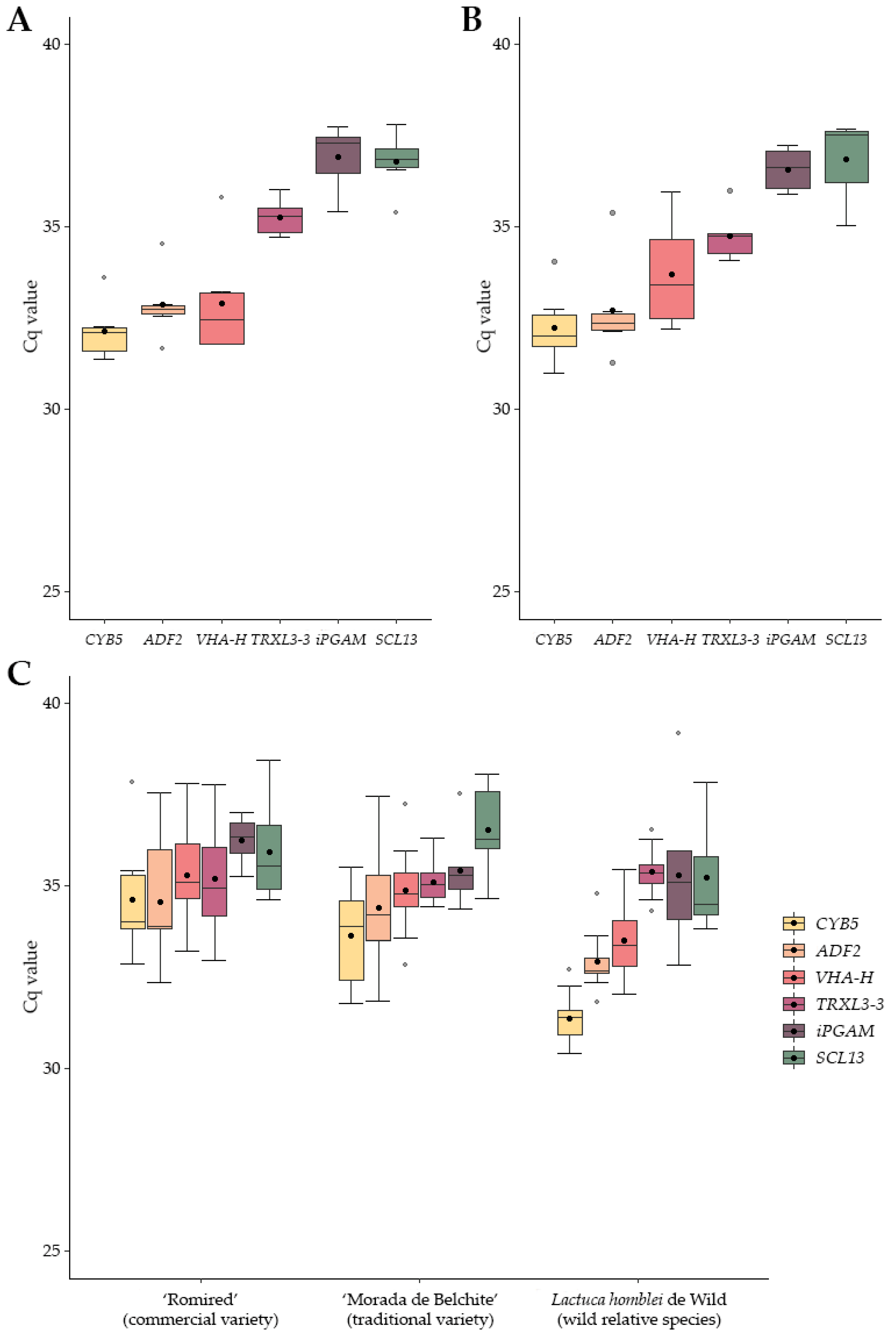
2.2. Expression Profile of Candidate RGs
2.3. Analysis of Gene Expression Stability in Accessions of Lactuca: Different Leaf Colour, Tissues, and Drought Stress Conditions
3. Discussion
4. Materials and Methods
4.1. Plant Material and Experimental Designs
4.2. RNA Extraction and RNA-Seq Analysis
4.3. Selection of Candidate RGs
4.4. mRNA Isolation, cDNA Synthesis and Real-Time qPCR
4.5. Stability Analysis of RGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Medina-Lozano, I.; Bertolín, J.R.; Díaz, A. Nutritional value of commercial and traditional lettuce (Lactuca sativa L.) and wild relatives: Vitamin C and anthocyanin content. Food Chem. 2021, 359, 129864. [Google Scholar] [CrossRef]
- Yousuf, B.; Gul, K.; Wani, A.A.; Singh, P. Health Benefits of Anthocyanins and Their Encapsulation for Potential Use in Food Systems: A Review. Crit. Rev. Food Sci. Nutr. 2016, 56, 2223–2230. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Escamilla, J.O.; Jimeńez-Hernández, F.E.; Alvarez-Parrilla, E.; De La Rosa, L.A.; Martĺnez-Ruiz, N.D.R.; González-Fernández, R.; Orozco-Lucero, E.; González-Aguilar, G.A.; García-Fajardo, J.A.; Rodrigo-García, J. Effect of Elicitation on Polyphenol and Carotenoid Metabolism in Butterhead Lettuce (Lactuca sativa var. capitata). ACS Omega 2020, 5, 11535–11546. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Tao, R.; Liu, W.; Yu, C.; Yue, Z.; He, S.; Lavelle, D.; Zhang, W.; Zhang, L.; An, G.; et al. Characterization of four polymorphic genes controlling red leaf colour in lettuce that have undergone disruptive selection since domestication. Plant Biotechnol. J. 2020, 18, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Wada, K.C.; Inagaki, N.; Sakai, H.; Yamashita, H.; Nakai, Y.; Fujimoto, Z.; Yonemaru, J.; Itoh, H. Genetic effects of Red Lettuce Leaf genes on red coloration in leaf lettuce under artificial lighting conditions. Plant-Environment Interact. 2022, 3, 179–192. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Xu, S.Z.; Cheng, Y.W.; Ya, H.Y.; Han, J.M. Transcriptome analysis and anthocyanin-related genes in red leaf lettuce. Genet. Mol. Res. 2016, 15, 10-4238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Su, W.; Tao, R.; Zhang, W.; Chen, J.; Wu, P.; Yan, C.; Jia, Y.; Larkin, R.M.; Lavelle, D.; et al. RNA sequencing provides insights into the evolution of lettuce and the regulation of flavonoid biosynthesis. Nat. Commun. 2017, 8, 2264. [Google Scholar] [CrossRef]
- Ren, W.; Zhao, L.; Zhang, L.; Wang, Y.; Cui, L.; Tang, Y.; Sun, X.; Tang, K. Molecular cloning and characterization of 4-hydroxyphenylpyruvate dioxygenase gene from Lactuca sativa. J. Plant Physiol. 2011, 168, 1076–1083. [Google Scholar] [CrossRef]
- Damerum, A.; Selmes, S.L.; Biggi, G.F.; Clarkson, G.J.J.; Rothwell, S.D.; Truco, M.J.; Michelmore, R.W.; Hancock, R.D.; Shellcock, C.; Chapman, M.A.; et al. Elucidating the genetic basis of antioxidant status in lettuce (Lactuca sativa). Hortic. Res. 2015, 2, 15055. [Google Scholar] [CrossRef]
- Park, S.; Shi, A.; Mou, B. Genome-wide identification and expression analysis of the CBF/DREB1 gene family in lettuce. Sci. Rep. 2020, 10, 5733. [Google Scholar] [CrossRef]
- Chen, L.; Xu, M.; Liu, C.; Hao, J.; Fan, S.; Han, Y. LsMYB15 Regulates Bolting in Leaf Lettuce (Lactuca sativa L.) Under High-Temperature Stress. Front. Plant Sci. 2022, 13, 921021. [Google Scholar] [CrossRef]
- Liu, R.; Su, Z.; Zhou, H.; Huang, Q.; Fan, S.; Liu, C.; Han, Y. LsHSP70 is induced by high temperature to interact with calmodulin, leading to higher bolting resistance in lettuce. Sci. Rep. 2020, 10, 15155. [Google Scholar] [CrossRef]
- Xiong, T.; Zhang, S.; Kang, Z.; Zhang, T.; Li, S. Dose-Dependent Physiological and Transcriptomic Responses of Lettuce (Lactuca sativa L.) to Copper Oxide Nanoparticles-Insights into the Phytotoxicity Mechanisms. Int. J. Mol. Sci. 2021, 22, 3688. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lu, C.; Tariq, M.; Xiao, Q.; Zhang, W.; Huang, K.; Lu, Q.; Lin, K.; Liu, Z. The response and tolerance mechanisms of lettuce (Lactuca sativa L.) exposed to nickel in a spiked soil system. Chemosphere 2019, 222, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Porcel, R.; Aroca, R.; Azcón, R.; Ruiz-Lozano, J.M. PIP Aquaporin Gene Expression in Arbuscular Mycorrhizal Glycine max and Lactuca sativa Plants in Relation to Drought Stress Tolerance. Plant Mol. Biol. 2006, 60, 389–404. [Google Scholar] [CrossRef]
- Ruiz-Lozano, J.M.; Aroca, R.; Zamarreño, Á.M.; Molina, S.; Andreo-Jiménez, B.; Porcel, R.; García-Mina, J.M.; Ruyter-Spira, C.; López-Ráez, J.A. Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant, Cell Environ. 2016, 39, 441–452. [Google Scholar] [CrossRef]
- Aksakal, O.; Tabay, D.; Esringu, A.; Icoglu Aksakal, F.; Esim, N. Effect of proline on biochemical and molecular mechanisms in lettuce (Lactuca sativa L.) exposed to UV-B radiation. Photochem. Photobiol. Sci. 2017, 16, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Navarro, E.; Serrano-Heras, G.; Castaño, M.J.; Solera, J. Real-time PCR detection chemistry. Clin. Chim. Acta 2015, 439, 231–250. [Google Scholar] [CrossRef] [PubMed]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef]
- Kiarash, J.G.; Dayton Wilde, H.; Amirmahani, F.; Mehdi Moemeni, M.; Zaboli, M.; Nazari, M.; Saeed Moosavi, S.; Jamalvandi, M. Selection and validation of reference genes for normalization of qRT-PCR gene expression in wheat (Triticum durum L.) under drought and salt stresses. J. Genet. 2018, 97, 1433–1444. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Shi, L.; Han, C.; Yu, J.; Li, D.; Zhang, Y. Validation of Reference Genes for Gene Expression Studies in Virus-Infected Nicotiana benthamiana Using Quantitative Real-Time PCR. PLoS ONE 2012, 7, e46451. [Google Scholar] [CrossRef] [PubMed]
- de Jesus Miranda, V.; Coelho, R.R.; Viana, A.A.B.; de Oliveira Neto, O.B.; Carneiro, R.M.D.G.; Rocha, T.L.; de Sa, M.F.G.; Fragoso, R.R. Validation of reference genes aiming accurate normalization of qPCR data in soybean upon nematode parasitism and insect attack. BMC Res. Notes 2013, 6, 196. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mauriat, M.; Guénin, S.; Pelloux, J.; Lefebvre, J.-F.; Louvet, R.; Rusterucci, C.; Moritz, T.; Guerineau, F.; Bellini, C.; et al. The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 2008, 6, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.T.; Poolakkalody, N.J.; Shah, J.M. Plant reference genes for development and stress response studies. J. Biosci. 2018, 43, 173–187. [Google Scholar] [CrossRef]
- Sgamma, T.; Pape, J.; Massiah, A.; Jackson, S. Selection of reference genes for diurnal and developmental time-course real-time PCR expression analyses in lettuce. Plant Methods 2016, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Borowski, J.M.; Galli, V.; da Silva Messias, R.; Perin, E.C.; Buss, J.H.; dos Anjos e Silva, S.D.; Rombaldi, C.V. Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta 2014, 239, 1187–1200. [Google Scholar] [CrossRef]
- Song, Y.; Hanner, R.H.; Meng, B. Genome-wide screening of novel RT-qPCR reference genes for study of GLRaV-3 infection in wine grapes and refinement of an RNA isolation protocol for grape berries. Plant Methods 2021, 17, 110. [Google Scholar] [CrossRef]
- Yim, A.K.Y.; Wong, J.W.H.; Ku, Y.S.; Qin, H.; Chan, T.F.; Lam, H.M. Using RNA-seq Data to Evaluate Reference Genes Suitable for Gene Expression Studies in Soybean. PLoS ONE 2015, 10, e0136343. [Google Scholar] [CrossRef]
- Zhou, Z.; Cong, P.; Tian, Y.; Zhu, Y. Using RNA-seq data to select reference genes for normalizing gene expression in apple roots. PLoS ONE 2017, 12, e0185288. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Rime Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T. Determination of most stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- PGR (Plant Genetic Resources) Lettuce. The Lettuce Gene Pool. Available online: https://www.pgrportal.nl/en/lettuce-genetic-resources-portal.htm (accessed on 21 November 2022).
- Garrido, J.; Aguilar, M.; Prieto, P. Identification and validation of reference genes for RT-qPCR normalization in wheat meiosis. Sci. Rep. 2020, 10, 2726. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; Van Den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Broad Institute. Picard Tools. Available online: http://broadinstitute.github.io/picard/ (accessed on 25 October 2022).
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Kubista, M.; Sindelka, R. The Prime Technique-Real-time PCR Data Analysis. G.I.T Lab. J. 2007, 9, 33–35. [Google Scholar]
 ) geNorm (M), (
) geNorm (M), ( ) NormFinder, (
) NormFinder, ( ) BestKeeper, and (
) BestKeeper, and ( ) ΔCt (SV) methods in three experiments: (A) comparison of leaf colour (green and red) in lettuce commercial varieties; (B) comparison of tissues (leaf and stem) in a wild relative species; and (C) under drought stress in a commercial variety, a traditional variety, and a wild relative. The most stable genes are represented on the left and the least stable on the right of the graph.
) ΔCt (SV) methods in three experiments: (A) comparison of leaf colour (green and red) in lettuce commercial varieties; (B) comparison of tissues (leaf and stem) in a wild relative species; and (C) under drought stress in a commercial variety, a traditional variety, and a wild relative. The most stable genes are represented on the left and the least stable on the right of the graph.
 ) geNorm (M), (
) geNorm (M), ( ) NormFinder, (
) NormFinder, ( ) BestKeeper, and (
) BestKeeper, and ( ) ΔCt (SV) methods in three experiments: (A) comparison of leaf colour (green and red) in lettuce commercial varieties; (B) comparison of tissues (leaf and stem) in a wild relative species; and (C) under drought stress in a commercial variety, a traditional variety, and a wild relative. The most stable genes are represented on the left and the least stable on the right of the graph.
) ΔCt (SV) methods in three experiments: (A) comparison of leaf colour (green and red) in lettuce commercial varieties; (B) comparison of tissues (leaf and stem) in a wild relative species; and (C) under drought stress in a commercial variety, a traditional variety, and a wild relative. The most stable genes are represented on the left and the least stable on the right of the graph.
| Name | Description | Primer Sequence (5′–3′) | Amplicon Length (bp) | Annealing Temperature (°C) |
|---|---|---|---|---|
| ADF2 | Actin-depolymerizing factor 2 | F-TTGGAGAACCAGCAGAAAC | 199 | 62 |
| R-CCATCAAGCTCTCTCTTGAAC | ||||
| CYB5 | Cytochrome B5 | F-GCACGCTACGAAAGAGG | 80 | 59 |
| R-CAGGATGATCATCTAGAAAAGG | ||||
| iPGAM | Probable 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | F-GGGAGATGTTTCAATTCCAAG | 162 | 62 |
| R-CCCATTAGAGAAAGATGAGCAG | ||||
| SCL13 | Scarecrow-like protein 13 | F-AGTCGGTTAGCACGGTTA | 100 | 56 |
| R-TTCGTGTTCGATTCTTGTT | ||||
| TRXL3-3 | Thioredoxin-like protein 3-3 | F-TGGTGTCGTGTTTGTGCAGAG | 112 | 62 |
| R-GTTGGGTTGTTTCTGGGCATT | ||||
| VHA-H | V-type proton ATPase subunit H | F-TGCAAGTGATGATGTTTTGA | 152 | 59 |
| R-TGCTTGAACAAATGAAGACC |
| Gene a | Green vs. Red | Leaf vs. Stem | Drought Stress | ||
|---|---|---|---|---|---|
| ‘Romired’ | ‘Morada de Belchite’ | L. homblei | |||
| ACT | 0.564 ns | 0.938 ns | 0.421 ns | 0.018 * | 0.036 * |
| α-TUB | 0.635 ns | 0.001 ** | 0.435 ns | 0.637 ns | 0.643 ns |
| EEF1-α | 0.019 * | 0.066 ns | 0.976 ns | 0.089 ns | 0.696 ns |
| GAPDH-2C | 0.028 * | 0.027 * | 0.407 ns | 0.302 ns | 0.593 ns |
| UBC32 | 0.159 ns | 0.159 ns | 0.398 ns | 0.276 ns | 0.010 * |
| UPL6 | 0.471 ns | 0.013 * | 0.003 ** | 0.013 * | 0.168 ns |
| ADF2 | 0.569 ns | 0.884 ns | 0.400 ns | 0.660 ns | 0.547 ns |
| CYB5 | 0.449 ns | 0.270 ns | 0.723 ns | 0.371 ns | 0.673 ns |
| iPGAM | 0.833 ns | 0.773 ns | 0.128 ns | 0.418 ns | 0.210 ns |
| SCL13 | 0.408 ns | 0.902 ns | 0.883 ns | 0.427 ns | 0.239 ns |
| TRXL3-3 | 0.487 ns | 0.406 ns | 0.843 ns | 0.161 ns | 0.470 ns |
| VHA-H | 0.198 ns | 0.576 ns | 0.116 ns | 0.195 ns | 0.741 ns |
| geNorm | NormFinder | BestKeeper | Delta Ct | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Experiment | Ranking | Gene | M | Gene | SV | Gene | SV | Gene | SV |
| Green vs. red | 1 | CYB5 | 0.33 | TRXL3-3 | 0.71 | TRXL3-3 | 0.40 | CYB5 | 0.83 |
| 2 | ADF2 | 0.33 | VHA-H | 0.77 | CYB5 | 0.53 | ADF2 | 0.83 | |
| 3 | TRXL3-3 | 0.54 | CYB5 | 0.81 | SCL13 | 0.53 | SCL13 | 0.92 | |
| 4 | VHA-H | 1.11 | ADF2 | 1.09 | ADF2 | 0.57 | TRXL3-3 | 1.10 | |
| 5 | iPGAM | 1.33 | iPGAM | 1.49 | iPGAM | 0.72 | iPGAM | 1.28 | |
| 6 | SCL13 | 1.90 | SCL13 | 2.88 | VHA-H | 1.12 | VHA-H | 1.41 | |
| Leaf vs. stem | 1 | CYB5 | 0.77 | TRXL3-3 | 0.30 | TRXL3-3 | 0.44 | TRXL3-3 | 0.93 |
| 2 | ADF2 | 0.77 | VHA-H | 0.30 | iPGAM | 0.47 | CYB5 | 1.03 | |
| 3 | TRXL3-3 | 1.01 | ADF2 | 1.18 | CYB5 | 0.77 | iPGAM | 1.08 | |
| 4 | VHA-H | 1.07 | CYB5 | 1.34 | ADF2 | 0.89 | ADF2 | 1.10 | |
| 5 | iPGAM | 1.22 | iPGAM | 1.55 | SCL13 | 0.92 | VHA-H | 1.38 | |
| 6 | SCL13 | 2.41 | SCL13 | 4.73 | VHA-H | 1.23 | SCL13 | 1.54 | |
| Drought stress | 1 | TRXL3-3 | 1.54 | TRXL3-3 | 0.77 | TRXL3-3 | 0.85 | SCL13 | 3.89 |
| 2 | ADF2 | 1.54 | ADF2 | 0.77 | SCL13 | 0.92 | TRXL3-3 | 3.90 | |
| 3 | CYB5 | 1.98 | iPGAM | 2.81 | iPGAM | 1.11 | iPGAM | 4.01 | |
| 4 | iPGAM | 3.40 | CYB5 | 2.91 | ADF2 | 1.23 | ADF2 | 4.04 | |
| 5 | SCL13 | 4.41 | SCL13 | 6.62 | CYB5 | 1.50 | CYB5 | 4.07 | |
| 6 | VHA-H | 6.82 | VHA-H | 11.27 | VHA-H | 10.35 | VHA-H | 14.08 | |
| Ranking | Green vs. Red | Leaf vs. Stem | Drought Stress |
|---|---|---|---|
| 1 | CYB5 | TRXL3-3 | TRXL3-3 |
| 2 | TRXL3-3 | CYB5 | ADF2 |
| 3 | ADF2 | ADF2 | SCL13 |
| 4 | VHA-H | VHA-H | iPGAM |
| 5 | SCL13 | iPGAM | CYB5 |
| 6 | iPGAM | SCL13 | VHA-H |
| Experiment | Accession Name | Species | Group | Leaf Colour | Year | Source a | Accession Number |
|---|---|---|---|---|---|---|---|
| Leaf colour (green vs. red) | ‘Begoña’ | Lactuca sativa L. | Commercial variety | Green | 2018/2019 | Ramiro Arnedo Semillas S.A. | - |
| ‘Romired’ | Lactuca sativa L. | Commercial variety | Red | CGN | CGN24713 | ||
| Tissue (leaf vs. stem) | Lactuca squarrosa | Lactuca squarrosa (Thunb.) Miq. | Wild crop relative | Semi-red (red stems) | 2020/2021 | BGHZ | BGHZ5124 |
| Drought stress (C vs. DI-1 vs. DI-2) b | ‘Romired’ | Lactuca sativa L. | Commercial variety | Red | 2020/2021 | CGN | CGN24713 |
| ‘Morada de Belchite’ | Lactuca sativa L. | Traditional variety | Semi-red | BGHZ | BGHZ0527 | ||
| Lactuca homblei | Lactuca homblei De Wild | Wild crop relative | Semi-red | BGHZ | BGHZ5322 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medina-Lozano, I.; Arnedo, M.S.; Grimplet, J.; Díaz, A. Selection of Novel Reference Genes by RNA-Seq and Their Evaluation for Normalising Real-Time qPCR Expression Data of Anthocyanin-Related Genes in Lettuce and Wild Relatives. Int. J. Mol. Sci. 2023, 24, 3052. https://doi.org/10.3390/ijms24033052
Medina-Lozano I, Arnedo MS, Grimplet J, Díaz A. Selection of Novel Reference Genes by RNA-Seq and Their Evaluation for Normalising Real-Time qPCR Expression Data of Anthocyanin-Related Genes in Lettuce and Wild Relatives. International Journal of Molecular Sciences. 2023; 24(3):3052. https://doi.org/10.3390/ijms24033052
Chicago/Turabian StyleMedina-Lozano, Inés, María Soledad Arnedo, Jérôme Grimplet, and Aurora Díaz. 2023. "Selection of Novel Reference Genes by RNA-Seq and Their Evaluation for Normalising Real-Time qPCR Expression Data of Anthocyanin-Related Genes in Lettuce and Wild Relatives" International Journal of Molecular Sciences 24, no. 3: 3052. https://doi.org/10.3390/ijms24033052
APA StyleMedina-Lozano, I., Arnedo, M. S., Grimplet, J., & Díaz, A. (2023). Selection of Novel Reference Genes by RNA-Seq and Their Evaluation for Normalising Real-Time qPCR Expression Data of Anthocyanin-Related Genes in Lettuce and Wild Relatives. International Journal of Molecular Sciences, 24(3), 3052. https://doi.org/10.3390/ijms24033052







