Fine Mapping and Identification of SmAPRR2 Regulating Rind Color in Eggplant (Solanum melongena L.)
Abstract
:1. Introduction
2. Results
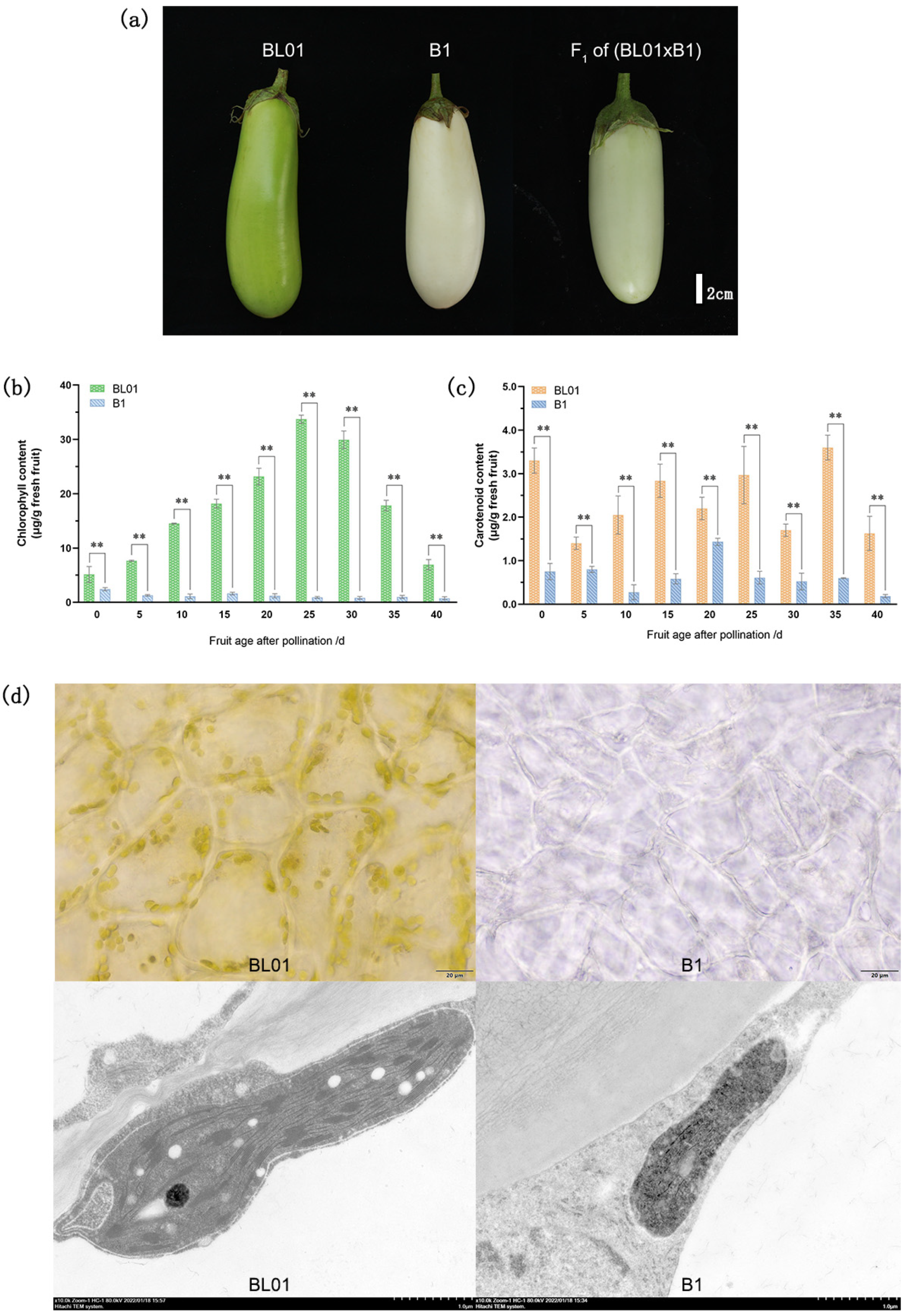
2.1. Inheritance and Phenotypic Characterization of Rind Color in Eggplant
2.2. Determination of Pigment Contents
2.3. Chloroplast Microscopic Observation
2.4. Candidate Gene Located on Chromosome 8 Identified by BSA-seq
2.5. Fine Mapping and MAS Application Strategy of SmAPRR2
2.6. Sequence Alignment and Expression Analysis of the SmAPRR2 Candidate Gene
2.7. SmAPRR2 Protein Domains
2.8. SmAPRR2 Protein Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Phenotypic Evaluation
4.2. DNA Extraction
4.3. Pigment Extraction and Measurement
4.4. Cytological Observation
4.5. BSA-seq Mapping Approach
4.6. Fine Mapping and Marker Development
4.7. Cloning and Sequencing of Candidate Gene
4.8. RNA Extraction and Gene Expression Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, J.; Zhou, X.H.; Bao, S.Y.; Zhuang, Y. Fruit color heredity and related characters of white eggplant. Acta Agric. Univ. Jiangxiensis 2016, 38, 255–259. [Google Scholar] [CrossRef]
- Liao, Y.; Sun, B.J.; Sun, G.W.; Liu, H.C.; Li, Z.L.; Li, Z.X.; Wang, G.P.; Chen, R.Y. AFLP and SCAR markers associated with peel color in eggplant (Solanum melongena). Agric. Sci. China 2009, 8, 1466–1474. [Google Scholar] [CrossRef]
- Daunay, M.C.; Aubert, S.; Frary, A.; Doganlar, S.; Jullian, E.; Lester, R.N.; Barendse, G.; Gerard, W.; Hennart, J.W.; Haanstra, J.; et al. Eggplant (Solanum melongena) fruit colour: Pigments, measurements and genetics. In Proceedings of the XII Meeting on Genetics & Breeding of Capsicum & Eggplant, Noordwijkerhout, The Netherlands, 17–19 May 2004. [Google Scholar]
- Hetherington, S.E.; Smillie, R.M.; Davies, W.J. Photosynthetic activities of vegetative and fruiting tissues of tomato. J. Exp. Bot. 1998, 49, 1173–1181. [Google Scholar] [CrossRef]
- Stenbaek, A.; Jensen, P.E. Redox regulation of chlorophyll biosynthesis. Phytochemistry 2010, 71, 853–859. [Google Scholar] [CrossRef]
- Beale, S.I. Enzymes of chlorophyll biosynthesis. Photosynth. Res. 1999, 60, 43–73. [Google Scholar] [CrossRef]
- Kobayashi, K.; Masuda, T. Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1811. [Google Scholar] [CrossRef]
- Bang, W.Y.; Jeong, I.S.; Kim, D.W.; Im, C.H.; Ji, C.; Hwang, S.M.; Kim, S.W.; Son, Y.S.; Jeong, J.; Shiina, T.; et al. Role of Arabidopsis CHL27 protein for photosynthesis, chloroplast development and gene expression profiling. Plant. Cell Physiol. 2008, 49, 1350–1363. [Google Scholar] [CrossRef]
- Pang, W.L.; Liu, F.Z.; Chen, Y.H.; Lian, Y. Genetic study on fruit color traits of eggplant. Acta Horticuhurae Sin. 2008, 7, 979–986. [Google Scholar] [CrossRef]
- Gonzali, S.; Mazzucato, A.; Perata, P. Purple as a tomato: Towards high anthocyanin tomatoes. Trends Plant Sci. 2009, 14, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.T.; Liu, Y.; Han, H.Q.; Chen, H.Y. Cloning and expression analysis of an anthocyanin-related transcription factor gene SmMYB in eggplant. Acta Hortic. Sin. 2013, 40, 467–478. [Google Scholar] [CrossRef]
- Gisbert, C.; Dumm, J.M.; Prohens, J.; Vilanova, S.; Stommel, J.R. A spontaneous eggplant (Solanum melongena L.) color mutant conditions anthocyanin-free fruit pigmentation. Hortscience 2016, 51, 793–798. [Google Scholar] [CrossRef]
- Jiang, M.M. The molecular mechanism of light signal induced anthocyanin biosynthesis in Solanum melongena. Ph.D. Thesis, Shanghai Jiao Tong University, Shanghai, China, 2016. [Google Scholar] [CrossRef]
- Barchi, L.; Lanteri, S.; Portis, E.; Vale, G.; Volante, A.; Pulcini, L.; Ciriaci, T.; Acciarri, N.; Barbierato, V.; Toppino, L.; et al. RAD tag derived marker based eggplant linkage map and the location of QTLs determining anthocyanin pigmentation. PloS ONE 2012, 7, e43740. [Google Scholar] [CrossRef] [PubMed]
- Tatebe, T. On inheritance of color in Solanum melongena Linn. Jpn. J. Genet. 1939, 15, 261–271. [Google Scholar] [CrossRef]
- Tatebe, T. Further studies on inheritance of color in Solanum Melongena Linn. Jpn. J. Genet. 1944, 20, 1–7. [Google Scholar] [CrossRef]
- Doganlar, S.; Frary, A.; Daunay, M.C.; Lester, R.N.; Tanksley, S.D. Conservation of gene function in the Solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics 2002, 161, 1713–1726. [Google Scholar] [CrossRef] [PubMed]
- Arnon, B.; Yelena, B.; Sagit, M.; Ilana, R.; Asaph, A.; Ilan, P. pc8.1, a major QTL for pigment content in pepper fruit, is associated with variation in plastid compartment size. Planta 2012, 235, 579–588. [Google Scholar] [CrossRef]
- Arnon, B.; Yelena, B.; Theresa, H.; Khalis, A.A.R.; Aharon, B.; Allen, D.; Ilan, P. CaGLK2 regulates natural variation of chlorophyll content and fruit color in pepper fruit. Theor. Appl. Genet. 2014, 127, 2139–2148. [Google Scholar] [CrossRef]
- Mark, T.W.; Peng, W.; Muris, K.; Richard, G.C.; Nigel, J.S.; Jane, A.L. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 2009, 21, 1109–1128. [Google Scholar] [CrossRef]
- Pan, Y.; Bradley, G.; Pyke, K.; Ball, G.; Lu, C.; Fray, R.; Marshall, A.; Jayasuta, S.; Baxter, C.; Wijk, R.; et al. Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 2013, 161, 1476–1485. [Google Scholar] [CrossRef] [Green Version]
- Nadakuduti, S.S.; Holdsworth, W.L.; Klein, C.L.; Barry, C.S. KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. Plant J. 2014, 78, 1022–1033. [Google Scholar] [CrossRef]
- Liu, H.; Jiao, J.; Liang, X.; Liu, J.; Meng, H.; Chen, S.; Li, Y.; Cheng, Z. Map-based cloning, identification and characterization of the w gene controlling white immature fruit color in cucumber (Cucumis sativus L.). Appl. Genet. 2016, 129, 1247–1256. [Google Scholar] [CrossRef]
- Jiao, J.Q.; Liu, H.Q.; Liu, J.; Cui, M.M.; Xu, J.; Meng, H.W.; Li, Y.H.; Chen, S.X.; Cheng, Z.H. Identification and functional characterization of APRR2 controlling green immature fruit color in cucumber (Cucumis sativus L.). Plant Growth Regul. 2017, 83, 233–243. [Google Scholar] [CrossRef]
- Yang, K.; Song, M.F.; Wei, Q.Z.; Wang, J.; Chen, J.F.; Lou, Q.F. Mapping and prediction of candidate gene w controlling white-green fruit color in cucumber. J. Nanjing Agric. Univ. 2018, 41, 1003–1008. [Google Scholar]
- Oren, E.; Tzuri, G.; Vexler, L.; Dafna, A.; Meir, A.; Faigenboim, A.; Kenigswald, M.; Portnoy, V.; Schaffer, A.A.; Levi, A.; et al. The multi-allelic APRR2 gene is associated with fruit pigment accumulation in melon and watermelon. J. Exp. Bot. 2019, 70, 3781–3794. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.L.; Liu, Z.G.; Cheng, Z.K.; Gou, J.Q.; Chen, J.Y.; Yu, W.J.; Wang, P. Identification and application of BhAPRR2 controlling peel Colour in wax gourd (Benincasa hispida). Front. Plant Sci. 2021, 12, 716772. [Google Scholar] [CrossRef] [PubMed]
- Montero-Pau, J.; Blanca, J.; Esteras, C.; Martinez-Perez, E.M.; Gomez, P.; Monforte, A.J.; Canizares, J.; Pico, B. An SNP-based saturated genetic map and QTL analysis of fruit-related traits in Zucchini using Genotyping-by-sequencing. BMC Genom. 2017, 18, 94. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Halsted, B.D. Colors in vegetable fruits: Tomato, eggplant and pepper, belonging to the same family, have similar groups of simple color factors—Combinations are easy to make. J. Hered. 1918, 9, 18–23. [Google Scholar] [CrossRef]
- Sambandam, C.N. Guide chart for color combinations in hybrid eggplants. Econ. Bot. 1967, 21, 309–311. [Google Scholar] [CrossRef]
- Liu, J.; Ai, X.; Wang, Y.; Lu, Q.; Li, T.; Wu, L.; Sun, L.; Shen, H. Fine mapping of the Ca3GT gene controlling anthocyanin biosynthesis in mature unripe fruit of Capsicum annuum L. Theor. Appl. Genet. 2020, 133, 2729–2742. [Google Scholar] [CrossRef]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef]
- Makino, S.; Kiba, T.; Imamura, A.; Hanaki, N.; Nakamura, A.; Suzuki, T.; Taniguchi, M.; Ueguchi, C.; Sugiyama, T.; Mizuno, T. Genes encoding pseudo-response regulators: Insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000, 41, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.B.; Jang, S.J.; Kang, M.Y.; Kim, S.; Kwon, J.K.; Kang, B.C. Candidate gene analysis reveals that the fruit color locus C1 corresponds to PRR2 in pepper (Capsicum frutescens). Front. Plant Sci. 2020, 11, 399. [Google Scholar] [CrossRef] [PubMed]
- Jacob-Wilk, D.; Holland, D.; Goldschmidt, E.E.; Riov, J.; Eyal, Y. Chlorophyll breakdown by chlorophyllase: Isolation and functional expression of the Chlase1 gene from ethylene-treated Citrus fruit and its regulation during development. Plant J. 1999, 20, 653–661. [Google Scholar] [CrossRef]
- Yamauchi, N.; Funamoto, Y.; Shigyo, M. Peroxidase-mediated chlorophyll degradation in horticultural crops. Phytochem. Rev. 2004, 3, 221–228. [Google Scholar] [CrossRef]
- Tang, H.Y.; Dong, X.; Wang, J.K.; Xia, J.H.; Xie, F.; Zhang, Y.; Yao, X.; Xu, Y.J.; Wang, Z.J. Fine mapping and candidate gene prediction for white immature fruit skin in cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2018, 19, 1493. [Google Scholar] [CrossRef] [PubMed]
- Andrea, A.; Giulio, M.; David, A.; Mariola, P.; Jaime, P.; Ezio, P.; Lorenzo, B.; Giovanni, G.; Santiago, V.; Pietro, G. Mutations in the SmAPRR2 transcription factor suppressing chlorophyll pigmentation in the eggplant fruit peel are key drivers of a diversified colour palette. Front. Plant. Sci. 2022, 13, 1025951. [Google Scholar] [CrossRef]
- Wilkie, S.; Clark, M.S.; Leroy, P.; Merlino, M.; Bernard, M. Genomic DNA isolation, southern blotting and hybridization. In Plant Molecular Biology; Springer: Berlin/Heidelberg, Germany, 1997; pp. 3–53. [Google Scholar] [CrossRef]
- Li, H.S. Principles and Techniques of Plant Physiological and Biochemical Experiments; Higher Education Press: Beijing, China, 2000. [Google Scholar] [CrossRef]
- Li, D.D.; Qian, J.; Li, W.J.; Jiang, Y.Q.; Gan, G.Y.; Li, W.L.; Chen, R.Y.; Yu, N.; Li, Y.; Wu, Y.G.; et al. Genome sequence and analysis of the eggplant (Solanum melongena L.). biorXiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.K.; Liu, Z.G.; Xu, Y.C.; Ma, L.L.; Chen, J.Y.; Gou, J.Q.; Su, L.W.; Wu, W.T.; Chen, Y.; Yu, W.; et al. Fine mapping and identification of the candidate gene BFS for fruit shape in wax gourd (Benincasa hispida). Theor. Appl. Genet. 2021, 134, 3983–3995. [Google Scholar] [CrossRef] [PubMed]
 represent stop codon.
represent stop codon.
 represent stop codon.
represent stop codon.




| Population | No. Plants Tested | Green:White | Expected Mendelian Distribution | χ2 | p |
|---|---|---|---|---|---|
| F1 a | 35 | 35:0 | - | - | - |
| BC1P1 b | 44 | 44:0 | - | - | - |
| BC1P2 c | 41 | 21:20 | 1:1 | 0.024 | 0.876 |
| F2 d | 2794 | 2094:700 | 3:1 | 0.004 | 0.948 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, H.; Wang, P.; Wang, W.; Peng, J.; Zheng, J.; Zhu, G.; Zhong, C.; Yu, W. Fine Mapping and Identification of SmAPRR2 Regulating Rind Color in Eggplant (Solanum melongena L.). Int. J. Mol. Sci. 2023, 24, 3059. https://doi.org/10.3390/ijms24043059
Fang H, Wang P, Wang W, Peng J, Zheng J, Zhu G, Zhong C, Yu W. Fine Mapping and Identification of SmAPRR2 Regulating Rind Color in Eggplant (Solanum melongena L.). International Journal of Molecular Sciences. 2023; 24(4):3059. https://doi.org/10.3390/ijms24043059
Chicago/Turabian StyleFang, Huarong, Peng Wang, Wanhao Wang, Jiechun Peng, Jieming Zheng, Guangwei Zhu, Chuan Zhong, and Wenjin Yu. 2023. "Fine Mapping and Identification of SmAPRR2 Regulating Rind Color in Eggplant (Solanum melongena L.)" International Journal of Molecular Sciences 24, no. 4: 3059. https://doi.org/10.3390/ijms24043059





