COBL, MKX and MYOC Are Potential Regulators of Brown Adipose Tissue Development Associated with Obesity-Related Metabolic Dysfunction in Children
Abstract
1. Introduction
2. Results
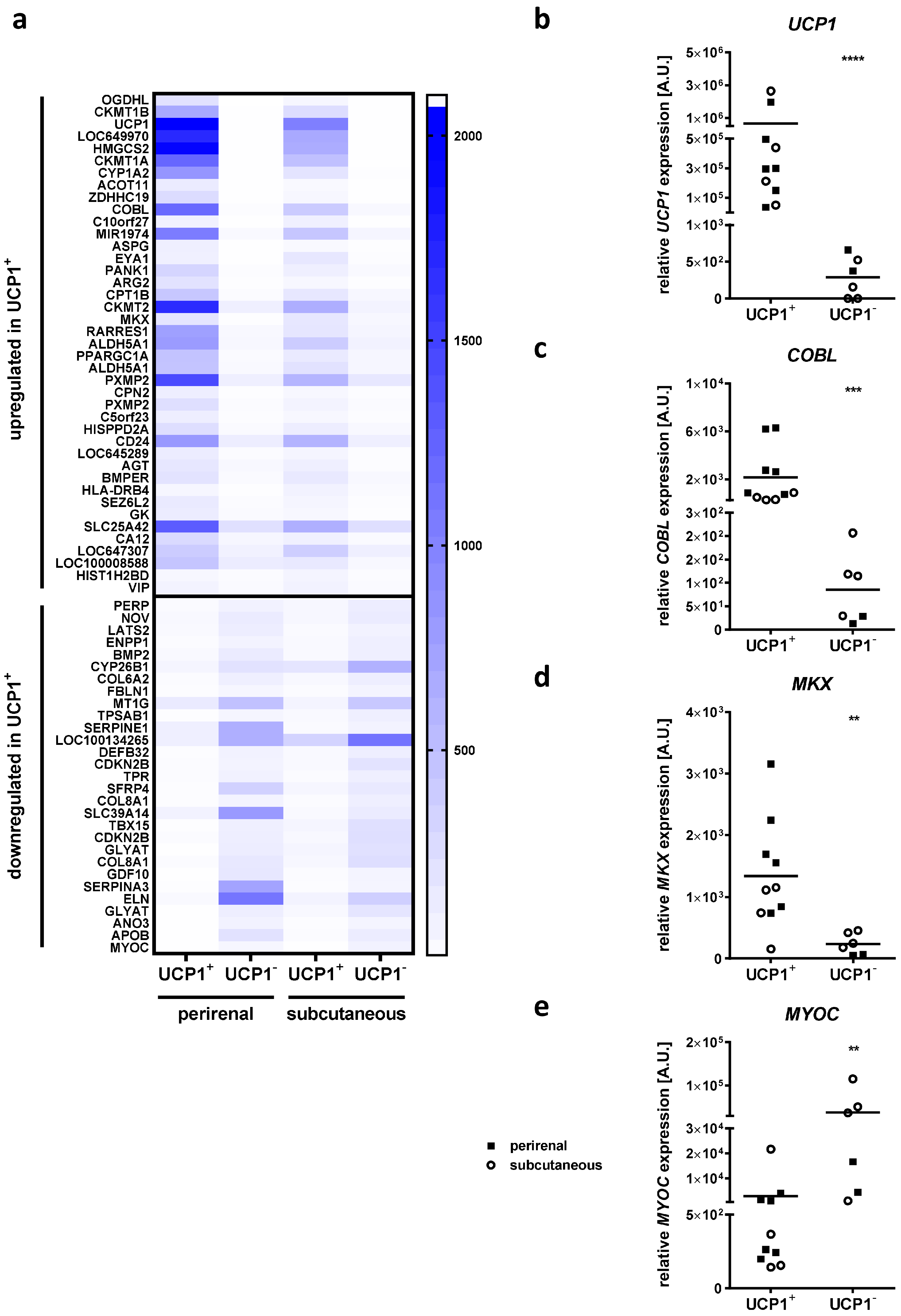
2.1. Identification of the Candidate Genes Potentially Relevant for BAT Development
2.2. Effect of Cold Exposure on Candidate Gene Expression in AT Depots of Mice
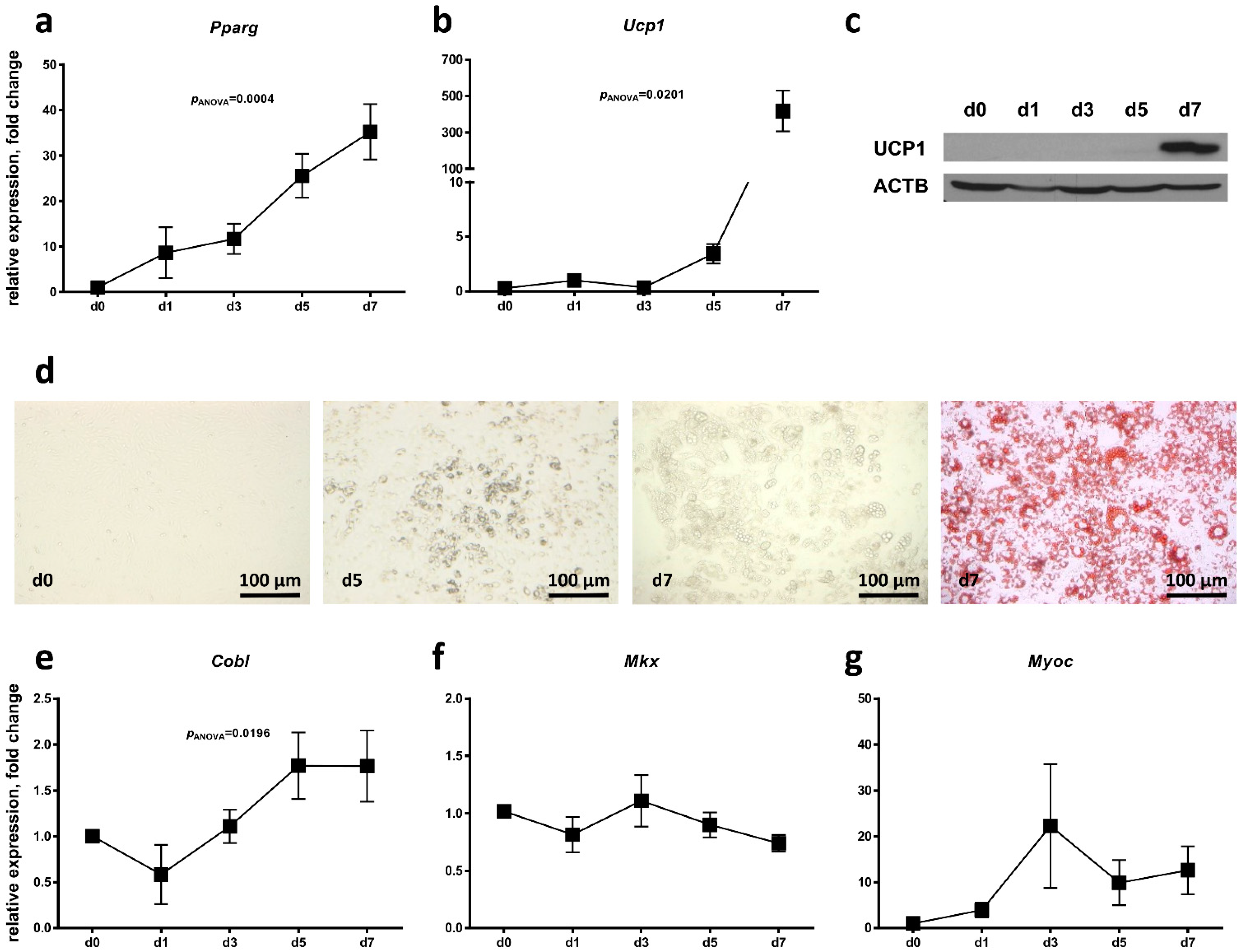
2.3. Regulation of Candidate Gene Expression during Brown Adipocyte Differentiation
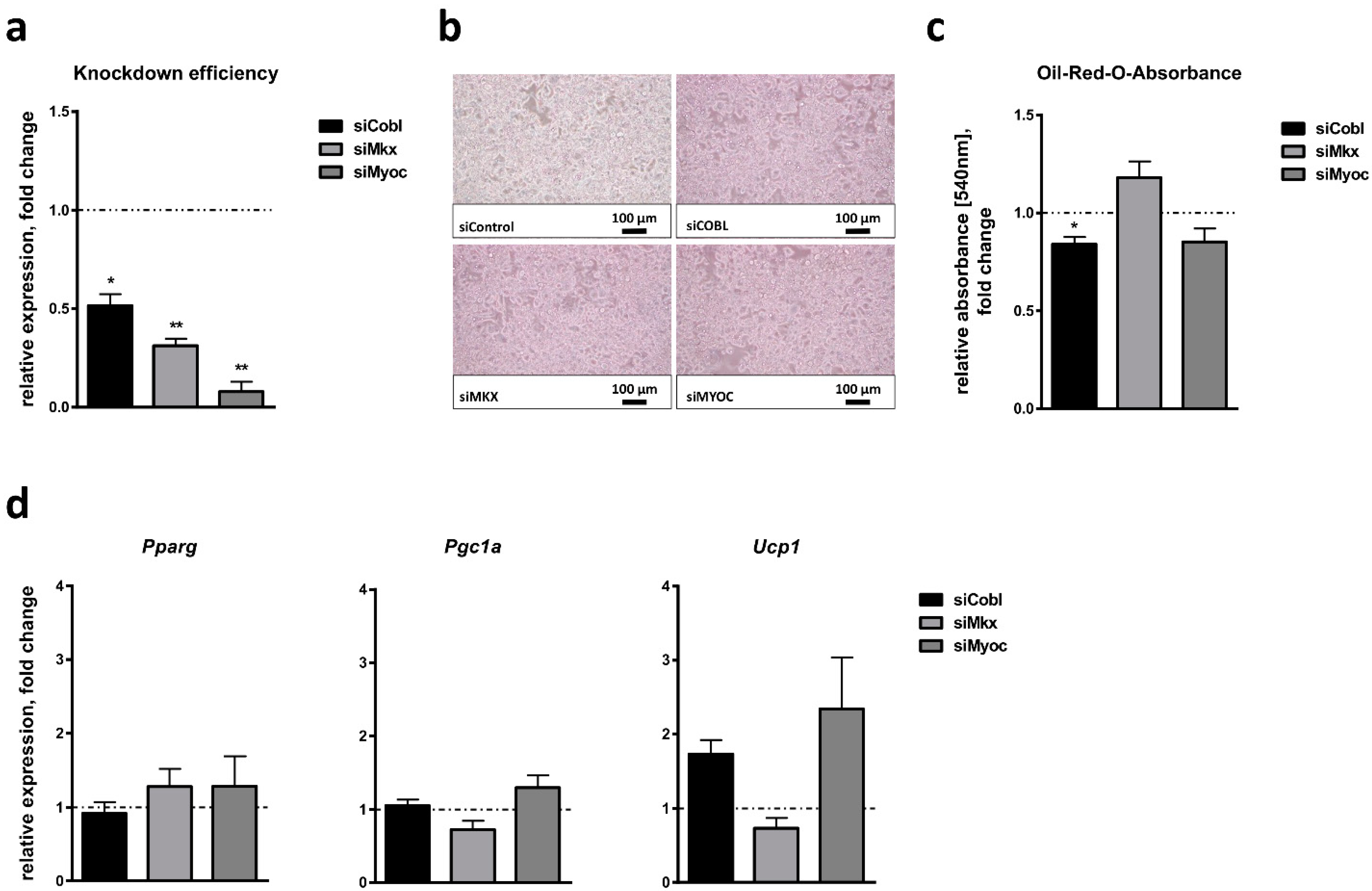
2.4. Effect of Candidate Gene Knockdown on Brown Adipocyte Differentiation
2.5. Regulation of Candidate Gene Expression during White Adipocyte Browning
2.6. Effect of Candidate Gene Knockdown on White Adipocyte Browning
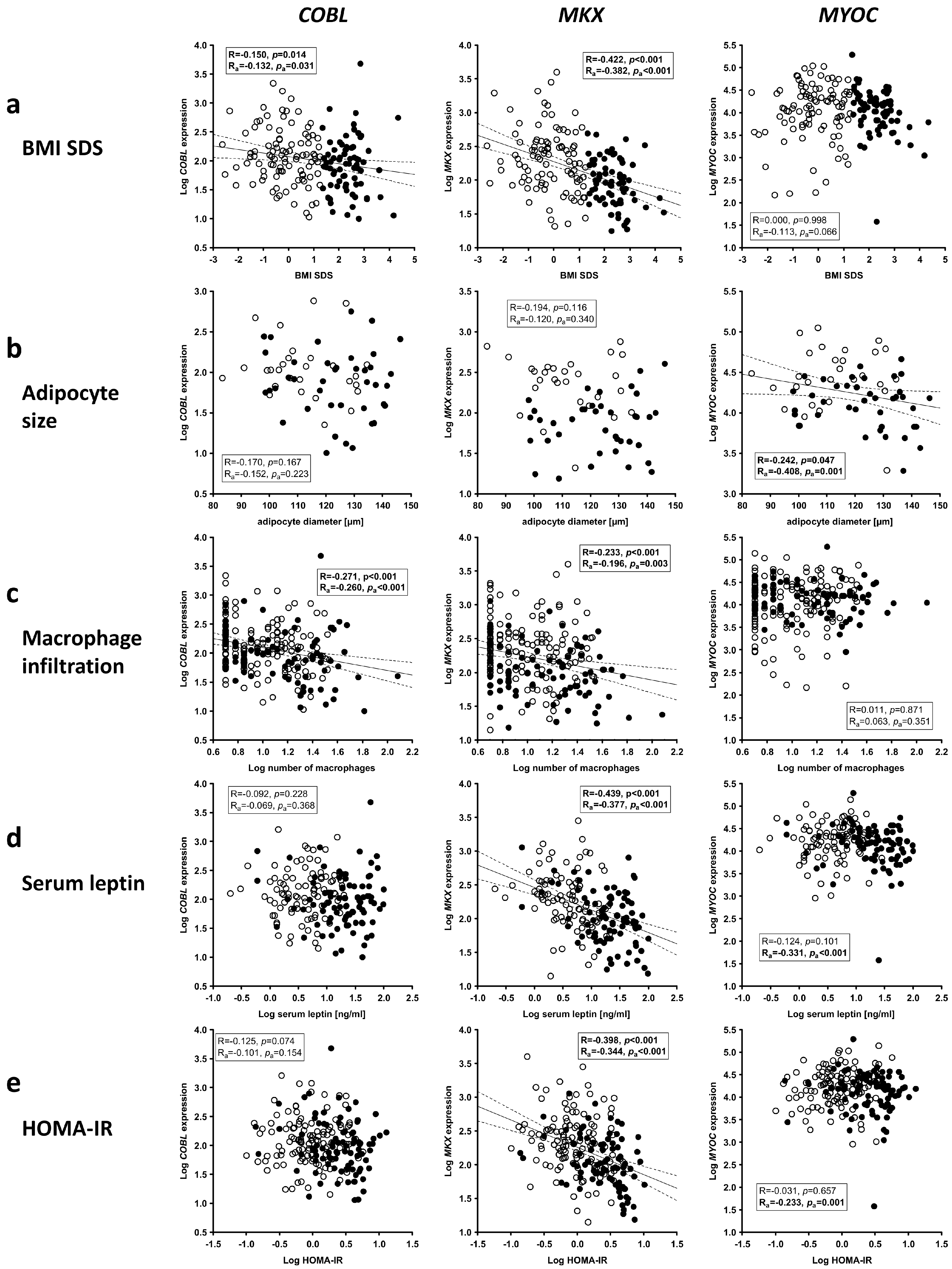
2.7. Association of Candidate Gene Expression in AT with Childhood Obesity
3. Discussion
4. Material and Methods
4.1. Human AT Biopsies (Leipzig AT Childhood Cohort)
4.2. Cold Exposure Experiments in Mice
4.3. Cell Culture
4.4. siRNA-Mediated Knockdown of Candidate Genes
4.5. RNA Isolation and Quantitative Real-Time PCR Analysis
4.6. Protein Isolation and Western Blot
4.7. Oil-Red-O Staining
4.8. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Geserick, M.; Vogel, M.; Gausche, R.; Lipek, T.; Spielau, U.; Keller, E.; Pfäffle, R.; Kiess, W.; Körner, A. Acceleration of BMI in Early Childhood and Risk of Sustained Obesity. N. Engl. J. Med. 2018, 379, 1303–1312. [Google Scholar] [CrossRef] [PubMed]
- Cummins, T.D.; Holden, C.R.; Sansbury, B.E.; Gibb, A.A.; Shah, J.; Zafar, N.; Tang, Y.; Hellmann, J.; Rai, S.N.; Spite, M.; et al. Metabolic remodeling of white adipose tissue in obesity. Am. J. Physiol. Endocrinol. Metab. 2014, 307, E262–E277. [Google Scholar] [CrossRef]
- Vázquez-Vela, M.E.F.; Torres, N.; Tovar, A.R. White Adipose Tissue as Endocrine Organ and Its Role in Obesity. Arch. Med. Res. 2008, 39, 715–728. [Google Scholar] [CrossRef] [PubMed]
- Kahn, C.R.; Wang, G.; Lee, K.Y. Altered adipose tissue and adipocyte function in the pathogenesis of metabolic syndrome. J. Clin. Investig. 2019, 129, 3990–4000. [Google Scholar] [CrossRef] [PubMed]
- Saely, C.H.; Geiger, K.; Drexel, H. Brown versus white adipose tissue: A mini-review. Gerontology 2012, 58, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Cypess, A.M. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef]
- Leitner, B.P.; Huang, S.; Brychta, R.J.; Duckworth, C.J.; Baskin, A.S.; McGehee, S.; Tal, I.; Dieckmann, W.; Gupta, G.; Kolodny, G.M.; et al. Mapping of human brown adipose tissue in lean and obese young men. Proc. Natl. Acad. Sci. USA 2017, 114, 8649–8654. [Google Scholar] [CrossRef]
- Tews, D.; Wabitsch, M. Brown Adipose Tissue in Children and Its Metabolic Function. Horm. Res. Paediatr. 2022, 95, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Maretich, P.; Kajimura, S. The Common and Distinct Features of Brown and Beige Adipocytes. Trends Endocrinol. Metab. TEM 2018, 29, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H. The mechanism of white and brown adipocyte differentiation. Diabetes Metab. J. 2013, 37, 85–90. [Google Scholar] [CrossRef]
- Herz, C.T.; Kiefer, F.W. Adipose tissue browning in mice and humans. J. Endocrinol. 2019, 241, R97–R109. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Wang, J.; Dai, H.; Duan, Y.; An, Y.; Shi, L.; Lv, Y.; Li, H.; Wang, C.; Ma, Q.; et al. Brown and beige adipose tissue: A novel therapeutic strategy for obesity and type 2 diabetes mellitus. Adipocyte 2021, 10, 48–65. [Google Scholar] [CrossRef]
- Tseng, Y.H.; Kokkotou, E.; Schulz, T.J.; Huang, T.L.; Winnay, J.N.; Taniguchi, C.M.; Tran, T.T.; Suzuki, R.; Espinoza, D.O.; Yamamoto, Y.; et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 2008, 454, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- van Marken Lichtenbelt, W.D.; Vanhommerig, J.W.; Smulders, N.M.; Drossaerts, J.M.; Kemerink, G.J.; Bouvy, N.D.; Schrauwen, P.; Teule, G.J. Cold-Activated Brown Adipose Tissue in Healthy Men. N. Engl. J. Med. 2009, 360, 1500–1508. [Google Scholar] [CrossRef]
- Chondronikola, M.; Volpi, E.; Borsheim, E.; Porter, C.; Annamalai, P.; Enerback, S.; Lidell, M.E.; Saraf, M.K.; Labbe, S.M.; Hurren, N.M.; et al. Brown adipose tissue improves whole-body glucose homeostasis and insulin sensitivity in humans. Diabetes 2014, 63, 4089–4099. [Google Scholar] [CrossRef] [PubMed]
- Drubach, L.A.; Palmer, E.L., III; Connolly, L.P.; Baker, A.; Zurakowski, D.; Cypess, A.M. Pediatric brown adipose tissue: Detection, epidemiology, and differences from adults. J. Pediatr. 2011, 159, 939–944. [Google Scholar] [CrossRef]
- Lundström, E.; Ljungberg, J.; Andersson, J.; Manell, H.; Strand, R.; Forslund, A.; Bergsten, P.; Weghuber, D.; Mörwald, K.; Zsoldos, F.; et al. Brown adipose tissue estimated with the magnetic resonance imaging fat fraction is associated with glucose metabolism in adolescents. Pediatr. Obes. 2019, 14, e12531. [Google Scholar] [CrossRef] [PubMed]
- Malpique, R.; Gallego-Escuredo, J.M.; Sebastiani, G.; Villarroya, J.; Lopez-Bermejo, A.; de Zegher, F.; Villarroya, F.; Ibanez, L. Brown adipose tissue in prepubertal children: Associations with sex, birthweight, and metabolic profile. Int. J. Obes. 2019, 43, 384–391. [Google Scholar] [CrossRef]
- Saito, M.; Okamatsu-Ogura, Y.; Matsushita, M.; Watanabe, K.; Yoneshiro, T.; Nio-Kobayashi, J.; Iwanaga, T.; Miyagawa, M.; Kameya, T.; Nakada, K.; et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: Effects of cold exposure and adiposity. Diabetes 2009, 58, 1526–1531. [Google Scholar] [CrossRef]
- Vijgen, G.H.; Bouvy, N.D.; Teule, G.J.; Brans, B.; Hoeks, J.; Schrauwen, P.; van Marken Lichtenbelt, W.D. Increase in brown adipose tissue activity after weight loss in morbidly obese subjects. J. Clin. Endocrinol. Metab. 2012, 97, E1229–E1233. [Google Scholar] [CrossRef]
- Soundarrajan, M.; Deng, J.; Kwasny, M.; Rubert, N.C.; Nelson, P.C.; El-Seoud, D.A.; Landsberg, L.; Neff, L.M. Activated brown adipose tissue and its relationship to adiposity and metabolic markers: An exploratory study. Adipocyte 2020, 9, 87–95. [Google Scholar] [CrossRef]
- White, J.D.; Dewal, R.S.; Stanford, K.I. The beneficial effects of brown adipose tissue transplantation. Mol. Asp. Med. 2019, 68, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Sidossis, L.; Kajimura, S. Brown and beige fat in humans: Thermogenic adipocytes that control energy and glucose homeostasis. J. Clin. Investig. 2015, 125, 478–486. [Google Scholar] [CrossRef]
- Landgraf, K.; Rockstroh, D.; Wagner, I.V.; Weise, S.; Tauscher, R.; Schwartze, J.T.; Löffler, D.; Bühligen, U.; Wojan, M.; Till, H.; et al. Evidence of early alterations in adipose tissue biology and function and its association with obesity-related inflammation and insulin resistance in children. Diabetes 2015, 64, 1249–1261. [Google Scholar] [CrossRef] [PubMed]
- Rockstroh, D.; Landgraf, K.; Wagner, I.V.; Gesing, J.; Tauscher, R.; Lakowa, N.; Kiess, W.; Bühligen, U.; Wojan, M.; Till, H.; et al. Direct evidence of brown adipocytes in different fat depots in children. PLoS ONE 2015, 10, e0117841. [Google Scholar] [CrossRef]
- Landgraf, K.; Kühnapfel, A.; Schlanstein, M.; Biemann, R.; Isermann, B.; Kempf, E.; Kirsten, H.; Scholz, M.; Körner, A. Transcriptome Analyses of Adipose Tissue Samples Identify EGFL6 as a Candidate Gene Involved in Obesity-Related Adipose Tissue Dysfunction in Children. Int. J. Mol. Sci. 2022, 23, 4349. [Google Scholar] [CrossRef]
- Haag, N.; Schwintzer, L.; Ahuja, R.; Koch, N.; Grimm, J.; Heuer, H.; Qualmann, B.; Kessels, M.M. The actin nucleator Cobl is crucial for Purkinje cell development and works in close conjunction with the F-actin binding protein Abp1. J. Neurosci. 2012, 32, 17842–17856. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, R.; Pinyol, R.; Reichenbach, N.; Custer, L.; Klingensmith, J.; Kessels, M.M.; Qualmann, B. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell 2007, 131, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Perdikari, A.; Leparc, G.G.; Balaz, M.; Pires, N.D.; Lidell, M.E.; Sun, W.; Fernandez-Albert, F.; Muller, S.; Akchiche, N.; Dong, H.; et al. BATLAS: Deconvoluting Brown Adipose Tissue. Cell Rep. 2018, 25, 784–797.e4. [Google Scholar] [CrossRef] [PubMed]
- Svensson, P.A.; Jernas, M.; Sjöholm, K.; Hoffmann, J.M.; Nilsson, B.E.; Hansson, M.; Carlsson, L.M. Gene expression in human brown adipose tissue. Int. J. Mol. Med. 2011, 27, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Ito, Y.; Shinohara, M.; Yamashita, S.; Ichinose, S.; Kishida, A.; Oyaizu, T.; Kayama, T.; Nakamichi, R.; Koda, N.; et al. Gene targeting of the transcription factor Mohawk in rats causes heterotopic ossification of Achilles tendon via failed tenogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, 7840–7845. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S.; Ito, Y.; Ueno-Kudoh, H.; Shimizu, H.; Uchibe, K.; Albini, S.; Mitsuoka, K.; Miyaki, S.; Kiso, M.; Nagai, A.; et al. A systems approach reveals that the myogenesis genome network is regulated by the transcriptional repressor RP58. Dev. Cell 2009, 17, 836–848. [Google Scholar] [CrossRef]
- Otabe, K.; Nakahara, H.; Hasegawa, A.; Matsukawa, T.; Ayabe, F.; Onizuka, N.; Inui, M.; Takada, S.; Ito, Y.; Sekiya, I.; et al. Transcription factor Mohawk controls tenogenic differentiation of bone marrow mesenchymal stem cells in vitro and in vivo. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2015, 33, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Crawford, A.; Souzeau, E.; Agar, A.; Ridge, B.; Dubowsky, A.; Burdon, K.P.; Craig, J.E. Identification of a novel MYOC mutation, p.(Trp373), in a family with open angle glaucoma. Gene 2014, 545, 271–275. [Google Scholar] [CrossRef]
- Kwon, H.S.; Johnson, T.V.; Tomarev, S.I. Myocilin stimulates osteogenic differentiation of mesenchymal stem cells through mitogen-activated protein kinase signaling. J. Biol. Chem. 2013, 288, 16882–16894. [Google Scholar] [CrossRef] [PubMed]
- Klaus, S.; Choy, L.; Champigny, O.; Cassard-Doulcier, A.M.; Ross, S.; Spiegelman, B.; Ricquier, D. Characterization of the novel brown adipocyte cell line HIB 1B. Adrenergic pathways involved in regulation of uncoupling protein gene expression. J. Cell Sci. 1994, 107 Pt 1, 313–319. [Google Scholar] [CrossRef]
- Fayyad, A.M.; Khan, A.A.; Abdallah, S.H.; Alomran, S.S.; Bajou, K.; Khattak, M.N.K. Rosiglitazone Enhances Browning Adipocytes in Association with MAPK and PI3-K Pathways During the Differentiation of Telomerase-Transformed Mesenchymal Stromal Cells into Adipocytes. Int. J. Mol. Sci. 2019, 20, 1618. [Google Scholar] [CrossRef]
- Nedergaard, J.; Petrovic, N.; Lindgren, E.M.; Jacobsson, A.; Cannon, B. PPARgamma in the control of brown adipocyte differentiation. Biochim. Biophys. Acta 2005, 1740, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Kaisanlahti, A.; Glumoff, T. Browning of white fat: Agents and implications for beige adipose tissue to type 2 diabetes. J. Physiol. Biochem. 2019, 75, 1–10. [Google Scholar] [CrossRef]
- Elsen, M.; Raschke, S.; Tennagels, N.; Schwahn, U.; Jelenik, T.; Roden, M.; Romacho, T.; Eckel, J. BMP4 and BMP7 induce the white-to-brown transition of primary human adipose stem cells. Am. J. Physiol. Cell Physiol. 2014, 306, C431–C440. [Google Scholar] [CrossRef]
- Gasca, S.; Hill, D.P.; Klingensmith, J.; Rossant, J. Characterization of a gene trap insertion into a novel gene, cordon-bleu, expressed in axial structures of the gastrulating mouse embryo. Dev. Genet. 1995, 17, 141–154. [Google Scholar] [CrossRef]
- Husson, C.; Renault, L.; Didry, D.; Pantaloni, D.; Carlier, M.F. Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol. Cell 2011, 43, 464–477. [Google Scholar] [CrossRef]
- Emont, M.P.; Jacobs, C.; Essene, A.L.; Pant, D.; Tenen, D.; Colleluori, G.; Di Vincenzo, A.; Jorgensen, A.M.; Dashti, H.; Stefek, A.; et al. A single-cell atlas of human and mouse white adipose tissue. Nature 2022, 603, 926–933. [Google Scholar] [CrossRef]
- Fang, L.; Kojima, K.; Zhou, L.; Crossman, D.K.; Mobley, J.A.; Grams, J. Analysis of the Human Proteome in Subcutaneous and Visceral Fat Depots in Diabetic and Non-diabetic Patients with Morbid Obesity. J. Proteom. Bioinform. 2015, 8, 133–141. [Google Scholar] [CrossRef]
- Anderson, D.M.; Arredondo, J.; Hahn, K.; Valente, G.; Martin, J.F.; Wilson-Rawls, J.; Rawls, A. Mohawk is a novel homeobox gene expressed in the developing mouse embryo. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2006, 235, 792–801. [Google Scholar] [CrossRef]
- Lee, K.I.; Gamini, R.; Olmer, M.; Ikuta, Y.; Hasei, J.; Baek, J.; Alvarez-Garcia, O.; Grogan, S.P.; D’Lima, D.D.; Asahara, H.; et al. Mohawk is a transcription factor that promotes meniscus cell phenotype and tissue repair and reduces osteoarthritis severity. Sci. Transl. Med. 2020, 12, aan7967. [Google Scholar] [CrossRef]
- Siersbaek, R.; Nielsen, R.; Mandrup, S. PPARgamma in adipocyte differentiation and metabolism--novel insights from genome-wide studies. FEBS Lett. 2010, 584, 3242–3249. [Google Scholar] [CrossRef]
- Nguyen, T.D.; Chen, P.; Huang, W.D.; Chen, H.; Johnson, D.; Polansky, J.R. Gene structure and properties of TIGR, an olfactomedin-related glycoprotein cloned from glucocorticoid-induced trabecular meshwork cells. J. Biol. Chem. 1998, 273, 6341–6350. [Google Scholar] [CrossRef] [PubMed]
- Ortego, J.; Escribano, J.; Coca-Prados, M. Cloning and characterization of subtracted cDNAs from a human ciliary body library encoding TIGR, a protein involved in juvenile open angle glaucoma with homology to myosin and olfactomedin. FEBS Lett. 1997, 413, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Fingert, J.H.; Ying, L.; Swiderski, R.E.; Nystuen, A.M.; Arbour, N.C.; Alward, W.L.; Sheffield, V.C.; Stone, E.M. Characterization and comparison of the human and mouse GLC1A glaucoma genes. Genome Res. 1998, 8, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Law, J.M.; Morris, D.E.; Robinson, L.; Randell, T.; Denvir, L.; Symonds, M.E.; Budge, H. Reduced brown adipose tissue-associated skin temperature following cold stimulation in children and adolescents with type 1 diabetes. Pediatr. Diabetes 2021, 22, 407–416. [Google Scholar] [CrossRef]
- Gomez-Garcia, I.; Trepiana, J.; Fernandez-Quintela, A.; Giralt, M.; Portillo, M.P. Sexual Dimorphism in Brown Adipose Tissue Activation and White Adipose Tissue Browning. Int. J. Mol. Sci. 2022, 23, 8250. [Google Scholar] [CrossRef]
- Landgraf, K.; Kloting, N.; Gericke, M.; Maixner, N.; Guiu-Jurado, E.; Scholz, M.; Witte, A.V.; Beyer, F.; Schwartze, J.T.; Lacher, M.; et al. The Obesity-Susceptibility Gene TMEM18 Promotes Adipogenesis through Activation of PPARG. Cell Rep. 2020, 33, 108295. [Google Scholar] [CrossRef]
- Kempf, E.; Vogel, M.; Vogel, T.; Kratzsch, J.; Landgraf, K.; Kühnapfel, A.; Gausche, R.; Grafe, D.; Sergeyev, E.; Pfäffle, R.; et al. Dynamic alterations in linear growth and endocrine parameters in children with obesity and height reference values. EClinicalMedicine 2021, 37, 100977. [Google Scholar] [CrossRef]
- Kromeyer-Hauschild, K.; Wabitsch, M.; Kunze, D.; Geller, F.; Geiss, H.C.; Ziegler, A.; Hesse, V.; von Hippel, A.; Johnsen, D.; Korte, W.; et al. Perzentilen für den body mass index für das Kindes- und Jugendalter unter Heranziehung verschiedener deutscher Stichproben. (Centiles for body mass index for children and adolescents derived from distinct independent German cohorts). Mon. Kinderheilkd 2001, 149, 807–818. [Google Scholar] [CrossRef]
- Weiner, J.; Rohde, K.; Krause, K.; Zieger, K.; Klöting, N.; Kralisch, S.; Kovacs, P.; Stumvoll, M.; Blüher, M.; Böttcher, Y.; et al. Brown adipose tissue (BAT) specific vaspin expression is increased after obesogenic diets and cold exposure and linked to acute changes in DNA-methylation. Mol. Metab. 2017, 6, 482–493. [Google Scholar] [CrossRef]
- Karamanlidis, G.; Karamitri, A.; Docherty, K.; Hazlerigg, D.G.; Lomax, M.A. C/EBPbeta reprograms white 3T3-L1 preadipocytes to a Brown adipocyte pattern of gene expression. J. Biol. Chem. 2007, 282, 24660–24669. [Google Scholar] [CrossRef]
- Zebisch, K.; Voigt, V.; Wabitsch, M.; Brandsch, M. Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Anal. Biochem. 2012, 425, 88–90. [Google Scholar] [CrossRef]
- Bernhard, F.; Landgraf, K.; Kloting, N.; Berthold, A.; Buttner, P.; Friebe, D.; Kiess, W.; Kovacs, P.; Bluher, M.; Korner, A. Functional relevance of genes implicated by obesity genome-wide association study signals for human adipocyte biology. Diabetologia 2013, 56, 311–322. [Google Scholar] [CrossRef]
- Christen, L.; Broghammer, H.; Rapöhn, I.; Möhlis, K.; Strehlau, C.; Ribas-Latre, A.; Gebhardt, C.; Roth, L.; Krause, K.; Landgraf, K.; et al. Myoglobin-mediated lipid shuttling increases adrenergic activation of brown and white adipocyte metabolism and is as a marker of thermogenic adipocytes in humans. Clin. Transl. Med. 2022, 12, e1108. [Google Scholar] [CrossRef]
- Weiner, J.; Roth, L.; Kranz, M.; Brust, P.; Boelen, A.; Klöting, N.; Heiker, J.T.; Blüher, M.; Tönjes, A.; Pfluger, P.T.; et al. Leptin counteracts hypothermia in hypothyroidism through its pyrexic effects and by stabilizing serum thyroid hormone levels. Mol. Metab. 2021, 54, 101348. [Google Scholar] [CrossRef]
- Zieger, K.; Weiner, J.; Kunath, A.; Gericke, M.; Krause, K.; Kern, M.; Stumvoll, M.; Klöting, N.; Blüher, M.; Heiker, J.T. Ablation of kallikrein 7 (KLK7) in adipose tissue ameliorates metabolic consequences of high fat diet-induced obesity by counteracting adipose tissue inflammation in vivo. Cell. Mol. Life Sci. CMLS 2018, 75, 727–742. [Google Scholar] [CrossRef]
- Weiner, J.; Kranz, M.; Klöting, N.; Kunath, A.; Steinhoff, K.; Rijntjes, E.; Köhrle, J.; Zeisig, V.; Hankir, M.; Gebhardt, C.; et al. Thyroid hormone status defines brown adipose tissue activity and browning of white adipose tissues in mice. Sci. Rep. 2016, 6, 38124. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdul Majeed, S.; Dunzendorfer, H.; Weiner, J.; Heiker, J.T.; Kiess, W.; Körner, A.; Landgraf, K. COBL, MKX and MYOC Are Potential Regulators of Brown Adipose Tissue Development Associated with Obesity-Related Metabolic Dysfunction in Children. Int. J. Mol. Sci. 2023, 24, 3085. https://doi.org/10.3390/ijms24043085
Abdul Majeed S, Dunzendorfer H, Weiner J, Heiker JT, Kiess W, Körner A, Landgraf K. COBL, MKX and MYOC Are Potential Regulators of Brown Adipose Tissue Development Associated with Obesity-Related Metabolic Dysfunction in Children. International Journal of Molecular Sciences. 2023; 24(4):3085. https://doi.org/10.3390/ijms24043085
Chicago/Turabian StyleAbdul Majeed, Sarah, Helene Dunzendorfer, Juliane Weiner, John T. Heiker, Wieland Kiess, Antje Körner, and Kathrin Landgraf. 2023. "COBL, MKX and MYOC Are Potential Regulators of Brown Adipose Tissue Development Associated with Obesity-Related Metabolic Dysfunction in Children" International Journal of Molecular Sciences 24, no. 4: 3085. https://doi.org/10.3390/ijms24043085
APA StyleAbdul Majeed, S., Dunzendorfer, H., Weiner, J., Heiker, J. T., Kiess, W., Körner, A., & Landgraf, K. (2023). COBL, MKX and MYOC Are Potential Regulators of Brown Adipose Tissue Development Associated with Obesity-Related Metabolic Dysfunction in Children. International Journal of Molecular Sciences, 24(4), 3085. https://doi.org/10.3390/ijms24043085






