Single-Cell Transcriptomic Profiling of the Mouse Testicular Germ Cells Reveals Important Role of Phosphorylated GRTH/DDX25 in Round Spermatid Differentiation and Acrosome Biogenesis during Spermiogenesis
Abstract
1. Introduction
2. Results
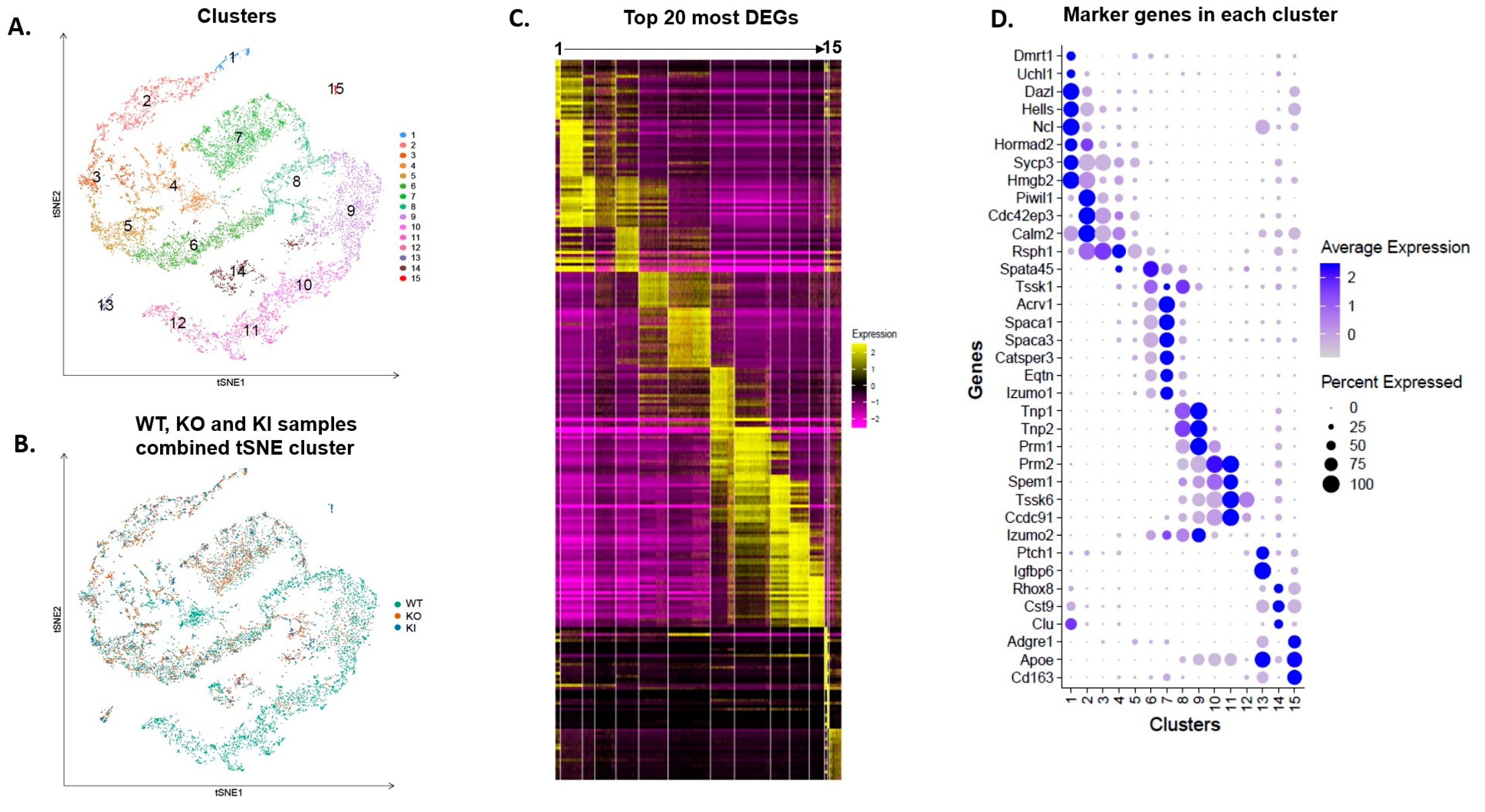
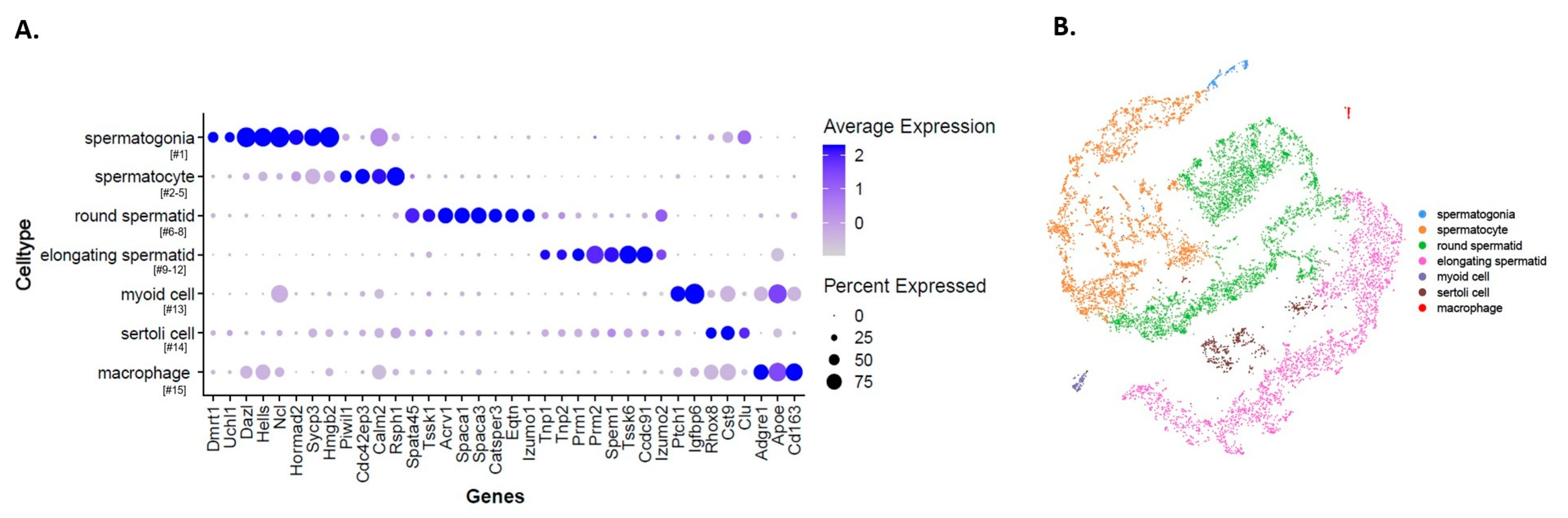
2.1. Single-Cell RNA-Sep Establishes Different Germ Cell Clusters in the Adult Mouse Testis
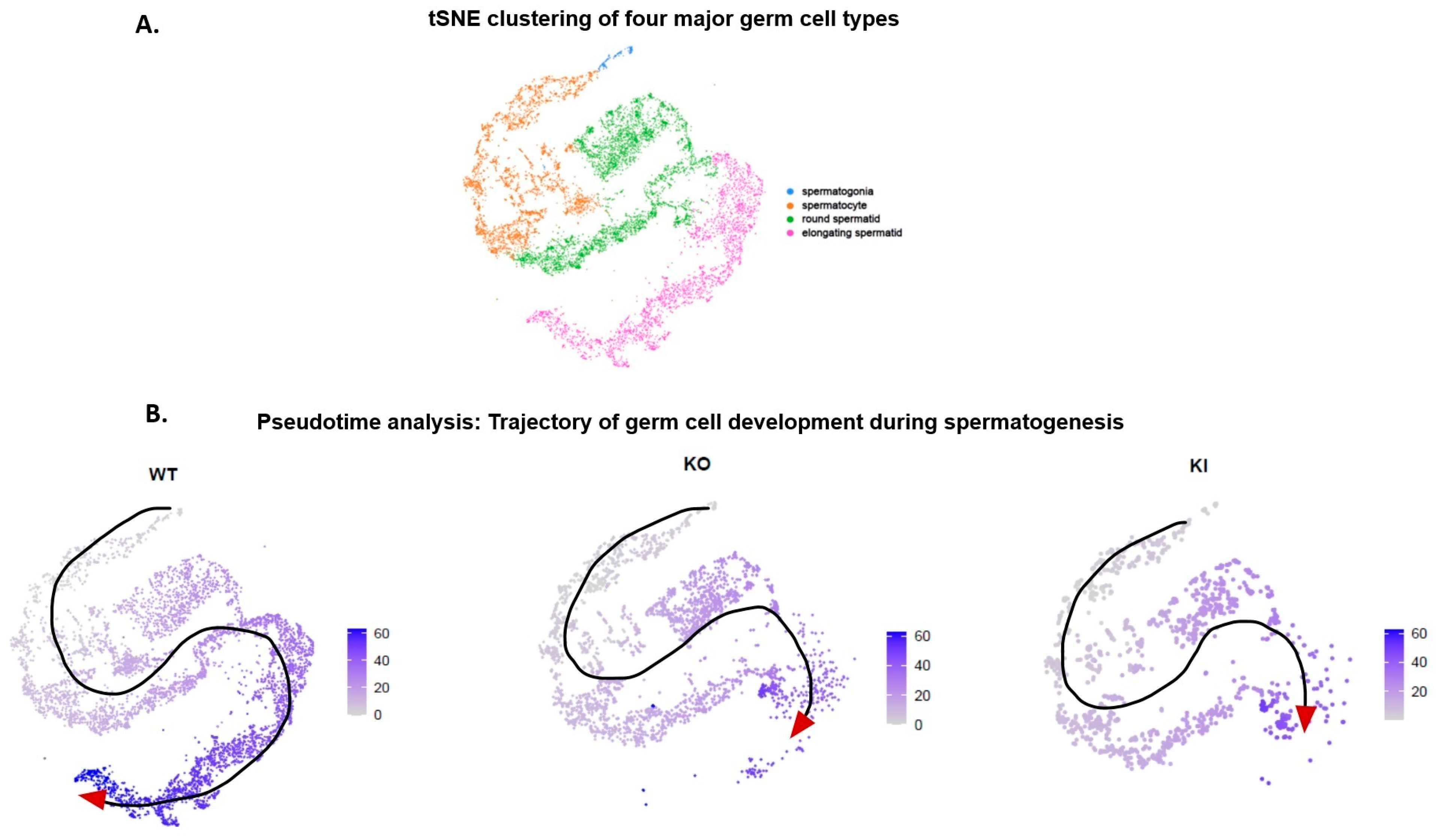
2.2. Pseudotime Trajectory Analysis of Germ Cell Development during Spermatogenesis
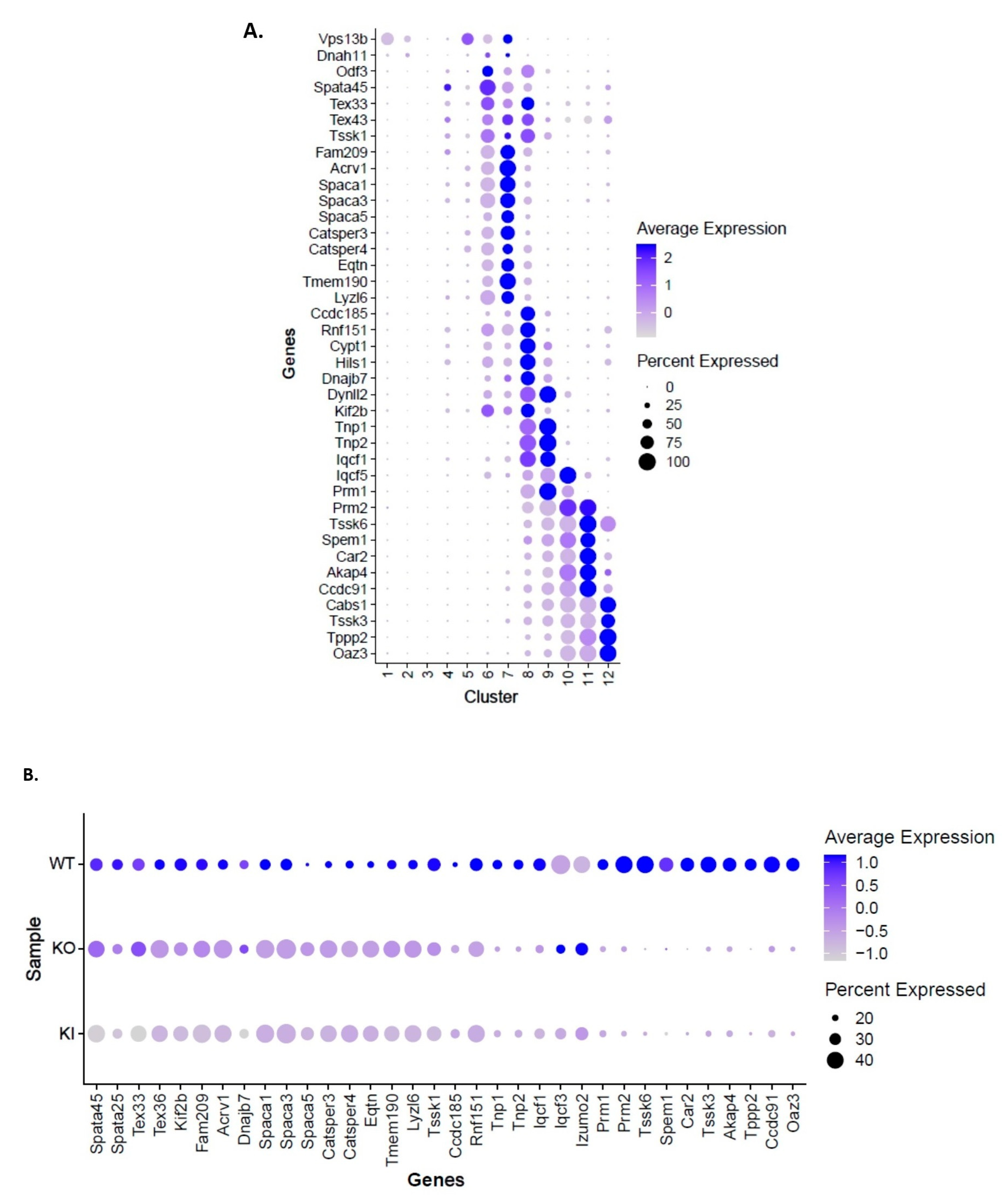
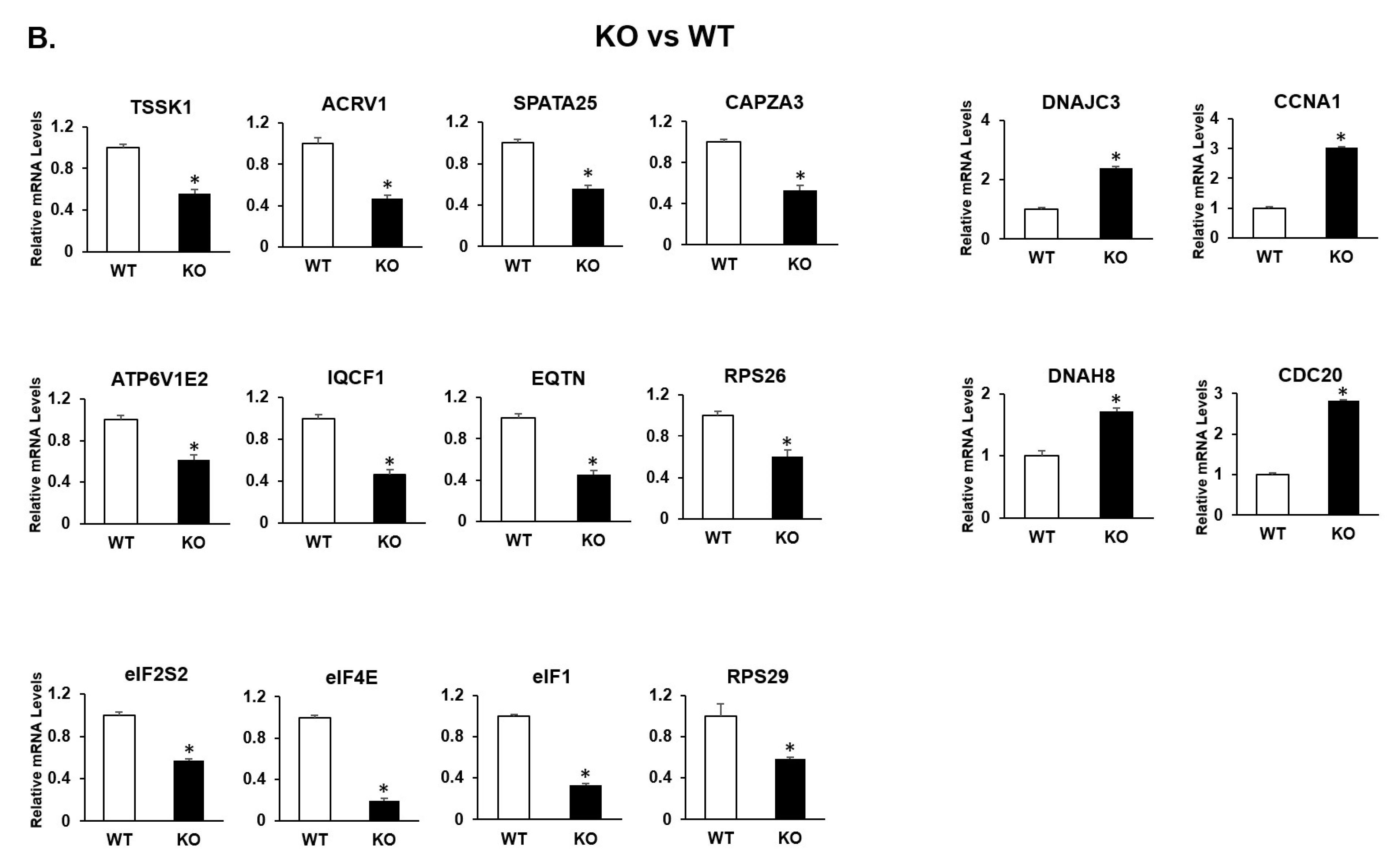
2.3. Transcriptional Profiles Are Significantly Altered in RS of KI and KO Mice during Spermiogenesis
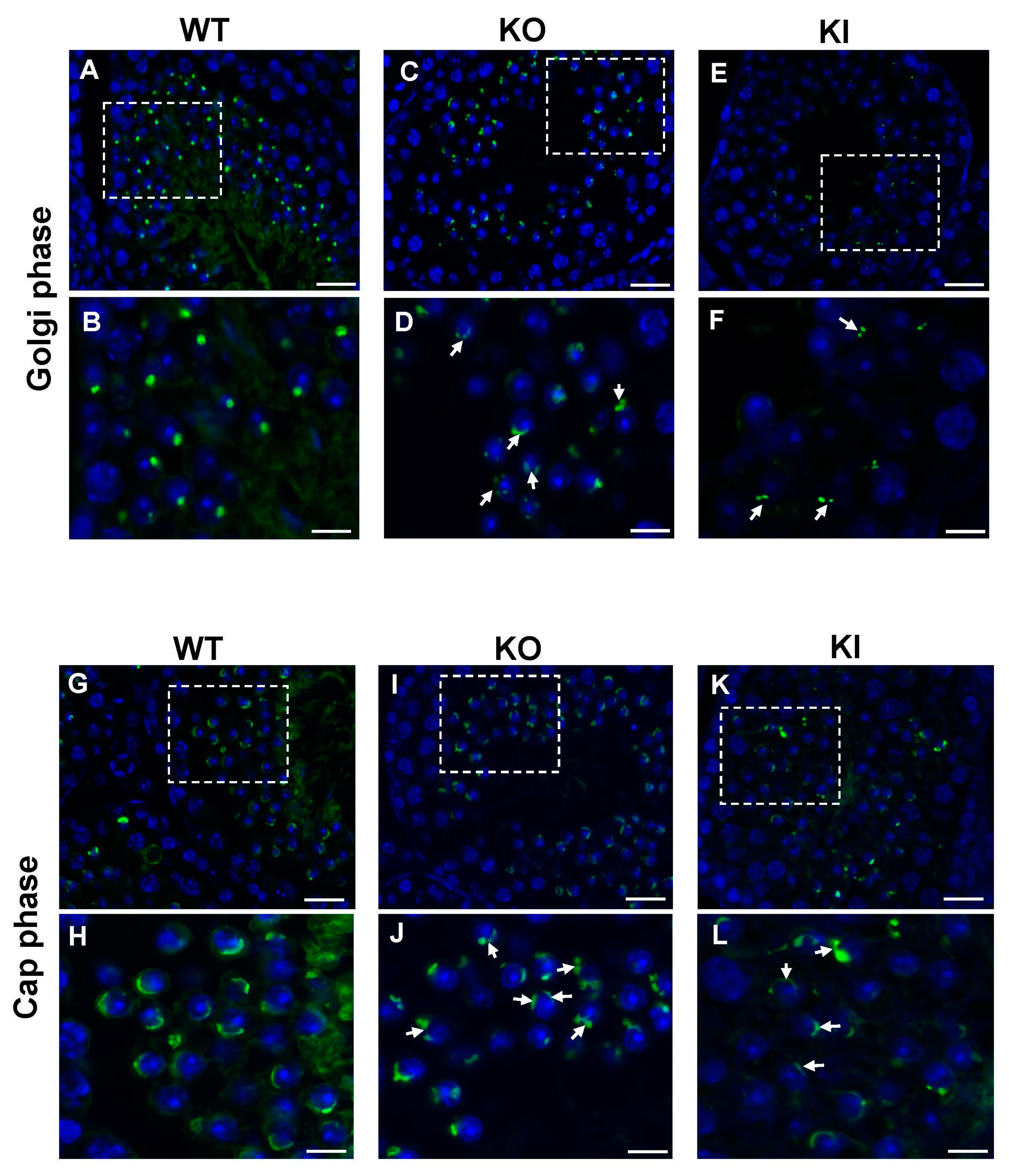
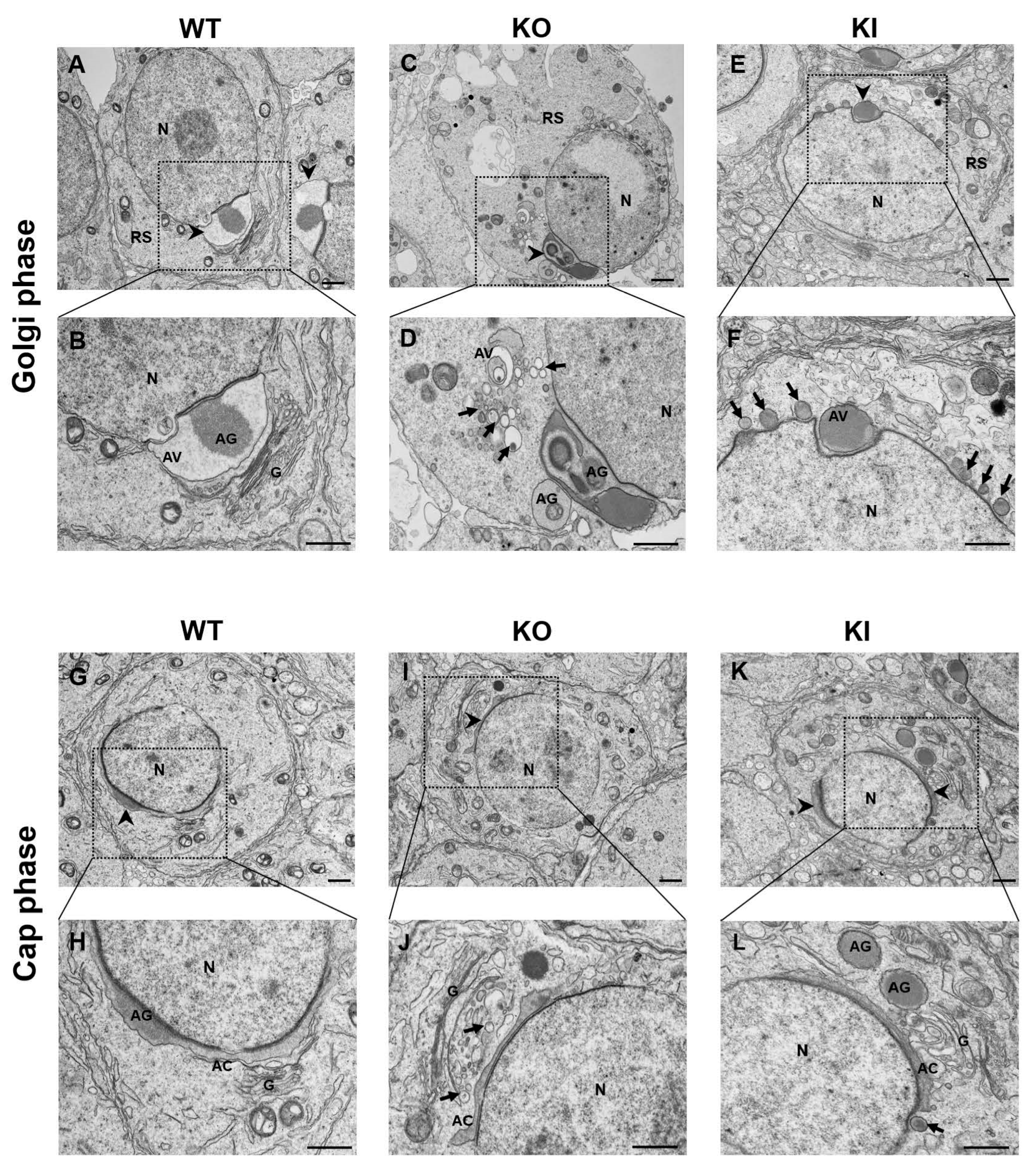
2.4. Morphological Changes in Acrosome Formation in Developing RS Revealed by TEM
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Isolation of Mice Testicular Cells for scRNA-seq
4.3. scRNA-seq Library Preparation, Sequencing and Transcript Counting
4.4. Cell Type Identification and Clustering Analysis
4.5. Pseudotime, Differential Gene Expression and GO Enrichment Analysis
4.6. Isolation of RS from Mice Seminiferous Tubules
4.7. Quantitative Real-Time PCR Analysis
4.8. Immunohistochemistry
4.9. Transmission Electron Microscopy
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clermont, Y. Spermatogenesis in man: A Study of the Spermatogonial Population. Fertil. Steril. 1966, 17, 705–721. [Google Scholar] [CrossRef]
- Griswold, M.D. Spermatogenesis: The commitment to meiosis. Physiol. Rev. 2015, 96, 1–17. [Google Scholar] [CrossRef]
- Kanatsu-Shinohara, M.; Shinohara, T. Spermatogonial stem cell self-renewal and development. Annu. Rev. Cell. Dev. Biol. 2013, 29, 163–187. [Google Scholar] [CrossRef]
- White-Cooper, H.; Bausek, N. Evolution and spermatogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 1465–1480. [Google Scholar] [CrossRef] [PubMed]
- Eddy, E.M. Male germ cell gene expression. Recent Prog. Horm. Res. 2002, 57, 103–128. [Google Scholar] [CrossRef] [PubMed]
- Hermann, B.P.; Cheng, K.; Singh, A.; Roa-De La Cruz, L.; Mutoji, K.N.; Chen, I.C.; Gildersleeve, H.; Lehle, J.D.; Mayo, M.; Westernstroer, B.; et al. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep. 2018, 25, 1650–1667.e1658. [Google Scholar] [CrossRef]
- Parvinen, M. The chromatoid body in spermatogenesis. Int. J. Androl. 2005, 28, 189–201. [Google Scholar] [CrossRef]
- Kotaja, N.; Sassone-Corsi, P. The chromatoid body: A germ-cell-specific RNA-processing centre. Nat. Rev. Mol. Cell. Biol. 2007, 8, 85–90. [Google Scholar] [CrossRef]
- Tang, P.Z.; Tsai-Morris, C.H.; Dufau, M.L. A novel gonadotropin-regulated testicular RNA helicase. A new member of the dead-box family. J. Biol. Chem. 1999, 274, 37932–37940. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Tsai-Morris, C.H.; Dufau, M.L. Cell-specific and hormone-regulated expression of gonadotropin-regulated testicular RNA helicase gene (GRTH/Ddx25) resulting from alternative utilization of translation initiation codons in the rat testis. J. Biol. Chem. 2003, 278, 27796–27803. [Google Scholar] [CrossRef]
- Tsai-Morris, C.H.; Sheng, Y.; Lee, E.; Lei, K.J.; Dufau, M.L. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is essential for spermatid development and completion of spermatogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 6373–6378. [Google Scholar] [CrossRef]
- Dufau, M.L.; Tsai-Morris, C.H. Gonadotropin-regulated testicular helicase (GRTH/DDX25): An essential regulator of spermatogenesis. Trends Endocrinol. Metab. 2007, 18, 314–320. [Google Scholar] [CrossRef]
- Tsai-Morris, C.H.; Sheng, Y.; Gutti, R.K.; Tang, P.Z.; Dufau, M.L. Gonadotropin-regulated testicular RNA helicase (GRTH/DDX25): A multifunctional protein essential for spermatogenesis. J. Androl. 2010, 31, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Dufau, M.L.; Kavarthapu, R. Gonadotropin regulation testicular RNA helicase, two decades of studies on its structure function and regulation from its discovery opens a window for development of a non-hormonal oral male Contraceptive. Front. Endocrinol. 2019, 10, 710. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Tsai-Morris, C.H.; Gutti, R.; Maeda, Y.; Dufau, M.L. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is a transport protein involved in gene-specific mRNA export and protein translation during spermatogenesis. J. Biol. Chem. 2006, 281, 35048–35056. [Google Scholar] [CrossRef] [PubMed]
- Tsai-Morris, C.H.; Sato, H.; Gutti, R.; Dufau, M.L. Role of gonadotropin regulated testicular RNA helicase (GRTH/Ddx25) on polysomal associated mRNAs in mouse testis. PLoS ONE 2012, 7, e32470. [Google Scholar] [CrossRef]
- Tsai-Morris, C.H.; Koh, E.; Sheng, Y.; Maeda, Y.; Gutti, R.; Namiki, M.; Dufau, M.L. Polymorphism of the GRTH/DDX25 gene in normal and infertile Japanese men: A missense mutation associated with loss of GRTH phosphorylation. Mol. Hum. Reprod. 2007, 13, 887–892. [Google Scholar] [CrossRef]
- Raju, M.; Hassan, S.A.; Kavarthapu, R.; Anbazhagan, R.; Dufau, M.L. Characterization of the Phosphorylation Site of GRTH/DDX25 and Protein Kinase A Binding Interface Provides Structural Basis for the Design of a Non-Hormonal Male Contraceptive. Sci. Rep. 2019, 9, 6705. [Google Scholar] [CrossRef]
- Kavarthapu, R.; Anbazhagan, R.; Raju, M.; Morris, C.T.; Pickel, J.; Dufau, M.L. Targeted knock-in mice with a human mutation in GRTH/DDX25 reveals the essential role of phosphorylated GRTH in spermatid development during spermatogenesis. Hum. Mol. Genet. 2019, 28, 2561–2572. [Google Scholar] [CrossRef] [PubMed]
- Bacher, R.; Kendziorski, C. Design and computational analysis of single-cell RNA sequencing experiments. Genome. Biol. 2016, 17, 63. [Google Scholar] [CrossRef] [PubMed]
- Green, C.D.; Ma, Q.; Manske, G.L.; Shami, A.N.; Zheng, X.; Marini, S.; Moritz, L.; Sultan, C.; Gurczynski, S.J.; Moore, B.B.; et al. A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev. Cell 2018, 46, 651–667.e610. [Google Scholar] [CrossRef] [PubMed]
- Lukassen, S.; Bosch, E.; Ekici, A.B.; Winterpacht, A. Characterization of germ cell differentiation in the male mouse through single-cell RNA sequencing. Sci. Rep. 2018, 8, 6521. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Hao, Z.; Jha, K.N.; Zhang, Z.; Urekar, C.; Digilio, L.; Pulido, S.; Strauss, I.I.I.J.F.; Flickinger, C.J.; Herr, J.C. Targeted deletion of Tssk1 and 2 causes male infertility due to haploinsufficiency. Dev. Biol. 2008, 319, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Osuru, H.P.; Monroe, J.E.; Chebolu, A.P.; Akamune, J.; Pramoonjago, P.; Ranpura, S.A.; Reddi, P.P. The acrosomal protein SP-10 (Acrv1) is an ideal marker for staging of the cycle of seminiferous epithelium in the mouse. Mol. Reprod. Dev. 2014, 81, 896–907. [Google Scholar] [CrossRef]
- Ito, C.; Yamatoya, K.; Yoshida, K.; Fujimura, L.; Sugiyama, H.; Suganami, A.; Tamura, Y.; Hatano, M.; Miyado, K.; Toshimori, K. Deletion of Eqtn in mice reduces male fertility and sperm-egg adhesion. Reproduction 2018, 156, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Xu, W.; Li, D.; Zhao, X.; Dai, J.; Wang, Z.; Yan, X.; Qin, M.; Zhang, Y.; Xu, C.; et al. A novel acrosomal protein, IQCF1, involved in sperm capacitation and the acrosome reaction. Andrology 2015, 3, 332–344. [Google Scholar] [CrossRef]
- Sosnik, J.; Buffone, M.G.; Visconti, P.E. Analysis of CAPZA3 localization reveals temporally discrete events during the acrosome reaction. J. Cell Physiol. 2010, 224, 575–580. [Google Scholar] [CrossRef]
- Pietrement, C.; Sun-Wada, G.-H.; Da Silva, N.; McKee, M.; Marshansky, V.; Brown, D.; Futai, M.; Breton, S. Distinct Expression Patterns of Different Subunit Isoforms of the V-ATPase in the Rat Epididymis. Biol. Reprod. 2006, 74, 185–194. [Google Scholar] [CrossRef]
- Zhou, Y.; Qin, D.; Tang, A.; Zhou, D.; Qin, J.; Yan, B.; Diao, R.; Jiang, Z.; Cai, Z.; Gui, Y. Developmental expression pattern of a novel gene, TSG23/Tsg23, suggests a role in spermatogenesis. Mol. Hum. Reprod. 2009, 15, 223–230. [Google Scholar] [CrossRef]
- Mondal, G.; Sengupta, S.; Panda, C.K.; Gollin, S.M.; Saunders, W.S.; Roychoudhury, S. Overexpression of Cdc20 leads to impairment of the spindle assembly checkpoint and aneuploidization in oral cancer. Carcinogenesis 2007, 28, 81–92. [Google Scholar] [CrossRef]
- Wolgemuth, D.J.; Lele, K.M.; Jobanputra, V.; Salazar, G. The A-type cyclins and the meiotic cell cycle in mammalian male germ cells. Int. J. Androl. 2004, 27, 192–199. [Google Scholar] [CrossRef]
- Rivera, A.; Mavila, A.; Bayless, K.J.; Davis, G.E.; Maxwell, S.A. Cyclin A1 is a p53-induced gene that mediates apoptosis, G2/M arrest, and mitotic catastrophe in renal, ovarian, and lung carcinoma cells. Cell Mol. Life Sci. 2006, 63, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Jennings, M.J.; Hathazi, D.; Nguyen, C.D.L.; Munro, B.; Münchberg, U.; Ahrends, R.; Schenck, A.; Eidhof, I.; Freier, E.; Synofzik, M.; et al. (2021) Intracellular Lipid Accumulation and Mitochondrial Dysfunction Accompanies Endoplasmic Reticulum Stress Caused by Loss of the Co-chaperone DNAJC3. Front. Cell Dev. Biol. 2021, 9, 710247. [Google Scholar] [CrossRef]
- Yang, Y.; Jiang, C.; Zhang, X.; Liu, X.; Li, J.; Qiao, X.; Liu, H.; Shen, Y. Loss-of-function mutation in DNAH8 induces asthenoteratospermia associated with multiple morphological abnormalities of the sperm flagella. Clin. Genet. 2020, 98, 396–401. [Google Scholar] [CrossRef]
- Khawar, M.B.; Gao, H.; Li, W. Mechanism of Acrosome Biogenesis in Mammals. Front. Cell Dev. Biol. 2019, 7, 195. [Google Scholar] [CrossRef]
- Ramalho-Santos, J.; Schatten, G.; Moreno, R.D. Control of membrane fusion during spermiogenesis and the acrosome reaction. Biol Reprod. 2007, 67, 1043–1051. [Google Scholar] [CrossRef]
- Li, Y.-C.; Hu, X.-Q.; Zhang, K.-Y.; Guo, J.; Hu, Z.-Y.; Tao, S.-X.; Xiao, L.-J.; Wang, Q.-Z.; Han, C.-S.; Liu, Y.-X. Afaf, a novel vesicle membrane protein, is related to acrosome formation in murine testis. FEBS Lett. 2006, 580, 4266–4273. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, Y.; Satouh, Y.; Inoue, N.; Isotani, A.; Ikawa, M.; Okabe, M. SPACA1-deficient male mice are infertile with abnormally shaped sperm heads reminiscent of globozoospermia. Development 2012, 139, 3583–3589. [Google Scholar] [CrossRef]
- Kavarthapu, R.; Anbazhagan, R.; Sharma, A.K.; Shiloach, J.; Dufau, M.L. Linking Phospho-Gonadotropin Regulated Testicular RNA Helicase (GRTH/DDX25) to Histone Ubiquitination and Acetylation Essential for Spermatid Development During Spermiogenesis. Front. Cell Dev. Biol. 2020, 8, 310. [Google Scholar] [CrossRef] [PubMed]
- Grive, K.J.; Hu, Y.; Shu, E.; Grimson, A.; Elemento, O.; Grenier, J.K.; Cohen, P.E. Dynamic transcriptome profiles within spermatogonial and spermatocyte populations during postnatal testis maturation revealed by single-cell sequencing. PLoS Genet. 2019, 15, e1007810. [Google Scholar] [CrossRef]
- Lukassen, S.; Bosch, E.; Ekici, A.B.; Winterpacht, A. Single-cell RNA sequencing of adult mouse testes. Sci. Data 2018, 5, 180192. [Google Scholar] [CrossRef] [PubMed]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; A Pliner, H.; Trapnell, C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 2017, 14, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Da Ros, M.; Lehtiniemi, T.; Olotu, O.; Meikar, O.; Kotaja, N. Enrichment of pachytene spermatocytes and spermatids from mouse testes using standard laboratory equipment. J. Vis. Exp. 2019, 151, e60271. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kavarthapu, R.; Anbazhagan, R.; Pal, S.; Dufau, M.L. Single-Cell Transcriptomic Profiling of the Mouse Testicular Germ Cells Reveals Important Role of Phosphorylated GRTH/DDX25 in Round Spermatid Differentiation and Acrosome Biogenesis during Spermiogenesis. Int. J. Mol. Sci. 2023, 24, 3127. https://doi.org/10.3390/ijms24043127
Kavarthapu R, Anbazhagan R, Pal S, Dufau ML. Single-Cell Transcriptomic Profiling of the Mouse Testicular Germ Cells Reveals Important Role of Phosphorylated GRTH/DDX25 in Round Spermatid Differentiation and Acrosome Biogenesis during Spermiogenesis. International Journal of Molecular Sciences. 2023; 24(4):3127. https://doi.org/10.3390/ijms24043127
Chicago/Turabian StyleKavarthapu, Raghuveer, Rajakumar Anbazhagan, Soumitra Pal, and Maria L. Dufau. 2023. "Single-Cell Transcriptomic Profiling of the Mouse Testicular Germ Cells Reveals Important Role of Phosphorylated GRTH/DDX25 in Round Spermatid Differentiation and Acrosome Biogenesis during Spermiogenesis" International Journal of Molecular Sciences 24, no. 4: 3127. https://doi.org/10.3390/ijms24043127
APA StyleKavarthapu, R., Anbazhagan, R., Pal, S., & Dufau, M. L. (2023). Single-Cell Transcriptomic Profiling of the Mouse Testicular Germ Cells Reveals Important Role of Phosphorylated GRTH/DDX25 in Round Spermatid Differentiation and Acrosome Biogenesis during Spermiogenesis. International Journal of Molecular Sciences, 24(4), 3127. https://doi.org/10.3390/ijms24043127





