Transcriptomic Analysis Reveals Dysregulation of the Mycobiome and Archaeome and Distinct Oncogenic Characteristics according to Subtype and Gender in Papillary Thyroid Carcinoma
Abstract
1. Introduction
2. Results
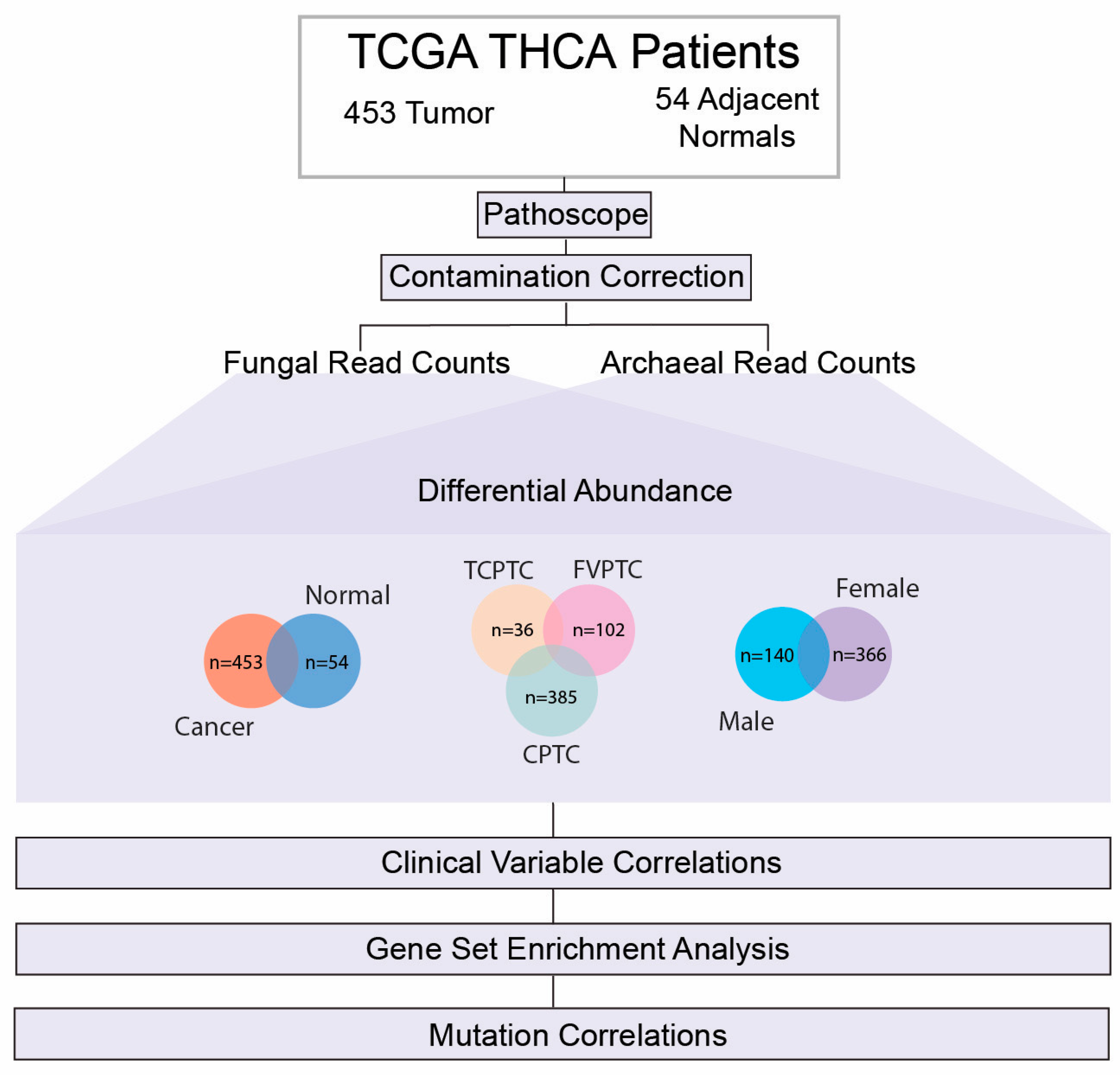
2.1. Data Acquisition and Extraction Identification of Microbial Reads
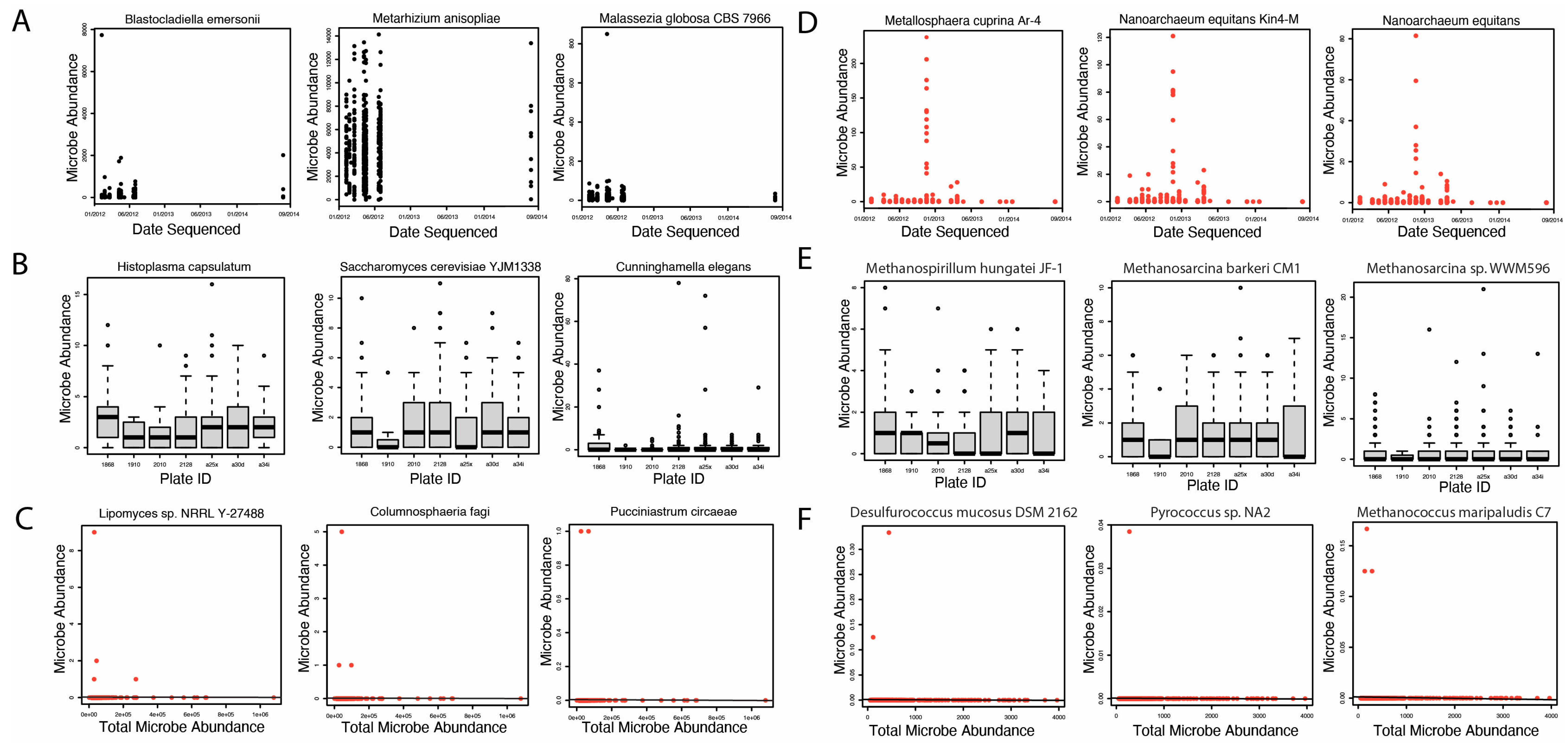
2.2. Removal of Potential Contaminants
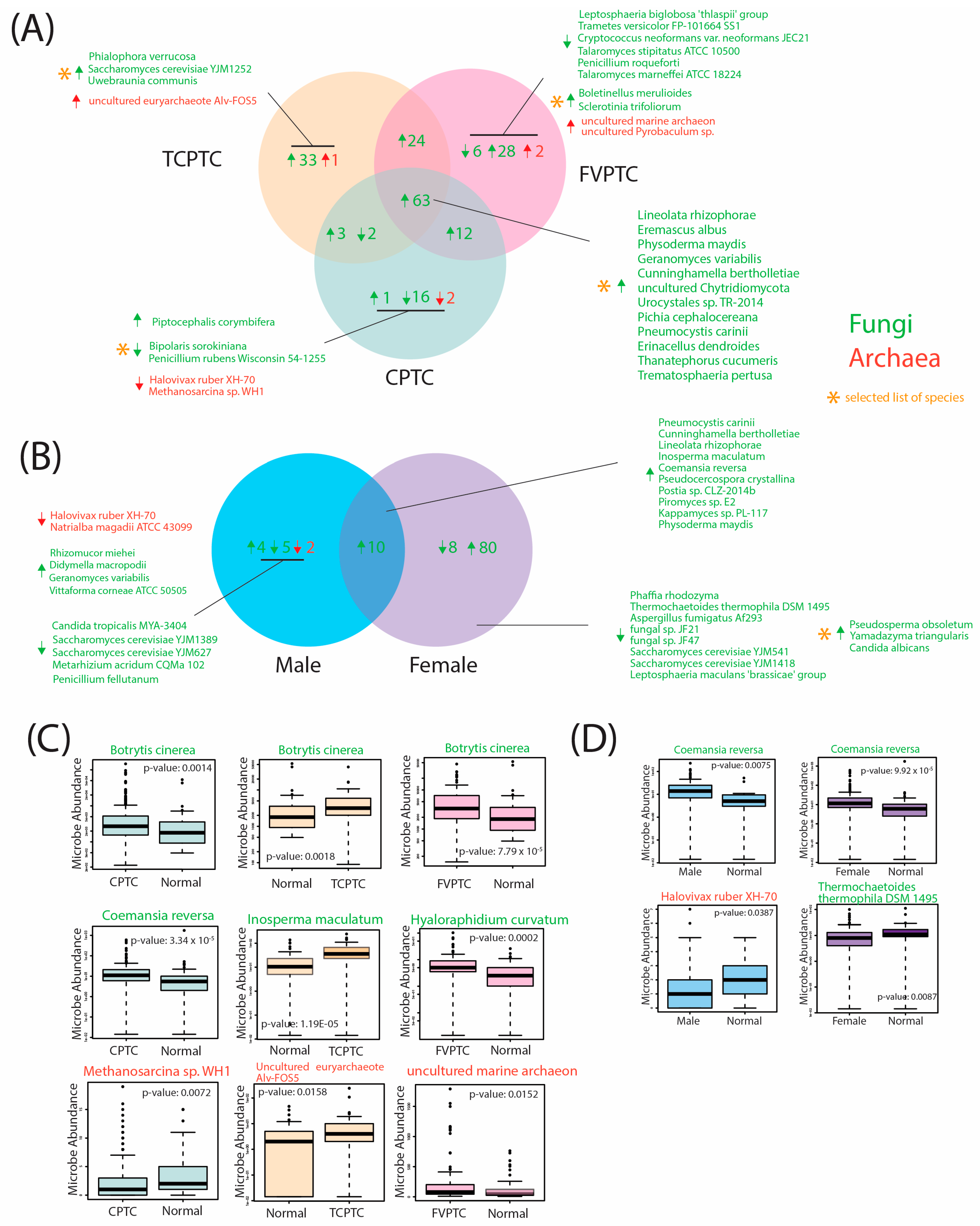
2.3. Differentially Abundant Fungal and Archaeal Species across PTC Subtype and Gender Comparisons
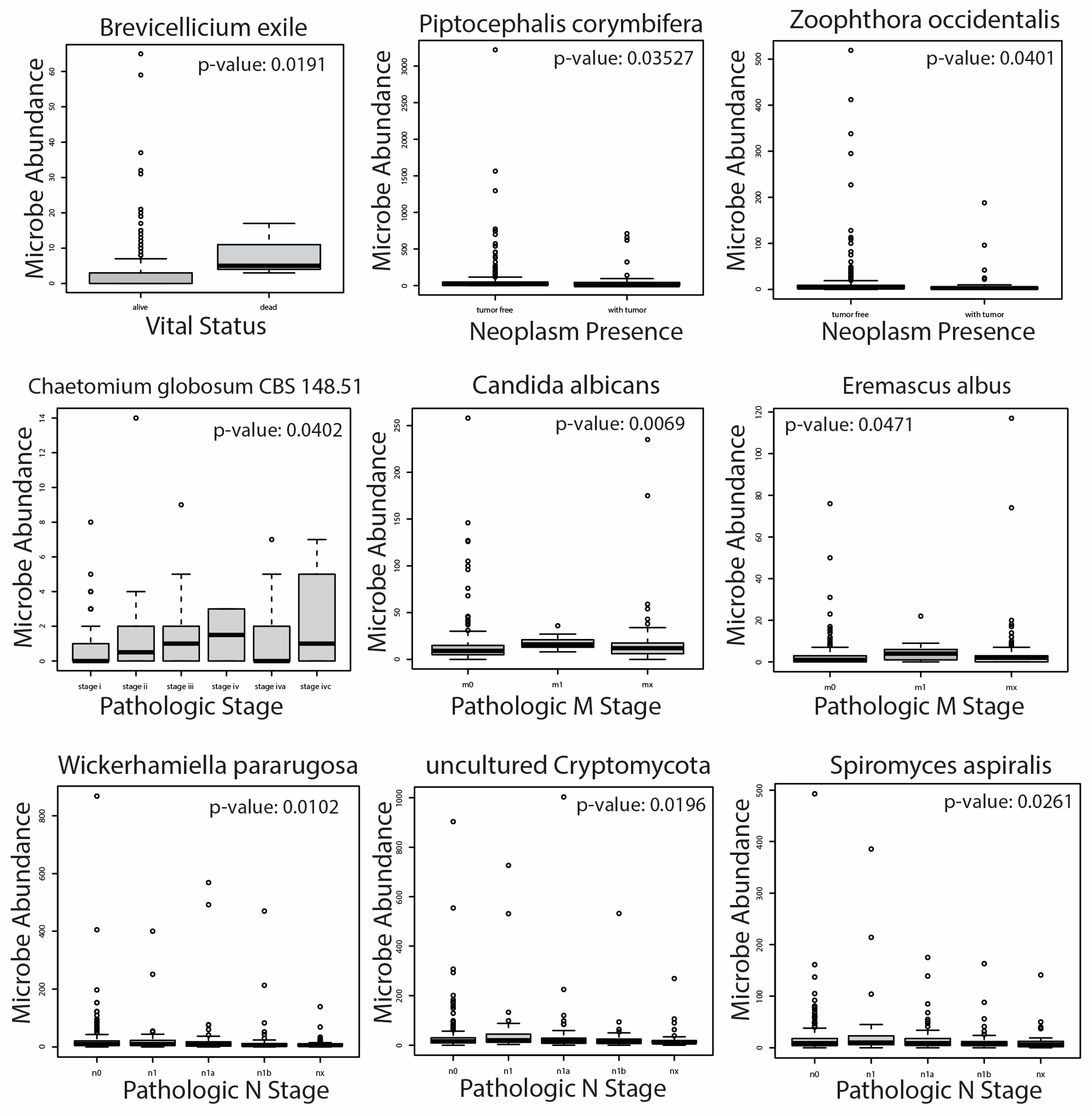
2.4. Correlation of Significantly Dysregulated Fungal and Archaeal Species to Clinical Variables
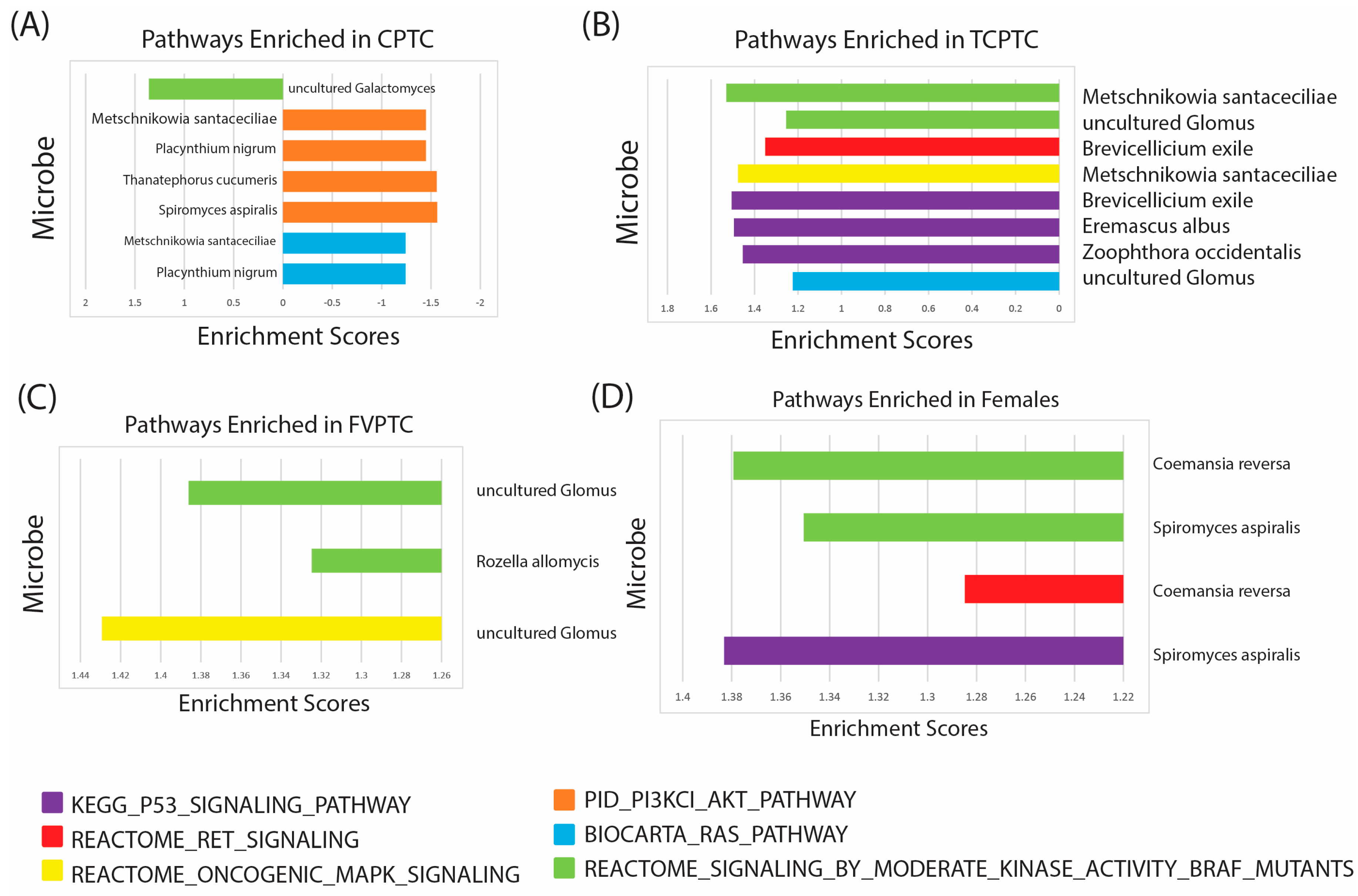
2.5. Microbe Abundance Correlation to PTC-Specific Oncogenic Pathways
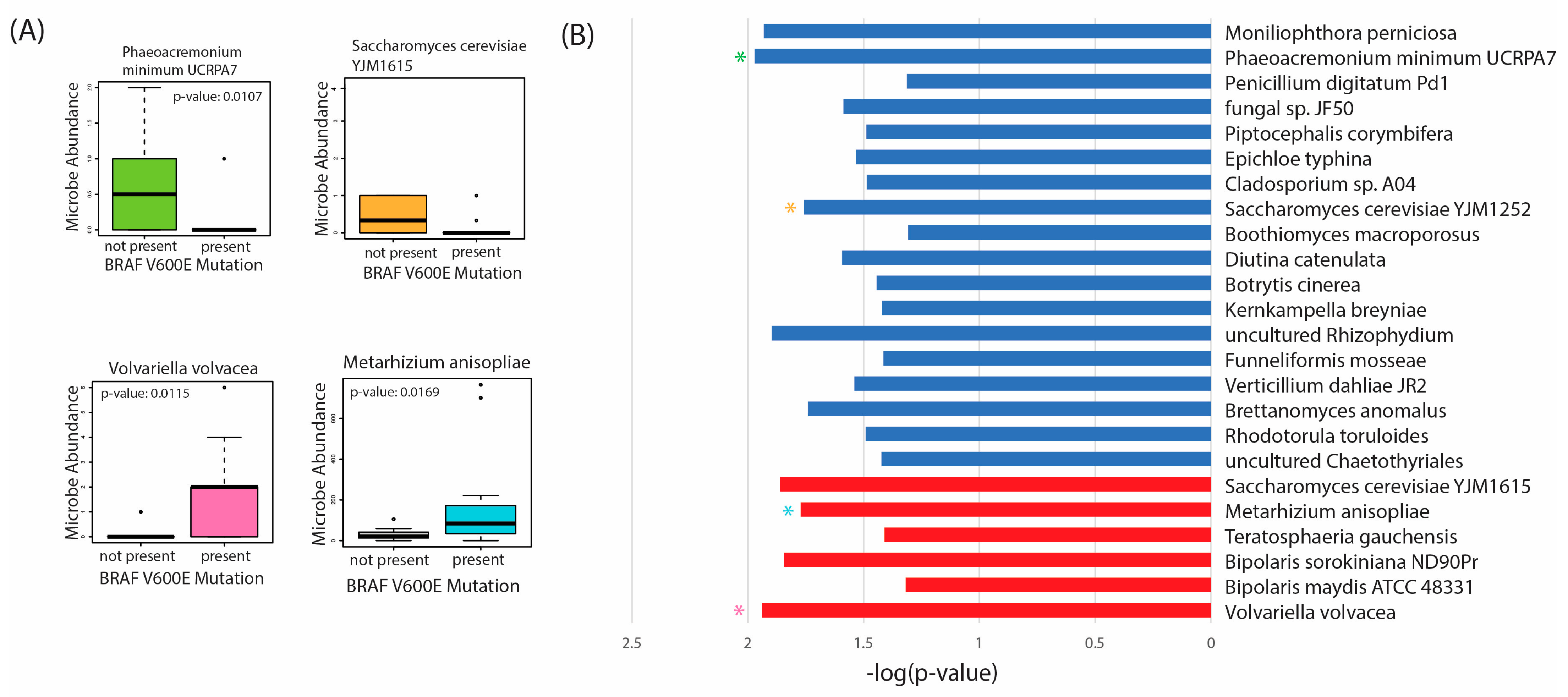
2.6. Differential Abundance according to BRAF 600VE Mutation Status
3. Discussion
4. Materials and Methods
4.1. Data Acquisition
4.2. Extraction and Normalization of Fungal and Archaeal Read Counts
4.3. Evaluation of Contamination
4.4. Differential Abundance between PTC, Gender, and Mutation Cohorts
4.5. Association of Microbial Abundance to Clinical Variable
4.6. Correlation of Microbial Abundance to Oncogenic PTC Signature Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cramer, J.D.; Fu, P.; Harth, K.C.; Margevicius, S.; Wilhelm, S.M. Analysis of the rising incidence of thyroid cancer using the Surveillance, Epidemiology and End Results national cancer data registry. Surgery 2010, 148, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Thyroid Cancer—Statistics. Available online: https://www.cancer.net/cancer-types/thyroid-cancer/statistics (accessed on 15 December 2022).
- Coca-Pelaz, A.; Shah, J.P.; Hernandez-Prera, J.C.; Ghossein, R.A.; Rodrigo, J.P.; Hartl, D.M.; Olsen, K.D.; Shaha, A.R.; Zafereo, M.; Suarez, C.; et al. Papillary thyroid cancer—Aggressive variants and impact on management: A narrative review. Adv. Ther. 2020, 37, 3112–3128. [Google Scholar] [CrossRef] [PubMed]
- Hescheler, D.A.; Riemann, B.; Hartmann, M.J.; Michel, M.; Faust, M.; Bruns, C.J.; Alakus, H.; Chiapponi, C. Targeted Therapy of Papillary Thyroid Cancer: A Comprehensive Genomic Analysis. Front. Endocrinol. 2021, 12, 1153. [Google Scholar] [CrossRef]
- Kitahara, C.M.; Sosa, J.A.; Shiels, M.S. Influence of Nomenclature Changes on Trends in Papillary Thyroid Cancer Incidence in the United States, 2000 to 2017. J. Clin. Endocrinol. Metab. 2020, 105, e4823–e4830. [Google Scholar] [CrossRef] [PubMed]
- Daniels, G.H. Follicular Variant of Papillary Thyroid Carcinoma: Hybrid or Mixture? Mary Ann Liebert, Inc.: New Rochelle, NY, USA, 2016; Volume 26, pp. 872–874. [Google Scholar]
- Villar-Taibo, R.; Peteiro-González, D.; Cabezas-Agrícola, J.M.; Aliyev, E.; Barreiro-Morandeira, F.; Ruiz-Ponte, C.; Cameselle-Teijeiro, J.M. Aggressiveness of the tall cell variant of papillary thyroid carcinoma is independent of the tumor size and patient age. Oncol. Lett. 2017, 13, 3501–3507. [Google Scholar] [CrossRef] [PubMed]
- Morris, L.G.; Shaha, A.R.; Tuttle, R.M.; Sikora, A.G.; Ganly, I. Tall-cell variant of papillary thyroid carcinoma: A matched-pair analysis of survival. Thyroid 2010, 20, 153–158. [Google Scholar] [CrossRef]
- Liu, Y.; Su, L.; Xiao, H. Review of Factors Related to the Thyroid Cancer Epidemic. Int. J. Endocrinol. 2017, 2017, 5308635. [Google Scholar] [CrossRef]
- Rahbari, R.; Zhang, L.; Kebebew, E. Thyroid cancer gender disparity. Future Oncol. 2010, 6, 1771–1779. [Google Scholar] [CrossRef]
- Jonklaas, J.; Nogueras-Gonzalez, G.; Munsell, M.; Litofsky, D.; Ain, K.B.; Bigos, S.T.; Brierley, J.D.; Cooper, D.S.; Haugen, B.R.; Ladenson, P.W.; et al. The Impact of Age and Gender on Papillary Thyroid Cancer Survival. J. Clin. Endocrinol. Metab. 2012, 97, E878–E887. [Google Scholar] [CrossRef]
- Liu, C.; Chen, T.; Zeng, W.; Wang, S.; Xiong, Y.; Liu, Z.; Huang, T. Reevaluating the prognostic significance of male gender for papillary thyroid carcinoma and microcarcinoma: A SEER database analysis. Sci. Rep. 2017, 7, 11412. [Google Scholar] [CrossRef] [PubMed]
- LeClair, K.; Bell, K.J.; Furuya-Kanamori, L.; Doi, S.A.; Francis, D.O.; Davies, L. Evaluation of gender inequity in thyroid cancer diagnosis: Differences by sex in US thyroid cancer incidence compared with a meta-analysis of subclinical thyroid cancer rates at autopsy. JAMA Intern. Med. 2021, 181, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the human microbiome. Nutr. Rev. 2012, 70 (Suppl. 1), S38–S44. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Tap, J.; Mondot, S.; Levenez, F.; Pelletier, E.; Caron, C.; Furet, J.P.; Ugarte, E.; Muñoz-Tamayo, R.; Paslier, D.L.; Nalin, R. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 2009, 11, 2574–2584. [Google Scholar] [CrossRef]
- Thangaraju, M.; Cresci, G.A.; Liu, K.; Ananth, S.; Gnanaprakasam, J.P.; Browning, D.D.; Mellinger, J.D.; Smith, S.B.; Digby, G.J.; Lambert, N.A. GPR109A is a G-protein–coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009, 69, 2826–2832. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St. Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wang, D.; Liang, D.; Wu, X.; Li, J.; Tang, L.; Li, Y. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef]
- Tsai, J.C.; Casteneda, G.; Lee, A.; Dereschuk, K.; Li, W.T.; Chakladar, J.; Lombardi, A.F.; Ongkeko, W.M.; Chang, E.Y. Identification and characterization of the intra-articular microbiome in the osteoarthritic knee. Int. J. Mol. Sci. 2020, 21, 8618. [Google Scholar] [CrossRef]
- Jiang, C.; Li, G.; Huang, P.; Liu, Z.; Zhao, B. The gut microbiota and Alzheimer’s disease. J. Alzheimer’s Dis. 2017, 58, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Varesi, A.; Pierella, E.; Romeo, M.; Piccini, G.B.; Alfano, C.; Bjørklund, G.; Oppong, A.; Ricevuti, G.; Esposito, C.; Chirumbolo, S. The potential role of gut microbiota in Alzheimer’s disease: From diagnosis to treatment. Nutrients 2022, 14, 668. [Google Scholar] [CrossRef] [PubMed]
- Craciun, C.-I.; Neag, M.-A.; Catinean, A.; Mitre, A.-O.; Rusu, A.; Bala, C.; Roman, G.; Buzoianu, A.-D.; Muntean, D.-M.; Craciun, A.-E. The Relationships between Gut Microbiota and Diabetes Mellitus, and Treatments for Diabetes Mellitus. Biomedicines 2022, 10, 308. [Google Scholar] [CrossRef] [PubMed]
- Frost, F.; Kacprowski, T.; Rühlemann, M.; Pietzner, M.; Bang, C.; Franke, A.; Nauck, M.; Völker, U.; Völzke, H.; Dörr, M. Long-term instability of the intestinal microbiome is associated with metabolic liver disease, low microbiota diversity, diabetes mellitus and impaired exocrine pancreatic function. Gut 2021, 70, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, R.; Anand, S.; Ounpuu, S.; Islam, S.; Zhang, X.; Rangarajan, S.; Chifamba, J.; Al-Hinai, A.; Keltai, M.; Yusuf, S. Dietary patterns and the risk of acute myocardial infarction in 52 countries: Results of the INTERHEART study. Circulation 2008, 118, 1929–1937. [Google Scholar] [CrossRef]
- Estruch, R.; Ros, E.; Salas-Salvadó, J.; Covas, M.-I.; Corella, D.; Arós, F.; Gómez-Gracia, E.; Ruiz-Gutiérrez, V.; Fiol, M.; Lapetra, J. Primary prevention of cardiovascular disease with a Mediterranean diet supplemented with extra-virgin olive oil or nuts. N. Engl. J. Med. 2018, 378, e34. [Google Scholar] [CrossRef]
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371, eabc4552. [Google Scholar] [CrossRef]
- Rossi, T.; Vergara, D.; Fanini, F.; Maffia, M.; Bravaccini, S.; Pirini, F. Microbiota-derived metabolites in tumor progression and metastasis. Int. J. Mol. Sci. 2020, 21, 5786. [Google Scholar] [CrossRef]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, B.; Wei, Y.; Kuang, D.-M. Influence of gut and intratumoral microbiota on the immune microenvironment and anti-cancer therapy. Pharmacol. Res. 2021, 174, 105966. [Google Scholar] [CrossRef]
- Chakladar, J.; John, D.; Magesh, S.; Uzelac, M.; Li, W.T.; Dereschuk, K.; Apostol, L.; Brumund, K.T.; Rodriguez, J.-W.; Ongkeko, W.M. The Intratumor Bacterial and Fungal Microbiome Is Characterized by HPV, Smoking, and Alcohol Consumption in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 13250. [Google Scholar] [CrossRef] [PubMed]
- Chakladar, J.; Kuo, S.Z.; Castaneda, G.; Li, W.T.; Gnanasekar, A.; Yu, M.A.; Chang, E.Y.; Wang, X.Q.; Ongkeko, W.M. The pancreatic microbiome is associated with carcinogenesis and worse prognosis in males and smokers. Cancers 2020, 12, 2672. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Gnanasekar, A.; Lee, A.; Li, W.T.; Haas, M.; Wang-Rodriguez, J.; Chang, E.Y.; Rajasekaran, M.; Ongkeko, W.M. Influence of Intratumor Microbiome on Clinical Outcome and Immune Processes in Prostate Cancer. Cancers 2020, 12, 2524. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.M.; Shende, N.; Li, W.T.; Castaneda, G.; Apostol, L.; Chang, E.Y.; Ongkeko, W.M. Comparative Analysis of Age- and Gender-Associated Microbiome in Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. Cancers 2020, 12, 1447. [Google Scholar] [CrossRef] [PubMed]
- Li, W.T.; Iyangar, A.S.; Reddy, R.; Chakladar, J.; Bhargava, V.; Sakamoto, K.; Ongkeko, W.M.; Rajasekaran, M. The Bladder Microbiome Is Associated with Epithelial–Mesenchymal Transition in Muscle Invasive Urothelial Bladder Carcinoma. Cancers 2021, 13, 3649. [Google Scholar] [CrossRef]
- Chakladar, J.; Li, W.T.; Bouvet, M.; Chang, E.Y.; Wang-Rodriguez, J.; Ongkeko, W.M. Papillary thyroid carcinoma variants are characterized by co-dysregulation of immune and cancer associated genes. Cancers 2019, 11, 1179. [Google Scholar] [CrossRef] [PubMed]
- Kebebew, E.; Weng, J.; Bauer, J.; Ranvier, G.; Clark, O.H.; Duh, Q.-Y.; Shibru, D.; Bastian, B.; Griffin, A. The prevalence and prognostic value of BRAF mutation in thyroid cancer. Ann. Surg. 2007, 246, 466. [Google Scholar] [CrossRef]
- Soares, P.; Trovisco, V.; Rocha, A.S.; Lima, J.; Castro, P.; Preto, A.; Maximo, V.; Botelho, T.; Seruca, R.; Sobrinho-Simoes, M. BRAF mutations and RET/PTC rearrangements are alternative events in the etiopathogenesis of PTC. Oncogene 2003, 22, 4578–4580. [Google Scholar] [CrossRef]
- Li, X.; Abdel-Mageed, A.B.; Kandil, E. BRAF mutation in papillary thyroid carcinoma. Int. J. Clin. Exp. Med. 2012, 5, 310–315. [Google Scholar]
- Mitsutake, N.; Miyagishi, M.; Mitsutake, S.; Akeno, N.; Mesa, C.; Knauf, J.A.; Zhang, L.; Taira, K.; Fagin, J.A. BRAF mediates RET/PTC-induced mitogen-activated protein kinase activation in thyroid cells: Functional support for requirement of the RET/PTC-RAS-BRAF pathway in papillary thyroid carcinogenesis. Endocrinology 2006, 147, 1014–1019. [Google Scholar] [CrossRef]
- Melillo, R.M.; Castellone, M.D.; Guarino, V.; De Falco, V.; Cirafici, A.M.; Salvatore, G.; Caiazzo, F.; Basolo, F.; Giannini, R.; Kruhoffer, M. The RET/PTC-RAS-BRAF linear signaling cascade mediates the motile and mitogenic phenotype of thyroid cancer cells. J. Clin. Investig. 2005, 115, 1068–1081. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.; Melillo, R.M.; Fusco, A. RET/PTC activation in papillary thyroid carcinoma: European Journal of Endocrinology Prize Lecture. Eur. J. Endocrinol. 2006, 155, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Zafon, C.; Obiols, G.; Castellvi, J.; Tallada, N.; Baena, J.; Simó, R.; Mesa, J. Clinical significance of RET/PTC and p53 protein expression in sporadic papillary thyroid carcinoma. Histopathology 2007, 50, 225–231. [Google Scholar] [CrossRef] [PubMed]
- McFadden, D.G.; Vernon, A.; Santiago, P.M.; Martinez-McFaline, R.; Bhutkar, A.; Crowley, D.M.; McMahon, M.; Sadow, P.M.; Jacks, T. p53 constrains progression to anaplastic thyroid carcinoma in a Braf-mutant mouse model of papillary thyroid cancer. Proc. Natl. Acad. Sci. USA 2014, 111, E1600–E1609. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef]
- Xing, M. Genetic alterations in the phosphatidylinositol-3 kinase/Akt pathway in thyroid cancer. Thyroid 2010, 20, 697–706. [Google Scholar] [CrossRef]
- Kim, S.J.; Lee, K.E.; Myong, J.P.; Park, J.H.; Jeon, Y.K.; Min, H.S.; Park, S.Y.; Jung, K.C.; Koo, D.H.; Youn, Y.K. BRAF V600E mutation is associated with tumor aggressiveness in papillary thyroid cancer. World J. Surg. 2012, 36, 310–317. [Google Scholar] [CrossRef]
- Zhu, G.; Deng, Y.; Pan, L.; Ouyang, W.; Feng, H.; Wu, J.; Chen, P.; Wang, J.; Chen, Y.; Luo, J. Clinical significance of the BRAFV600E mutation in PTC and its effect on radioiodine therapy. Endocr. Connect. 2019, 8, 754–763. [Google Scholar] [CrossRef]
- Gnanasekar, A.; Castaneda, G.; Iyangar, A.; Magesh, S.; Perez, D.; Chakladar, J.; Li, W.T.; Bouvet, M.; Chang, E.Y.; Ongkeko, W.M. The intratumor microbiome predicts prognosis across gender and subtypes in papillary thyroid carcinoma. Comput. Struct. Biotechnol. J. 2021, 19, 1986–1997. [Google Scholar] [CrossRef]
- Narunsky-Haziza, L.; Sepich-Poore, G.D.; Livyatan, I.; Asraf, O.; Martino, C.; Nejman, D.; Gavert, N.; Stajich, J.E.; Amit, G.; González, A.; et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell 2022, 185, 3789–3806. [Google Scholar] [CrossRef]
- Kim, J.Y.; Whon, T.W.; Lim, M.Y.; Kim, Y.B.; Kim, N.; Kwon, M.-S.; Kim, J.; Lee, S.H.; Choi, H.-J.; Nam, I.-H. The human gut archaeome: Identification of diverse haloarchaea in Korean subjects. Microbiome 2020, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Chibani, C.M.; Mahnert, A.; Borrel, G.; Almeida, A.; Werner, A.; Brugère, J.-F.; Gribaldo, S.; Finn, R.D.; Schmitz, R.A.; Moissl-Eichinger, C. A catalogue of 1,167 genomes from the human gut archaeome. Nat. Microbiol. 2022, 7, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Koskinen, K.; Pausan, M.R.; Perras, A.K.; Beck, M.; Bang, C.; Mora, M.; Schilhabel, A.; Schmitz, R.; Moissl-Eichinger, C. First Insights into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin. mBio 2017, 8, e00824-17. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Kandalai, S.; Tang, X.; Zheng, Q. Contributions of Human-Associated Archaeal Metabolites to Tumor Microenvironment and Carcinogenesis. Microbiol. Spectr. 2022, 10, e0236721. [Google Scholar] [CrossRef]
- Abdi, H.; Kordi-Tamandani, D.M.; Lagzian, M.; Bakhshipour, A. Archaeome in Colorectal Cancer: High Abundance of Methanogenic Archaea in Colorectal Cancer Patients. Int. J. Cancer Manag. 2022, 15, e117843. [Google Scholar] [CrossRef]
- Uzelac, M.; Li, Y.; Chakladar, J.; Li, W.T.; Ongkeko, W.M. Archaea Microbiome Dysregulated Genes and Pathways as Molecular Targets for Lung Adenocarcinoma and Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 11566. [Google Scholar] [CrossRef]
- Rascio, F.; Spadaccino, F.; Rocchetti, M.T.; Castellano, G.; Stallone, G.; Netti, G.S.; Ranieri, E. The Pathogenic Role of PI3K/AKT Pathway in Cancer Onset and Drug Resistance: An Updated Review. Cancers 2021, 13, 3949. [Google Scholar] [CrossRef]
- Janku, F.; Yap, T.A.; Meric-Bernstam, F. Targeting the PI3K pathway in cancer: Are we making headway? Nat. Rev. Clin. Oncol. 2018, 15, 273–291. [Google Scholar] [CrossRef]
- Nikiforov, Y.E. RET/PTC rearrangement in thyroid tumors. Endocr. Pathol. 2002, 13, 3–16. [Google Scholar] [CrossRef]
- Stanciu, M.; Ristea, R.P.; Popescu, M.; Vasile, C.M.; Popa, F.L. Thyroid Carcinoma Showing Thymus-like Differentiation (CASTLE): A Case Report. Life 2022, 12, 1314. [Google Scholar] [CrossRef]
- Sherman, E.J.; Harris, J.; Bible, K.C.; Xia, P.; Ghossein, R.A.; Chung, C.H.; Riaz, N.; Gunn, G.B.; Foote, R.L.; Yom, S.S.; et al. Radiotherapy and paclitaxel plus pazopanib or placebo in anaplastic thyroid cancer (NRG/RTOG 0912): A randomised, double-blind, placebo-controlled, multicentre, phase 2 trial. Lancet Oncol. 2023, 24, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Saltiki, K.; Simeakis, G.; Karapanou, O.; Paschou, S.A.; Alevizaki, M. Metastatic medullary thyroid carcinoma (MTC): Disease course, treatment modalities and factors predisposing for drug resistance. Endocrine 2023, 1–10. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
John, D.; Yalamarty, R.; Barakchi, A.; Chen, T.; Chakladar, J.; Li, W.T.; Ongkeko, W.M. Transcriptomic Analysis Reveals Dysregulation of the Mycobiome and Archaeome and Distinct Oncogenic Characteristics according to Subtype and Gender in Papillary Thyroid Carcinoma. Int. J. Mol. Sci. 2023, 24, 3148. https://doi.org/10.3390/ijms24043148
John D, Yalamarty R, Barakchi A, Chen T, Chakladar J, Li WT, Ongkeko WM. Transcriptomic Analysis Reveals Dysregulation of the Mycobiome and Archaeome and Distinct Oncogenic Characteristics according to Subtype and Gender in Papillary Thyroid Carcinoma. International Journal of Molecular Sciences. 2023; 24(4):3148. https://doi.org/10.3390/ijms24043148
Chicago/Turabian StyleJohn, Daniel, Rishabh Yalamarty, Armon Barakchi, Tianyi Chen, Jaideep Chakladar, Wei Tse Li, and Weg M. Ongkeko. 2023. "Transcriptomic Analysis Reveals Dysregulation of the Mycobiome and Archaeome and Distinct Oncogenic Characteristics according to Subtype and Gender in Papillary Thyroid Carcinoma" International Journal of Molecular Sciences 24, no. 4: 3148. https://doi.org/10.3390/ijms24043148
APA StyleJohn, D., Yalamarty, R., Barakchi, A., Chen, T., Chakladar, J., Li, W. T., & Ongkeko, W. M. (2023). Transcriptomic Analysis Reveals Dysregulation of the Mycobiome and Archaeome and Distinct Oncogenic Characteristics according to Subtype and Gender in Papillary Thyroid Carcinoma. International Journal of Molecular Sciences, 24(4), 3148. https://doi.org/10.3390/ijms24043148





