Coculture with Neural Stem Cells May Shift the Transcription Profile of Glioblastoma Multiforme towards Cancer-Specific Stemness
Abstract
1. Introduction
2. Results
2.1. EV Internalization
2.2. Relative Expression of Genes in Coculture
2.3. Gene Expression in GBM CSCs
3. Discussion
4. Materials and Methods
4.1. Cell Culture
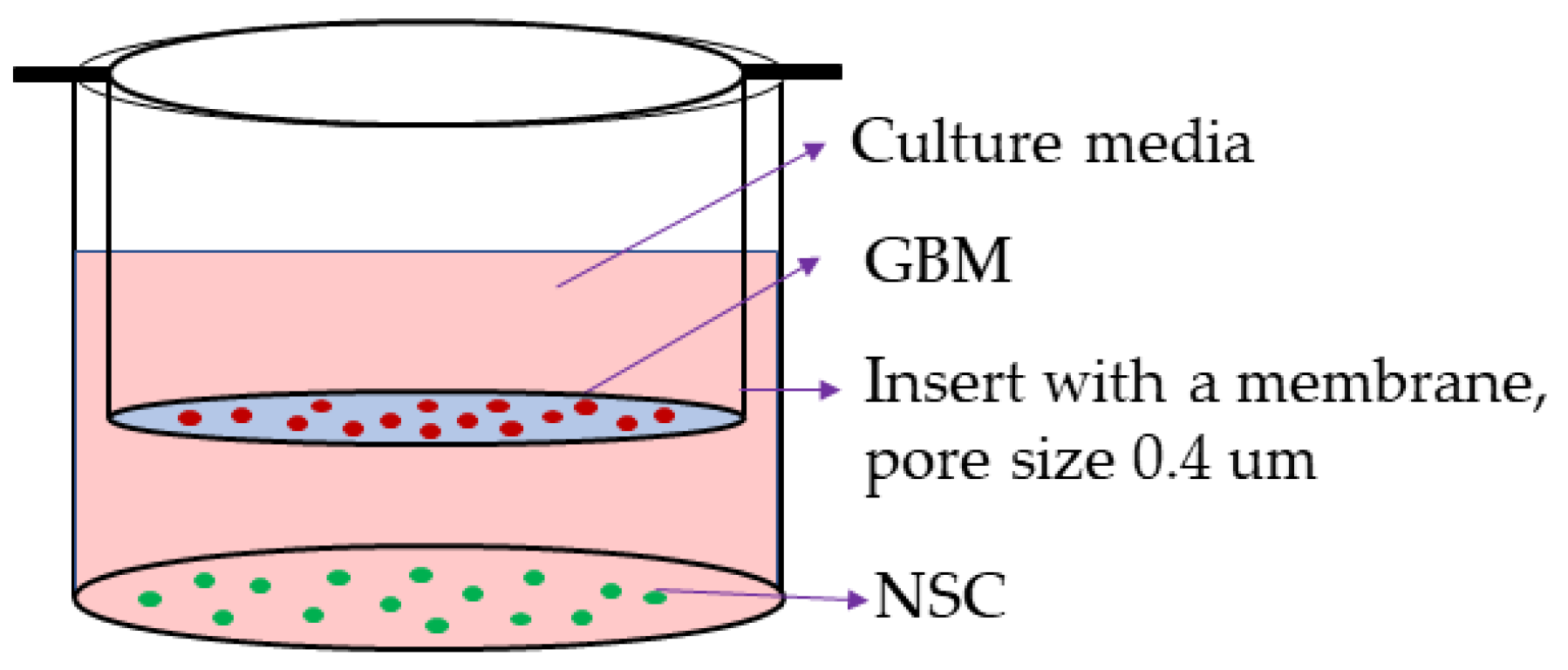
4.2. Coculture
4.3. RNA Extraction
4.4. cDNA Synthesis
4.5. qPCR
4.6. Statistical Analysis
4.7. EV Uptake and Internalization
4.8. Cell Preparation for Confocal Microscopy
4.9. Confocal Microscopy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Field, M.; Alvarez, A.; Bushnev, S.; Sugaya, K. Embryonic stem cell markers distinguishing cancer stem cells from normal human neuronal stem cell populations in malignant glioma patients. Clin. Neurosurg. 2010, 57, 151–159. [Google Scholar]
- Shergalis, A.; Bankhead, A.; Luesakul, U.; Muangsin, N.; Neamati, N. Current challenges and opportunities in treating glioblastoma. Pharmacol. Rev. 2018, 70, 412–445. [Google Scholar] [CrossRef]
- Sundar, S.J.; Hsieh, J.K.; Manjila, S.; Lathia, J.D.; Sloan, A. The role of cancer stem cells in glioblastoma. Neurosurg. Focus 2014, 37, E6. [Google Scholar] [CrossRef]
- Hadjimichael, C.; Chanoumidou, K.; Papadopoulou, N.; Arampatzi, P.; Papamatheakis, J.; Kretsovali, A. Common stemness regulators of embryonic and cancer stem cells. World J. Stem Cells 2015, 7, 1150. [Google Scholar]
- Alves, A.L.V.; Gomes, I.N.; Carloni, A.C.; Rosa, M.N.; da Silva, L.S.; Evangelista, A.F.; Reis, R.M.; Silva, V.A.O. Role of glioblastoma stem cells in cancer therapeutic resistance: A perspective on antineoplastic agents from natural sources and chemical derivatives. Stem Cell Res. Ther. 2021, 12, 206. [Google Scholar] [CrossRef]
- Zhou, W.; Yao, Y.; Scott, A.J.; Wilder-Romans, K.; Dresser, J.J.; Werner, C.K.; Sun, H.; Pratt, D.; Sajjakulnukit, P.; Zhao, S.G.; et al. Purine metabolism regulates DNA repair and therapy resistance in glioblastoma. Nat. Commun. 2020, 11, 3811. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Ajani, J.A.; Song, S. Drug resistance and Cancer stem cells. Cell Commun. Signal. 2021, 19, 19. [Google Scholar] [CrossRef]
- Begicevic, R.-R.; Falasca, M. ABC transporters in cancer stem cells: Beyond chemoresistance. Int. J. Mol. Sci. 2017, 18, 2362. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, M.; Shi, L.; Yang, X.; Chen, L.; Cao, N.; Lei, A.; Cao, Y. Neural stemness contributes to cell tumorigenicity. Cell Biosci. 2021, 11, 21. [Google Scholar] [CrossRef]
- Bradshaw, A.; Wickremsekera, A.; Tan, S.T.; Peng, L.; Davis, P.F.; Itinteang, T. Cancer stem cell hierarchy in glioblastoma multiforme. Front. Surg. 2016, 3, 21. [Google Scholar] [CrossRef]
- Matarredona, E.R.; Pastor, A.M. Neural stem cells of the subventricular zone as the origin of human glioblastoma stem cells. Therapeutic implications. Front. Oncol. 2019, 9, 779. [Google Scholar] [CrossRef]
- Broekman, M.L.; Maas, S.L.; Abels, E.R.; Mempel, T.R.; Krichevsky, A.M.; Breakefield, X.O. Multidimensional communication in the microenvirons of glioblastoma. Nat. Rev. Neurol. 2018, 14, 482–495. [Google Scholar] [CrossRef]
- Yekula, A.; Yekula, A.; Muralidharan, K.; Kang, K.; Carter, B.S.; Balaj, L. Extracellular vesicles in glioblastoma tumor microenvironment. Front. Immunol. 2020, 10, 3137. [Google Scholar] [CrossRef]
- Figueroa, J.M.; Carter, B.S. Detection of glioblastoma in biofluids. J. Neurosurg. 2017, 129, 334–340. [Google Scholar] [CrossRef]
- Dahlberg, D.; Rummel, J.; Distante, S.; De Souza, G.A.; Stensland, M.E.; Mariussen, E.; Roothwelt, H.; Voie, O.; Hassel, B. Glioblastoma microenvironment contains multiple hormonal and non-hormonal growth-stimulating factors. Fluids Barriers CNS 2022, 19, 45. [Google Scholar] [CrossRef]
- Thirant, C.; Gavard, J.; Junier, M.P.; Chneiweiss, H. Critical multiple angiogenic factors secreted by glioblastoma stem-like cells underline the need for combinatorial anti-angiogenic therapeutic strategies. PROTEOMICS–Clin. Appl. 2013, 7, 79–90. [Google Scholar] [CrossRef]
- Treps, L.; Perret, R.; Edmond, S.; Ricard, D.; Gavard, J. Glioblastoma stem-like cells secrete the pro-angiogenic VEGF-A factor in extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1359479. [Google Scholar] [CrossRef]
- Sharma, N.; Colangelo, N.W.; de Toledo, S.M.; Azzam, E.I. Diffusible factors secreted by glioblastoma and medulloblastoma cells induce oxidative stress in bystander neural stem progenitors. ASN Neuro 2016, 8, 1759091416662808. [Google Scholar] [CrossRef]
- Iglesia, R.P.; Prado, M.B.; Alves, R.N.; Escobar, M.I.M.; Fernandes, C.F.d.L.; Fortes, A.C.d.S.; Souza, M.C.D.S.; Boccacino, J.M.; Cangiano, G.; Soares, S.R.; et al. Unconventional protein secretion in brain tumors biology: Enlightening the mechanisms for tumor survival and progression. Front. Cell Dev. Biol. 2022, 10, 1260. [Google Scholar] [CrossRef]
- Willis, C.M.; Nicaise, A.M.; Hamel, R.; Pappa, V.; Peruzzotti-Jametti, L.; Pluchino, S. Harnessing the neural stem cell secretome for regenerative neuroimmunology. Front. Cell. Neurosci. 2020, 14, 590960. [Google Scholar] [CrossRef]
- Červenka, J.; Tylečková, J.; Skalníková, H.K.; Kepková, K.V.; Poliakh, I.; Valeková, I.; Pfeiferová, L.; Kolář, M.; Vaškovičová, M.; Pánková, T.; et al. Proteomic characterization of human neural stem cells and their secretome during in vitro differentiation. Front. Cell. Neurosci. 2021, 14, 612560. [Google Scholar] [CrossRef]
- Maacha, S.; Bhat, A.A.; Jimenez, L.; Raza, A.; Haris, M.; Uddin, S.; Grivel, J.-C. Extracellular vesicles-mediated intercellular communication: Roles in the tumor microenvironment and anti-cancer drug resistance. Mol. Cancer 2019, 18, 55. [Google Scholar] [CrossRef]
- Ciregia, F.; Urbani, A.; Palmisano, G. Extracellular vesicles in brain tumors and neurodegenerative diseases. Front. Mol. Neurosci. 2017, 10, 276. [Google Scholar] [CrossRef]
- Wu, M.; Wang, G.; Hu, W.; Yao, Y.; Yu, X.-F. Emerging roles and therapeutic value of exosomes in cancer metastasis. Mol. Cancer 2019, 18, 53. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, Y.; Wu, N. Role of Tumor-Derived Extracellular Vesicles in Glioblastoma. Cells 2021, 10, 512. [Google Scholar] [CrossRef]
- Marostica, G.; Gelibter, S.; Gironi, M.; Nigro, A.; Furlan, R. Extracellular Vesicles in Neuroinflammation. Front. Cell Dev. Biol. 2021, 8, 1841. [Google Scholar] [CrossRef]
- Matarredona, E.R.; Pastor, A.M. Extracellular vesicle-mediated communication between the glioblastoma and its microenvironment. Cells 2019, 9, 96. [Google Scholar] [CrossRef]
- Ramirez, S.H.; Andrews, A.M.; Paul, D.; Pachter, J.S. Extracellular vesicles: Mediators and biomarkers of pathology along CNS barriers. Fluids Barriers CNS 2018, 15, 19. [Google Scholar] [CrossRef]
- Banks, W.A.; Sharma, P.; Bullock, K.M.; Hansen, K.M.; Ludwig, N.; Whiteside, T.L. Transport of extracellular vesicles across the blood-brain barrier: Brain pharmacokinetics and effects of inflammation. Int. J. Mol. Sci. 2020, 21, 4407. [Google Scholar] [CrossRef]
- Tao, S.-C.; Guo, S.-C. Role of extracellular vesicles in tumour microenvironment. Cell Commun. Signal. 2020, 18, 163. [Google Scholar] [CrossRef]
- Vogel, A.; Upadhya, R.; Shetty, A.K. Neural stem cell derived extracellular vesicles: Attributes and prospects for treating neurodegenerative disorders. EBioMedicine 2018, 38, 273–282. [Google Scholar] [CrossRef]
- Yuan, P.; Ding, L.; Chen, H.; Wang, Y.; Li, C.; Zhao, S.; Yang, X.; Ma, Y.; Zhu, J.; Qi, X.; et al. Neural stem cell-derived exosomes regulate neural stem cell differentiation through miR-9-Hes1 axis. Front. Cell Dev. Biol. 2021, 9, 1181. [Google Scholar] [CrossRef]
- Dörsam, B.; Bösl, T.; Reiners, K.S.; Barnert, S.; Schubert, R.; Shatnyeva, O.; Zigrino, P.; Engert, A.; Hansen, H.P.; Von Strandmann, E.P. Hodgkin lymphoma-derived extracellular vesicles change the secretome of fibroblasts toward a CAF phenotype. Front. Immunol. 2018, 9, 1358. [Google Scholar] [CrossRef]
- De Chaumont, F.; Dallongeville, S.; Chenouard, N.; Hervé, N.; Pop, S.; Provoost, T.; Meas-Yedid, V.; Pankajakshan, P.; LeComte, T.; Le Montagner, Y.; et al. Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods 2012, 9, 690–696. [Google Scholar] [CrossRef]
- Zbinden, M.; Duquet, A.; Lorente-Trigos, A.; Ngwabyt, S.N.; Borges, I.; Ruiz i Altaba, A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. The EMBO J. 2010, 29, 2659–2674. [Google Scholar] [CrossRef]
- Galderisi, U.; Cipollaro, M.; Giordano, A. Stem cells and brain cancer. Cell Death Differ. 2006, 13, 5–11. [Google Scholar] [CrossRef]
- Tan, B.T.; Park, C.Y.; Ailles, L.E.; Weissman, I.L. The cancer stem cell hypothesis: A work in progress. Lab. Investig. 2006, 86, 1203–1207. [Google Scholar] [CrossRef]
- Bakhshinyan, D.; Savage, N.; Salim, S.K.; Venugopal, C.; Singh, S.K. The Strange Case of Jekyll and Hyde: Parallels Between Neural Stem Cells and Glioblastoma-Initiating Cells. Front. Oncol. 2021, 10, 2983. [Google Scholar] [CrossRef]
- Mooney, K.L.; Choy, W.; Sidhu, S.; Pelargos, P.; Bui, T.T.; Voth, B.; Barnette, N.; Yang, I. The role of CD44 in glioblastoma multiforme. J. Clin. Neurosci. 2016, 34, 1–5. [Google Scholar] [CrossRef]
- Si, D.; Yin, F.; Peng, J.; Zhang, G. High expression of CD44 predicts a poor prognosis in glioblastomas. Cancer Manag. Res. 2020, 12, 769. [Google Scholar] [CrossRef]
- Stevanovic, M.; Kovacevic-Grujicic, N.; Mojsin, M.; Milivojevic, M.; Drakulic, D. SOX transcription factors and glioma stem cells: Choosing between stemness and differentiation. World J. Stem Cells 2021, 13, 1417. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Li, J.; Coulouarn, C.; Lin, T.; Sulpice, L.; Bergeat, D.; De La Torre, C.; Liebe, R.; Gretz, N.; Ebert, M.P.A.; et al. SOX9 expression decreases survival of patients with intrahepatic cholangiocarcinoma by conferring chemoresistance. Br. J. Cancer 2018, 119, 1358–1366. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, H.; Xu, S.; Liu, Z.; Cheng, Q. The adaptive transition of glioblastoma stem cells and its implications on treatments. Signal Transduct. Target. Ther. 2021, 6, 124. [Google Scholar] [CrossRef]
- Katsetos, C.D.; Del Valle, L.; Geddes, J.F.; Assimakopoulou, M.; Legido, A.; Boyd, J.C.; Balin, B.; Parikh, N.A.; Maraziotis, T.; de Chadarevian, J.-P.; et al. Aberrant localization of the neuronal class III β-tubulin in astrocytomas: A marker for anaplastic potential. Arch. Pathol. Lab. Med. 2001, 125, 613–624. [Google Scholar] [CrossRef]
- Katsetos, C.D.; Dráberová, E.; Legido, A.; Dumontet, C.; Dráber, P. Tubulin targets in the pathobiology and therapy of glioblastoma multiforme. I. class III β-tubulin. J. Cell. Physiol. 2009, 221, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; De Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Egaña, L.; Auzmendi-Iriarte, J.; Andermatten, J.; Villanua, J.; Ruiz, I.; Elua-Pinin, A.; Aldaz, P.; Querejeta, A.; Sarasqueta, C.; Zubia, F.; et al. Methylation of MGMT promoter does not predict response to temozolomide in patients with glioblastoma in Donostia Hospital. Sci. Rep. 2020, 10, 18445. [Google Scholar] [CrossRef]
- Caldera, V.; Mellai, M.; Annovazzi, L.; Monzeglio, O.; Piazzi, A.; Schiffer, D. MGMT hypermethylation and MDR system in glioblastoma cancer stem cells. Cancer Genom. Proteom. 2012, 9, 171–178. [Google Scholar]
- Bleau, A.-M.; Huse, J.T.; Holland, E.C. The ABCG2 resistance network of glioblastoma. Cell Cycle 2009, 8, 2937–2945. [Google Scholar] [CrossRef]
- Xia, Q.; Liu, L.; Li, Y.; Zhang, P.; Han, D.; Dong, L. Therapeutic perspective of temozolomide resistance in glioblastoma treatment. Cancer Investig. 2021, 39, 627–644. [Google Scholar] [CrossRef]
- Wang, H.; Lathia, J.D.; Wu, Q.; Wang, J.; Li, Z.; Heddleston, J.M.; Eyler, C.E.; Elderbroom, J.; Gallagher, J.; Schuschu, J.; et al. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells 2009, 27, 2393–2404. [Google Scholar] [CrossRef] [PubMed]
- Saidi, A.; Hagedorn, M.; Allain, N.; Verpelli, C.; Sala, C.; Bello, L.; Bikfalvi, A.; Javerzat, S. Combined targeting of interleukin-6 and vascular endothelial growth factor potently inhibits glioma growth and invasiveness. Int. J. Cancer 2009, 125, 1054–1064. [Google Scholar] [CrossRef]
- Chen, N.; Peng, C.; Li, D. Epigenetic Underpinnings of Inflammation: A Key to Unlock the Tumor Microenvironment in Glioblastoma. Front. Immunol. 2022, 13, 869307. [Google Scholar] [CrossRef]
- Park, S.-Y.; Kang, M.-J.; Han, J.-S. Interleukin-1 beta promotes neuronal differentiation through the Wnt5a/RhoA/JNK pathway in cortical neural precursor cells. Mol. Brain 2018, 11, 39. [Google Scholar] [CrossRef]
- Fontana, A.; Hengartner, H.; De Tribolet, N.; Weber, E. Glioblastoma cells release interleukin 1 and factors inhibiting interleukin 2-mediated effects. J. Immunol. 1984, 132, 1837–1844. [Google Scholar] [CrossRef]
- Heddleston, J.M.; Li, Z.; McLendon, R.E.; Hjelmeland, A.B.; Rich, J.N. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 2009, 8, 3274–3284. [Google Scholar] [CrossRef]
- Hjelmeland, A.B.; Wu, Q.; Heddleston, J.; Choudhary, G.; MacSwords, J.; Lathia, J.; McLendon, R.; Lindner, D.; Sloan, A.; Rich, J.N. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 2011, 18, 829–840. [Google Scholar] [CrossRef]
- Papale, M.; Buccarelli, M.; Mollinari, C.; Russo, M.A.; Pallini, R.; Ricci-Vitiani, L.; Tafani, M. Hypoxia, inflammation and necrosis as determinants of glioblastoma cancer stem cells progression. Int. J. Mol. Sci. 2020, 21, 2660. [Google Scholar] [CrossRef]
- Alvarez, A.A.; Field, M.; Bushnev, S.; Longo, M.S.; Sugaya, K. The effects of histone deacetylase inhibitors on glioblastoma-derived stem cells. J. Mol. Neurosci. 2015, 55, 7–20. [Google Scholar] [CrossRef]
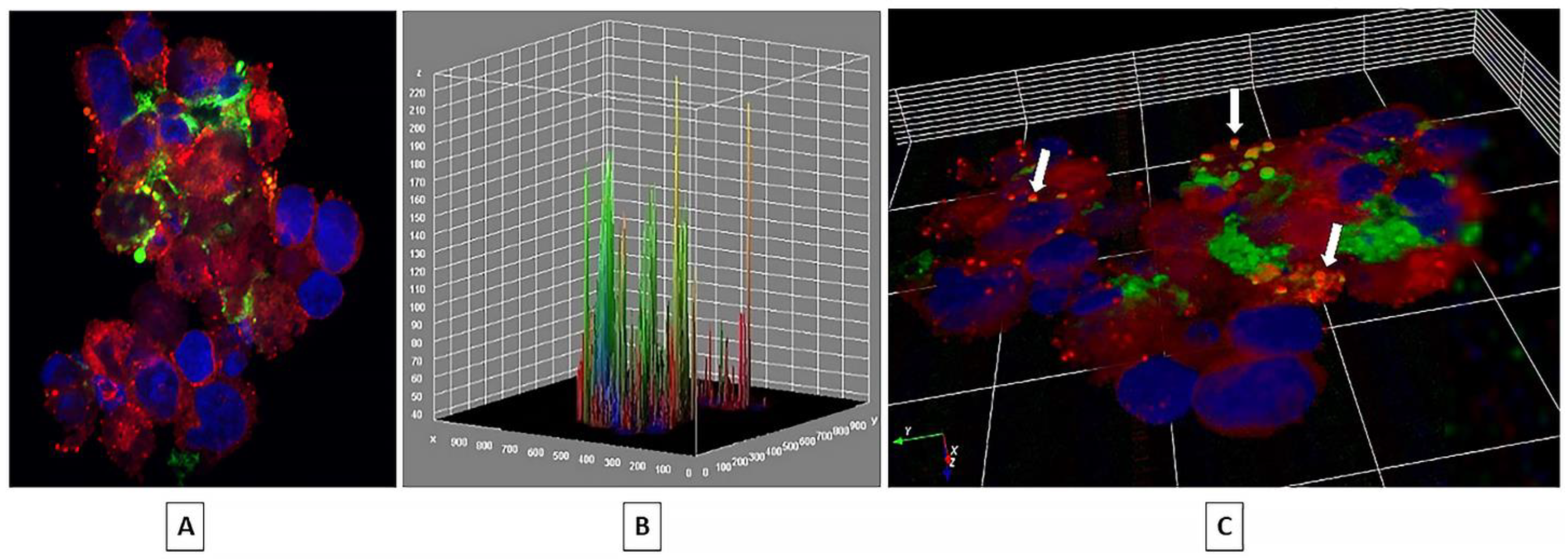
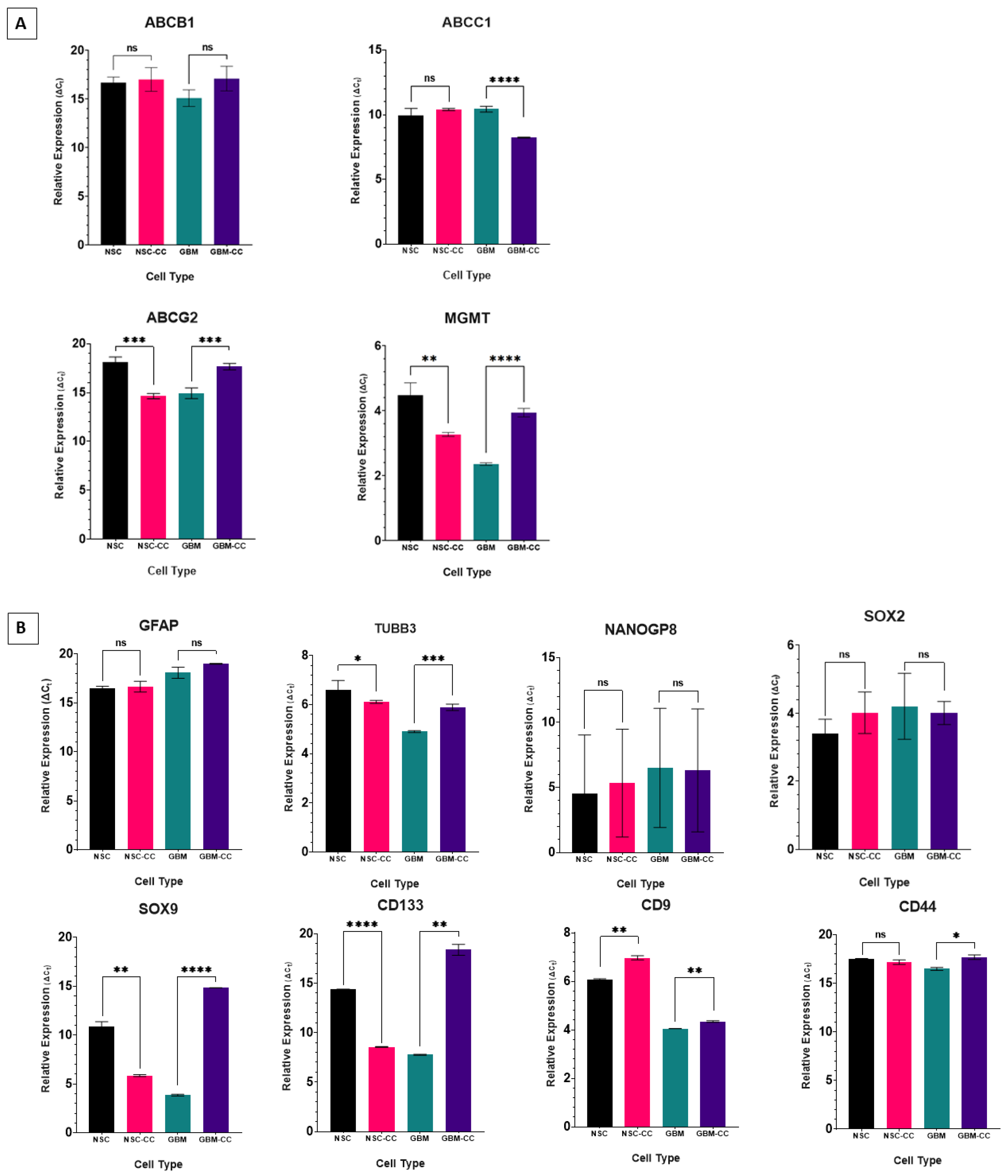

| Relative Expression (ΔCT) | ||||||
|---|---|---|---|---|---|---|
| One-Way ANOVA NSC | t-Test CNTRL VS CC | |||||
| GENE | NSC-CC | GBM | GBM-CC | NSC-CC | GBM-CC | |
| ABC transporters | ABCB1 | +/− | +/− | +/− | +/− | +/− |
| ABCC1 | +/− | +/− | − − − | +/− | − − − − | |
| ABCG2 | − − − − | − − − − | +/− | − − − | + + + | |
| MGMT | − − − | − − − − | − | − − | + + + + | |
| Stemness/CSC | TUBB3 | − | − − − − | − − | − | + + + |
| CD9 | + + + | − − − − | − − − − | + + | + + | |
| CD133 | − − − − | − − − − | + + + | − − − − | + + | |
| CD44 | +/− | − | +/− | +/− | + + | |
| GFAP | +/− | + + | + + + | +/− | +/− | |
| NANOGP8 | +/− | +/− | +/− | +/− | +/− | |
| SOX2 | +/− | +/− | +/− | +/− | +/− | |
| SOX9 | − − − | − − − − | + + + | − − | + + + + | |
| Forward: 5′->3′ | Reverse: 5′->3′ | |
|---|---|---|
| ABCB1 | CCCATCATTGCAATAGCAGG | GTTCAAACTTCTGCTCCTGA |
| ABCC1 | AACCTGGACCCATTCAGCC | GACTGGATGAGGTCGTCCGT |
| ABCG2 | ATGTCAACTCCTCCTTCTAC | AATGATCTGAGCTATAGAGGC |
| MGMT | TTCACCATCCCGTTTTCCAG | ATTGCCTCTCATTGCTCCTC |
| TUBB3 | CTCAGGGGCCTTTGGACATC | CAGGCAGTCGCAGTTTTCAC |
| CD9 | GGACGTACTCGAAACCTTCACC | GCGGATAGCACAGCACAAGA |
| CD133 | ACCAGGTAAGAACCCGGATCAA | CAAGAATTCCGCCTCCTAGCACT |
| CD44 | CCAGAAGGAACAGTGGTTTGGC | ACTGTCCTCTGGGCTTGGTGTT |
| GFAP | ACCTGCAGATTCGAGAAACC | CTCCTTAATGACCTCTCCATCC |
| NANOGP8 | TTTGTGGGCCTGAAGAAAACT | AGGGCTGTCCTGAATAAGCAG |
| SOX2 | TACAGCATGTCCTACTCGCAG | GAGGAAGAGGTAACCACAGGG |
| SOX9 | GCTCTGGAGACTTCTGAACGA | CCGTTCTTCACCGACTTCCT |
| ACTB | AGAGCTACGAGCTGCCTGAC | AGCACTGTGTTGGCGTACAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vaidya, M.; Sreerama, S.; Gonzalez-Vega, M.; Smith, J.; Field, M.; Sugaya, K. Coculture with Neural Stem Cells May Shift the Transcription Profile of Glioblastoma Multiforme towards Cancer-Specific Stemness. Int. J. Mol. Sci. 2023, 24, 3242. https://doi.org/10.3390/ijms24043242
Vaidya M, Sreerama S, Gonzalez-Vega M, Smith J, Field M, Sugaya K. Coculture with Neural Stem Cells May Shift the Transcription Profile of Glioblastoma Multiforme towards Cancer-Specific Stemness. International Journal of Molecular Sciences. 2023; 24(4):3242. https://doi.org/10.3390/ijms24043242
Chicago/Turabian StyleVaidya, Manjusha, Sandeep Sreerama, Maxine Gonzalez-Vega, Jonhoi Smith, Melvin Field, and Kiminobu Sugaya. 2023. "Coculture with Neural Stem Cells May Shift the Transcription Profile of Glioblastoma Multiforme towards Cancer-Specific Stemness" International Journal of Molecular Sciences 24, no. 4: 3242. https://doi.org/10.3390/ijms24043242
APA StyleVaidya, M., Sreerama, S., Gonzalez-Vega, M., Smith, J., Field, M., & Sugaya, K. (2023). Coculture with Neural Stem Cells May Shift the Transcription Profile of Glioblastoma Multiforme towards Cancer-Specific Stemness. International Journal of Molecular Sciences, 24(4), 3242. https://doi.org/10.3390/ijms24043242







