Integrated Transcriptome and Metabolome Dissecting Interaction between Vitis vinifera L. and Grapevine Fabavirus
Abstract
:1. Introduction
2. Results
2.1. GFabV Concentration in Infected Grapevines
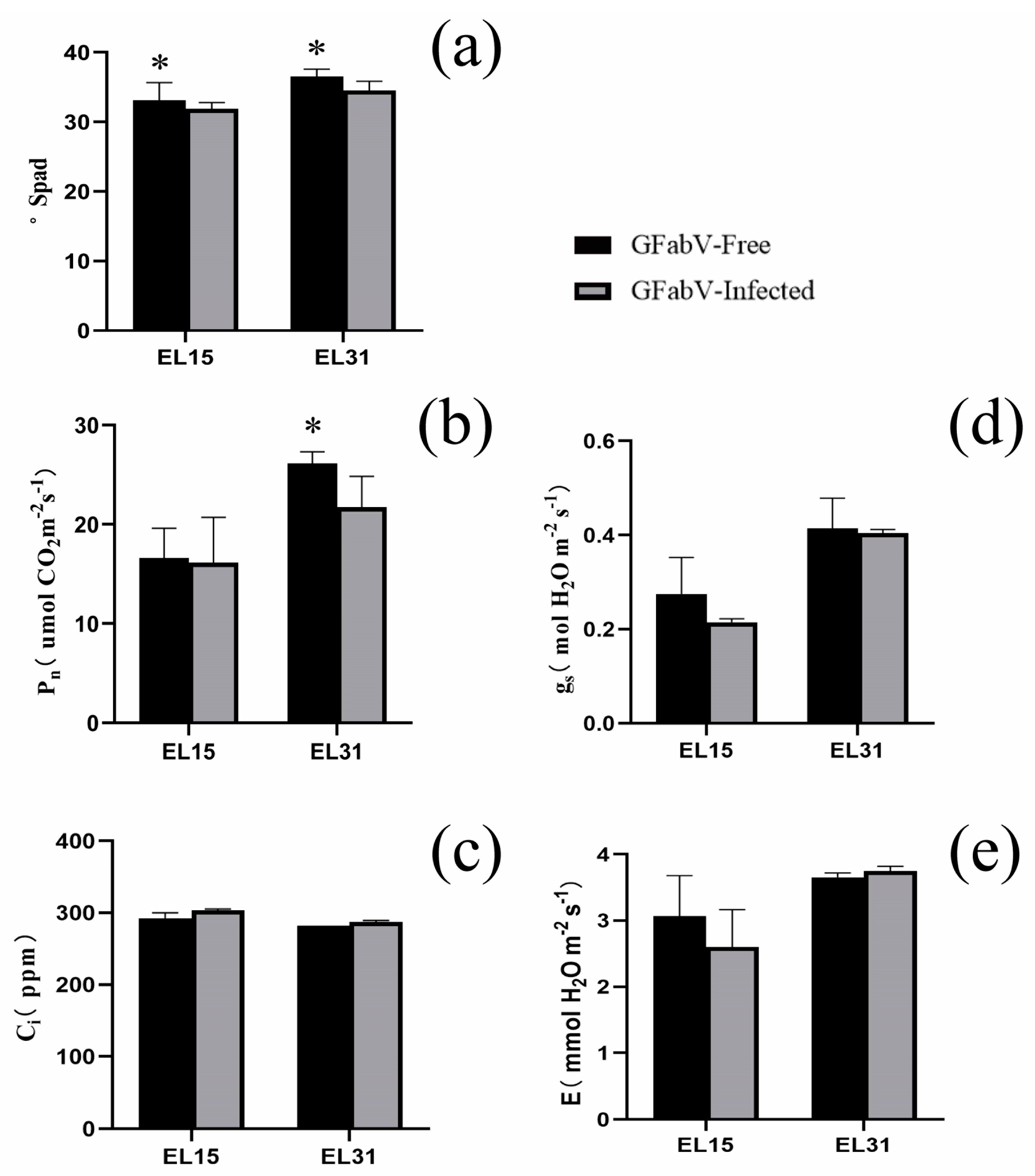
2.2. Physiological and Agronomic Performance of GFabV-Infected Grapevines
2.3. Changes in the Aroma Compounds between GFabV-Infected and GFabV-Free Grapevines
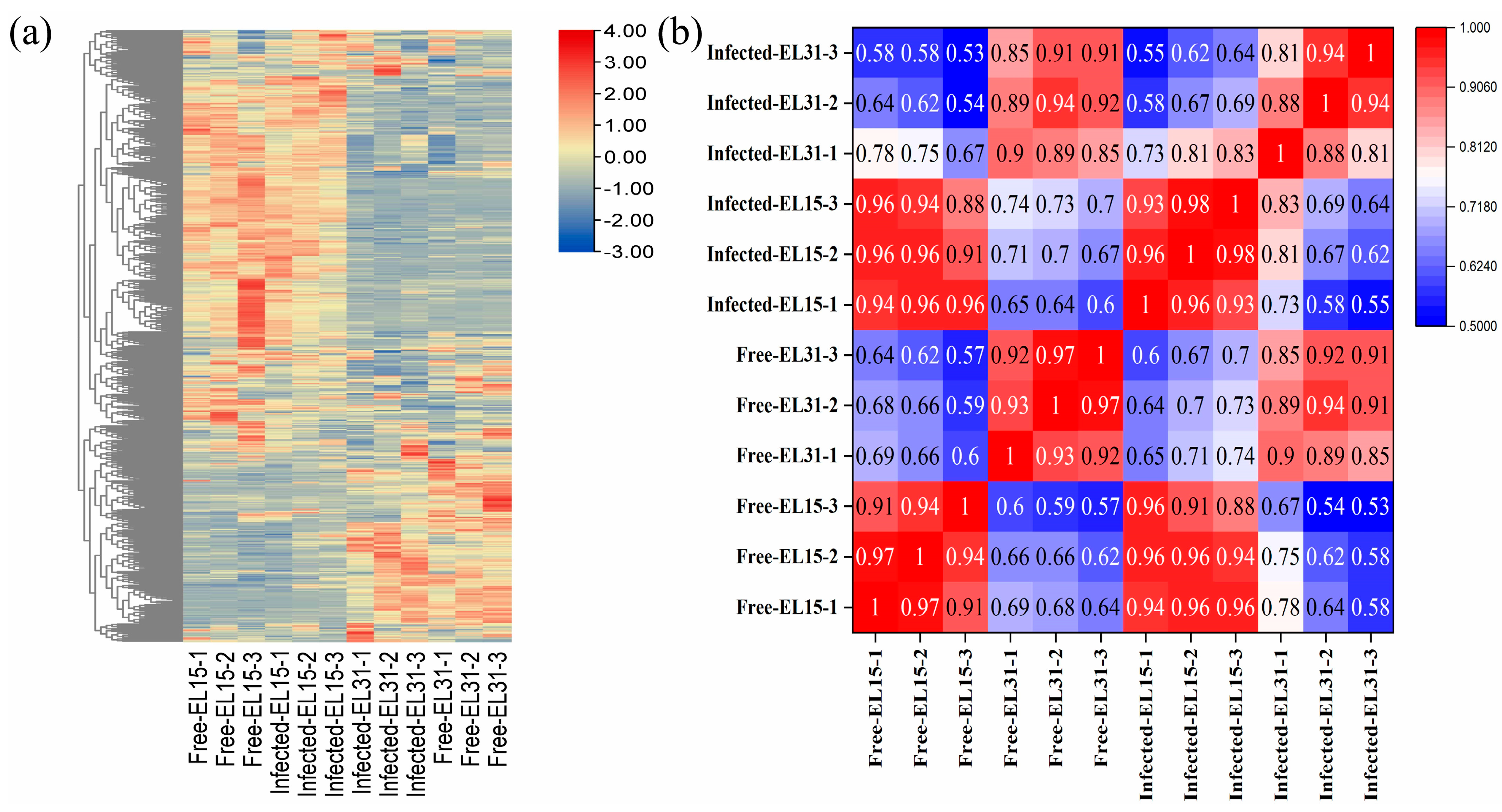
2.4. RNA–Seq Analysis
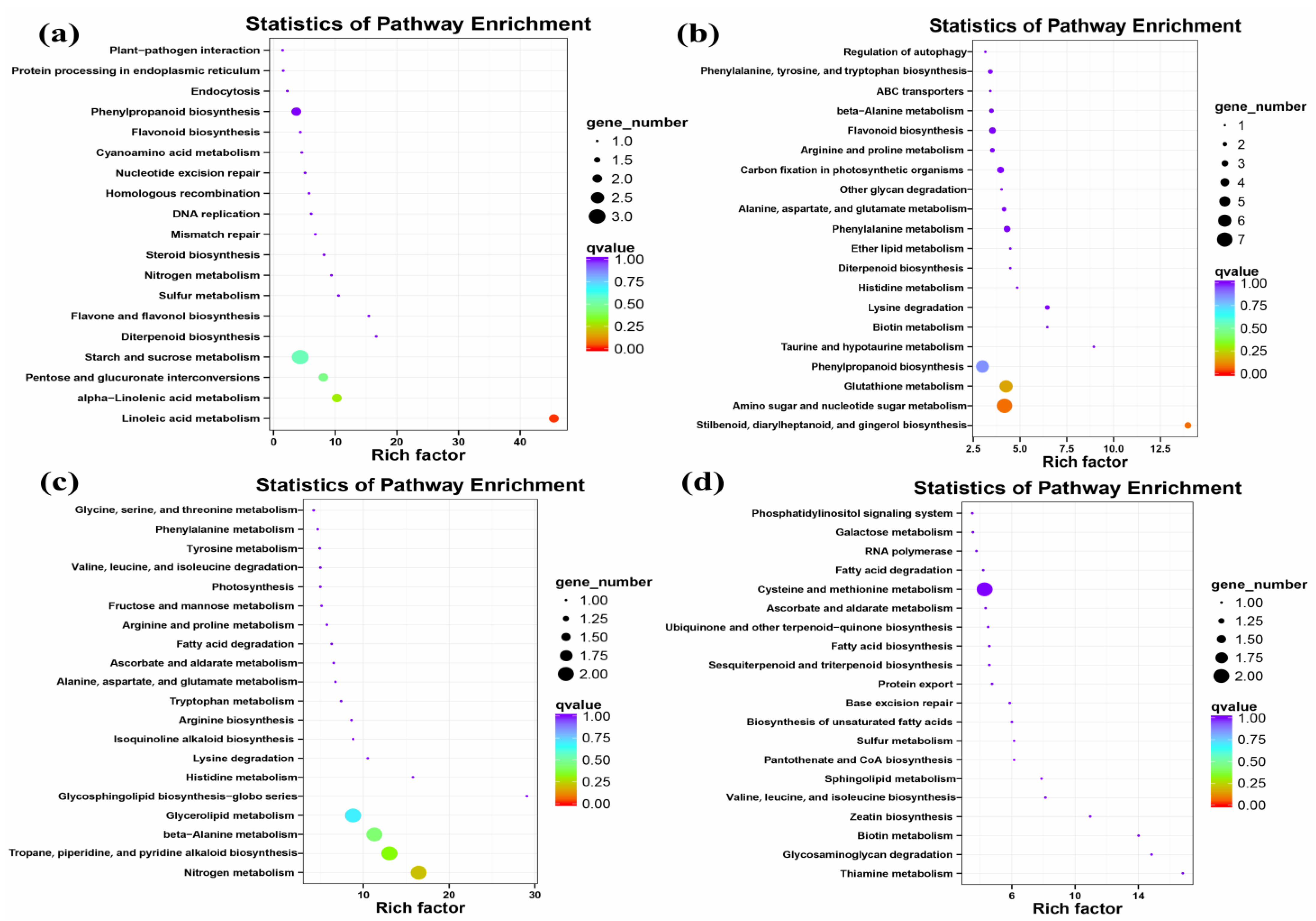
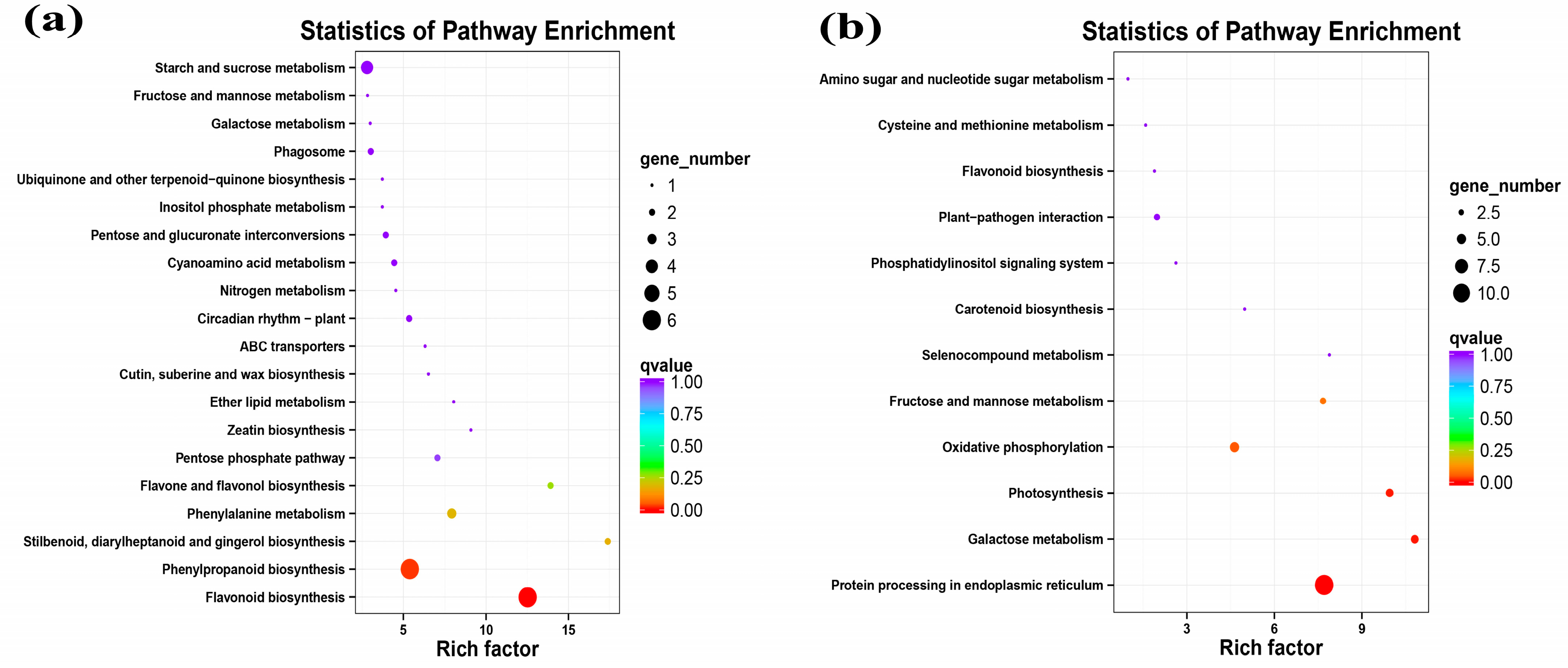
2.4.1. Global Gene Expression Changes in GFabV-Infected Leaves
2.4.2. Global Gene Expression Changes in GFabV-Infected Berries
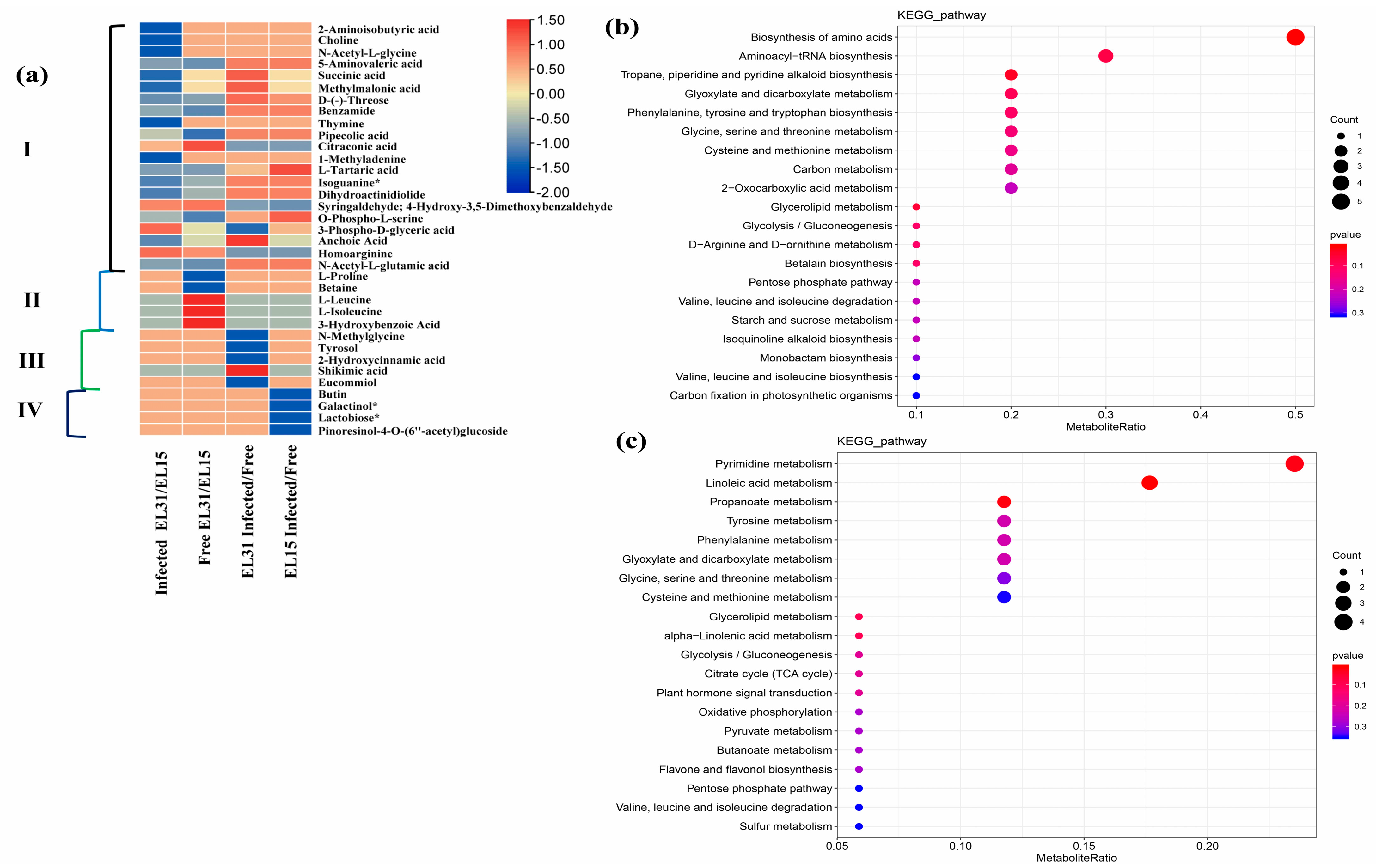
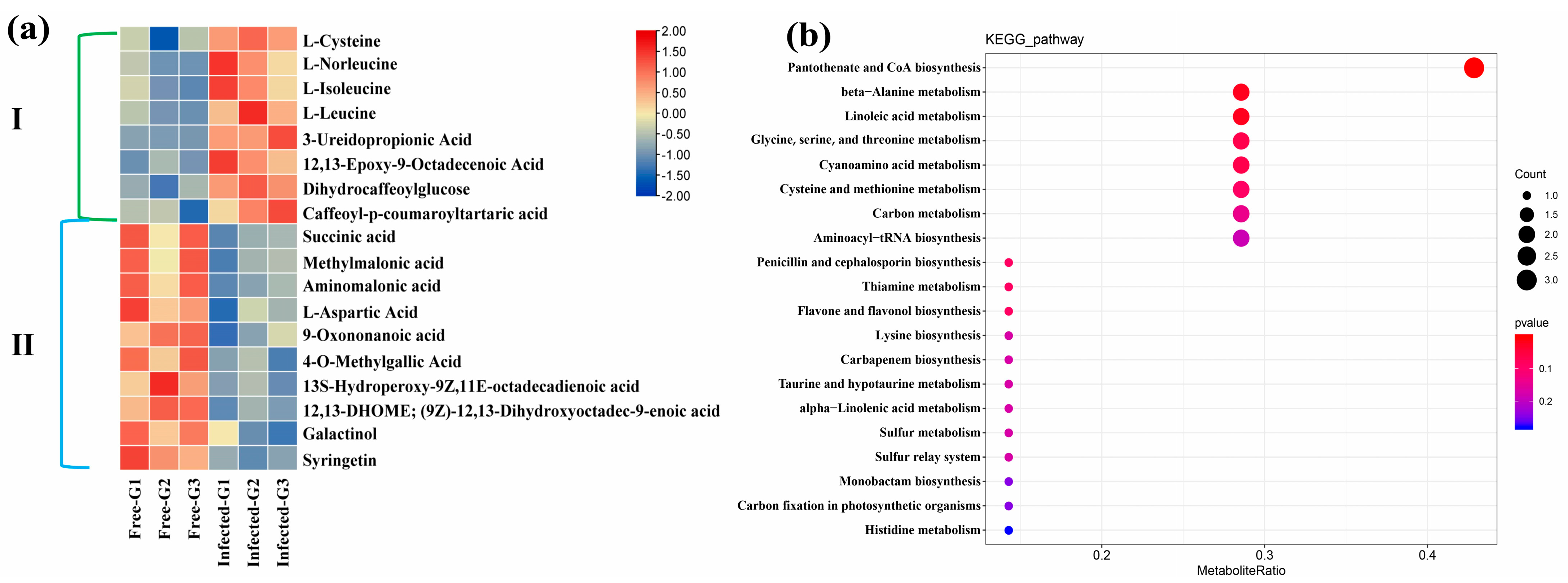
2.5. Metabolomic Analysis
2.6. Combined Analysis of Transcriptome and Metabolome
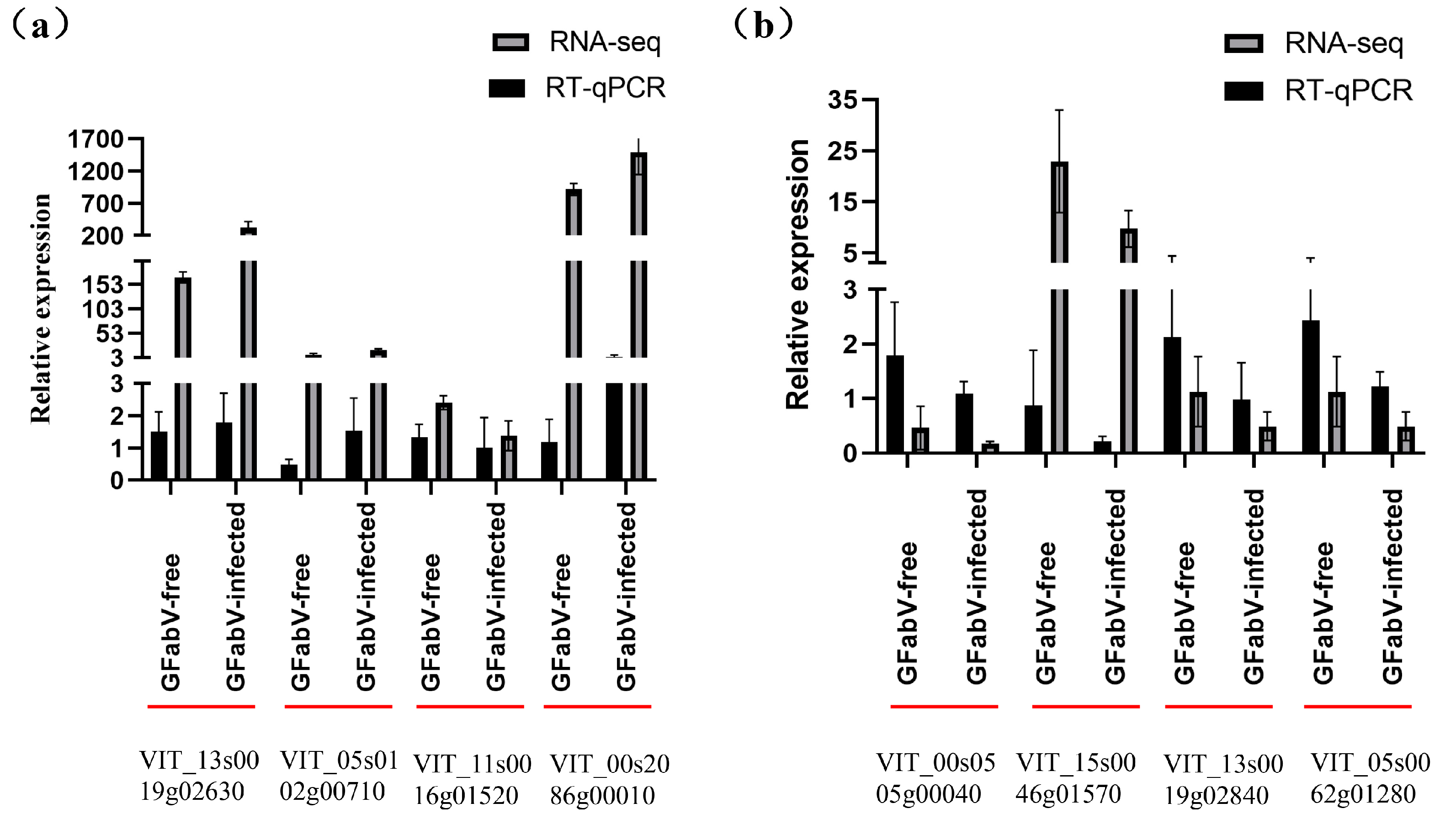
2.7. Validation of Differential Gene Expression Using RT–qPCR
3. Discussion
3.1. GFabV Concentration in Infected Grapevines
3.2. Physiological and Agronomic Performance of GFabV-Infected Grapevines
3.3. Differences in Aroma Compounds between GFabV-Infected and GFabV-Free Grapevines
3.4. Photosynthesis and Carbohydrate Metabolism in GFabV-Infected Grapevines
3.5. Changes in Hormone Genes under GFabV Infection in Grapevines
3.6. Transcription of Defense-Related Genes in Grapevines under GFabV Infection
3.7. Response of GFabV-Infected Grapevine to Several Stresses
3.8. Changes in Berries under GFabV Infection
4. Materials and Methods
4.1. Plant Materials
4.2. Agronomical and Physiological Parameters
4.3. GC–MS Analysis
4.4. RNA–Seq Experiment and Data Analysis
4.5. Real-Time RT–PCR
4.6. UPLC–MS/MS Analysis for Widely-Targeted Metabolome Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yadav, M.; Jain, S.; Bhardwaj, A.; Nagpal, R.; Puniya, M.; Tomar, R.; Singh, V.; Parkash, O.; Prasad, G.B.K.S.; Marotta, F.; et al. Biological and Medicinal Properties of Grapes and Their Bioactive Constituents: An Update. J. Med. Food 2009, 12, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Umer, M.; Liu, J.W.; You, H.F.; Xu, C.; Dong, K.L.; Luo, N.; Kong, L.H.; Li, X.P.; Hong, N.; Wang, G.P.; et al. Genomic, Morphological and Biological Traits of the Viruses Infecting Major Fruit Trees. Viruses 2019, 11, 515. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.D.; Dong, Y.F.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Li, Z.N.; Zhou, J. First Report of Grapevine Pinot Gris Virus in Grapevines in China. Plant Dis. 2016, 100, 540. [Google Scholar] [CrossRef]
- Rumbaugh, A.C.; Sudarshana, M.R.; Oberholster, A. Grapevine Red Blotch Disease Etiology and Its Impact on Grapevine Physiology and Berry and Wine Composition. Horticulturae 2021, 7, 552. [Google Scholar] [CrossRef]
- Zhang, Y.; Singh, K.; Kaur, R.; Qiu, W. Association of a Novel DNA Virus with the Grapevine Vein-Clearing and Vine Decline Syndrome. Phytopathology 2011, 101, 1081–1090. [Google Scholar] [CrossRef]
- Fan, X.D.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Li, Z.N.; Dong, Y.F. First Report of Grapevine Fabavirus in Grapevines in China. Plant Dis. 2017, 101, 847. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Olufemi, J.A.; Nathaniel, M.W.; Deborah, G.; Rowhani, A. Near-Complete Genome Sequence of Grapevine Fabavirus, a Novel Putative Member of the Genus Fabavirus. Genome Announc. 2016, 4, e00703-16. [Google Scholar] [CrossRef]
- Jo, Y.; Song, M.K.; Choi, H.; Park, J.S.; Lee, J.W.; Cho, W.K. First Report of Grapevine Fabavirus in Diverse Vitis Species in Korea. Plant Dis. 2017, 101, 1829. [Google Scholar] [CrossRef]
- Fan, X.D.; Zhang, M.Y.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Dong, Y.F. Prevalence and Genetic Diversity of Grapevine Fabavirus Isolates from Different Grapevine Cultivars and Regions in China. J. Integr. Agric. 2020, 19, 768–774. [Google Scholar] [CrossRef]
- Chiaki, Y.; Ito, T.; Sato, A.; Sugiura, H.; Nishimura, R. Dwarfing Caused by Viral Pathogens and Leaf Malformations in ‘Shine Muscat’ Grapevine. J. Gen. Plant Pathol. 2020, 86, 34–38. [Google Scholar] [CrossRef]
- Zhang, B.D.; Fan, X.D.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Li, C.; Jia, X.J.; Dong, Y.F. Sequence analysis of complete genome of two grapevine fabavirus isolates from China. Acta Phytopathol. Sin. 2022, 52, 601–612. (In Chinese) [Google Scholar] [CrossRef]
- Zhang, M.Y.; Zhang, B.D.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Fan, X.D.; Dong, Y.F. Complete Nucleotide Sequence of a Divergent Variant of Grapevine Fabavirus from Northeastern China. J. Plant Pathol. 2022, 104, 3–8. [Google Scholar] [CrossRef]
- Vega, A.; Gutiérrez, R.A.; Peña-Neira, A.; Cramer, G.R. Patricio Arce-Johnson Compatible GLRaV-3 Viral Infections Affect Berry Ripening Decreasing Sugar Accumulation and Anthocyanin Biosynthesis in Vitis vinifera. Plant Mol. Biol. 2011, 77, 261. [Google Scholar] [CrossRef] [PubMed]
- Rienth, M.; Ghaffari, S.; Reynard, J.S. Impact of grapevine leafroll virus infections on vine physiology and the berry transcriptome. In Proceedings of the 21st GiESCO International Meeting, Thessaloniki, Greece, 23–28 June 2019. [Google Scholar]
- Gambino, G.; Cuozzo, D.; Fasoli, M.; Pagliarani, C.; Vitali, M.; Boccacci, P.; Pezzotti, M.; Mannini, F. Co-Evolution between Grapevine Rupestris Stem Pitting-Associated Virus and Vitis vinifera L. Leads to Decreased Defence Responses and Increased Transcription of Genes Related to Photosynthesis. J. Exp. Bot. 2012, 63, 5919–5933. [Google Scholar] [CrossRef]
- Chitarra, W.; Cuozzo, D.; Ferrandino, A.; Secchi, F.; Palmano, S.; Perrone, I.; Boccacci, P.; Pagliarani, C.; Gribaudo, I.; Mannini, F.; et al. Dissecting Interplays between Vitis vinifera L. and Grapevine Virus B (GVB) under Field Conditions. Mol. Plant Pathol. 2018, 19, 2651–2666. [Google Scholar] [CrossRef]
- Martin, I.R.; Vigne, E.; Velt, A.; Hily, J.M.; Garcia, S.; Baltenweck, R.; Komar, V.; Rustenholz, C.; Hugueney, P.; Lemaire, O.; et al. Severe Stunting Symptoms upon Nepovirus Infection Are Reminiscent of a Chronic Hypersensitive-like Response in a Perennial Woody Fruit Crop. Viruses 2021, 13, 2138. [Google Scholar] [CrossRef]
- Čepin, U.; Gutiérrez-Aguirre, I.; Balažic, L.; Pompe-Novak, M.; Gruden, K.; Ravnikar, M. A One-Step Reverse Transcription Real-Time PCR Assay for the Detection and Quantitation of Grapevine Fanleaf Virus. J. Virol. Methods 2010, 170, 47–56. [Google Scholar] [CrossRef]
- Sampol, B.; Bota, J.; Riera, D.; Medrano, H.; Flexas, J. Analysis of the Virus-Induced Inhibition of Photosynthesis in Malmsey Grapevines. New Phytol. 2003, 160, 403–412. [Google Scholar] [CrossRef]
- Guidoni, S.; Mannini, F.; Ferrandino, A.; Argamante, N.; Stefano, R.D. The Effect of Grapevine Leafroll and Rugose Wood Sanitation on Agronomic Performance and Berry and Leaf Phenolic Content of a Nebbiolo Clone (Vitis vinifera L.). Am. J. Enol. Vitic. 1997, 48, 438–442. [Google Scholar] [CrossRef]
- Sicher, R.C.; Bunce, J.A. Adjustments of Net Photosynthesis in Solanum Tuberosum in Response to Reciprocal Changes in Ambient and Elevated Growth CO2 Partial Pressures. Physiol. Plant. 2001, 112, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Song, L.L.; Zhao, H.Q.; Zhu, X.Q.; Dong, G.X.; Xie, R. Effects of high temperature stress on photosynthesis and chlorophyll fluorescence of rice. Anhui Nongye Kexue 2011, 39, 13348–13353. [Google Scholar]
- Gao, M.; Qi, Y.; Song, W.; Xu, H. Effects of Di-n-Butyl Phthalate and Di (2-Ethylhexyl) Phthalate on the Growth, Photosynthesis, and Chlorophyll Fluorescence of Wheat Seedlings. Chemosphere 2016, 151, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Montero, R.; El Aou Ouad, H.; Pacifico, D.; Marzachì, C.; Castillo, N.; García, E.; Del Saz, N.F.; Florez-Sarasa, I.; Flexas, J.; Bota, J. Effects of Grapevine Leafroll-Associated Virus 3 on the Physiology in Asymptomatic Plants of Vitis vinifera. Ann. Appl. Biol. 2017, 171, 155–171. [Google Scholar] [CrossRef]
- Zhang, H.N.; Wang, Y.C.; Ma, H.; Yuan, X.D.; Ma, Y.K. GC-MS analysis of volatile aroma components in summer black grapes. Jiangsu Agric. Sci. 2014, 42, 294–297. [Google Scholar] [CrossRef]
- Matijašević, S.; Popović-Djordjević, J.; Ristić, R.; Ćirković, D.; Ćirković, B.; Popović, T. Volatile aroma compounds of brandy ‘Lozovača′ produced from Muscat table grapevine cultivars (Vitis vinifera L.). Molecules 2019, 24, 2485. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.; Sinha, A.K.; Roitsch, T. Plant Physiology Meets Phytopathology: Plant Primary Metabolism and Plant-Pathogen Interactions. J. Exp. Bot. 2007, 58, 4019–4026. [Google Scholar] [CrossRef]
- Parry, M.A.J.; Andralojc, P.J.; Mitchell, R.A.C.; Madgwick, P.J.; Keys, A.J. Manipulation of Rubisco: The Amount, Activity, Function and Regulation. J. Exp. Bot. 2003, 54, 1321–1333. [Google Scholar] [CrossRef]
- Biemelt, S.; Sonnewald, U. Plant-Microbe Interactions to Probe Regulation of Plant Carbon Metabolism. J. Plant Physiol. 2006, 163, 307–318. [Google Scholar] [CrossRef]
- Llave, C. Dynamic Cross-Talk between Host Primary Metabolism and Viruses during Infections in Plants. Curr. Opin. Virol. 2016, 19, 50–55. [Google Scholar] [CrossRef]
- Tecsi, L.I.; Smith, A.M.; Maule, A.J.; Leegood, R.C. A Spatial Analysis of Physiological Changes Associated with Infection of Cotyledons of Marrow Plants with Cucumber Mosaic Virus. Plant Physiol. 1996, 111, 975–985. [Google Scholar] [CrossRef]
- Balachandran, S.; Hurry, V.M.; Kelley, S.E.; Osmond, C.B.; Robinson, S.A.; Rohozinski, J.; Seaton, G.G.R.; Sims, D.A. Concepts of Plant Biotic Stress. Some Insights into the Stress Physiology of Virus-Infected Plants, from the Perspective of Photosynthesis. Physiol. Plant. 1997, 100, 203–213. [Google Scholar] [CrossRef]
- Herbers, K.; Takahata, Y.; Melzer, M.; Mock, H.-P.; Hajirezaei, M.; Sonnewald, U. Regulation of Carbohydrate Partitioning during the Interaction of Potato Virus Y with Tobacco. Mol. Plant Pathol. 2000, 1, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Van Wees, S.C.M.; De Swart, E.A.M.; Van Pelt, J.A.; Van Loon, L.C.; Pieterse, C.M.J. Enhancement of Induced Disease Resistance by Simultaneous Activation of Salicylate- and Jasmonate-Dependent Defense Pathways in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 8711–8716. [Google Scholar] [CrossRef] [PubMed]
- Caarls, L.; Pieterse, C.M.J.; Van Wees, S.C.M. How Salicylic Acid Takes Transcriptional Control over Jasmonic Acid Signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef]
- Fischer, U.; Dröge-Laser, W. Overexpression of NtERF5, a New Member of the Tobacco Ethylene Response Transcription Factor Family Enhances Resistance to Tobacco Mosaic Virus. MPMI 2004, 17, 1162–1171. [Google Scholar] [CrossRef]
- Agudelo-Romero, P.; Erban, A.; Rego, C.; Carbonell-Bejerano, P.; Nascimento, T.; Sousa, L.; Martínez-Zapater, J.M.; Kopka, J.; Fortes, A.M. Transcriptome and Metabolome Reprogramming in Vitis vinifera cv. Trincadeira Berries upon Infection with Botrytis Cinerea. J. Exp. Bot. 2015, 66, 1769–1785. [Google Scholar] [CrossRef]
- DeYoung, B.J.; Innes, R.W. Plant NBS-LRR Proteins in Pathogen Sensing and Host Defense. Nat. Immunol. 2006, 7, 1243–1249. [Google Scholar] [CrossRef]
- Figueiredo, A.; Fortes, A.M.; Ferreira, S.; Sebastiana, M.; Choi, Y.H.; Sousa, L.; Acioli-Santos, B.; Pessoa, F.; Verpoorte, R.; Pais, M.S. Transcriptional and Metabolic Profiling of Grape (Vitis vinifera L.) Leaves Unravel Possible Innate Resistance against Pathogenic Fungi. J. Exp. Bot. 2008, 59, 3371–3381. [Google Scholar] [CrossRef]
- Aseel, D.G.; Rashad, Y.M.; Hammad, S.M. Arbuscular Mycorrhizal Fungi Trigger Transcriptional Expression of Flavonoid and Chlorogenic Acid Biosynthetic Pathways Genes in Tomato against Tomato Mosaic Virus. Sci. Rep. 2019, 9, 9692. [Google Scholar] [CrossRef]
- Li, N.; Xu, C.; Li-Beisson, Y.; Philippar, K. Fatty Acid and Lipid Transport in Plant Cells. Trends Plant Sci. 2016, 21, 145–158. [Google Scholar] [CrossRef]
- Kachroo, A.; Kachroo, P. Fatty Acid-Derived Signals in Plant Defense. Annu. Rev. Phytopathol. 2009, 47, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Qiao, F.; Kong, L.-A.; Peng, H.; Huang, W.-K.; Wu, D.-Q.; Liu, S.-M.; Clarke, J.L.; Qiu, D.-W.; Peng, D.-L. Transcriptional Profiling of Wheat (Triticum Aestivum L.) during a Compatible Interaction with the Cereal Cyst Nematode Heterodera Avenae. Sci. Rep. 2019, 9, 2184. [Google Scholar] [CrossRef] [PubMed]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-Transferase Enzymes in Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef] [PubMed]
- Margis-Pinheiro, M.; Martin, C.; Didierjean, L.; Burkard, G. Differential Expression of Bean Chitinase Genes by Virus Infection, Chemical Treatment and UV Irradiation. Plant Mol. Biol. 1993, 22, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Brar, A.; Yadav, M.; Chawade, A.; Vivekanand, V.; Pareek, N. Chitinases—Potential Candidates for Enhanced Plant Resistance towards Fungal Pathogens. Agriculture 2018, 8, 88. [Google Scholar] [CrossRef]
- Bishop, J.G.; Dean, A.M.; Mitchell-Olds, T. Rapid Evolution in Plant Chitinases: Molecular Targets of Selection in Plant-Pathogen Coevolution. Proc. Natl. Acad. Sci. USA 2000, 97, 5322–5327. [Google Scholar] [CrossRef] [PubMed]
- Kabir, S.R.; Rahman, M.M.; Tasnim, S.; Karim, M.R.; Khatun, N.; Hasan, I.; Amin, R.; Islam, S.S.; Nurujjaman, M.; Kabir, A.H.; et al. Purification and Characterization of a Novel Chitinase from Trichosanthes Dioica Seed with Antifungal Activity. Int. J. Biol. Macromol. 2016, 84, 62–68. [Google Scholar] [CrossRef]
- Espinoza, C.; Medina, C.; Somerville, S.; Arce-Johnson, P. Senescence-Associated Genes Induced during Compatible Viral Interactions with Grapevine and Arabidopsis. J. Exp. Bot. 2007, 58, 3197–3212. [Google Scholar] [CrossRef]
- Coombe, B.G. Research on Development and Ripening of the Grape Berry. Am. J. Enol. Vitic. 1992, 43, 101–110. [Google Scholar] [CrossRef]
- Montero, R.; Mundy, D.; Albright, A.; Grose, C.; Trought, M.C.T.; Cohen, D.; Chooi, K.M.; MacDiarmid, R.; Flexas, J.; Bota, J. Effects of Grapevine Leafroll Associated Virus 3 (GLRaV-3) and Duration of Infection on Fruit Composition and Wine Chemical Profile of Vitis vinifera L. cv. Sauvignon Blanc. Food Chem. 2016, 197, 1177–1183. [Google Scholar] [CrossRef]
- Havaux, M. Carotenoids as Membrane Stabilizers in Chloroplasts. Trends Plant Sci. 1998, 3, 147–151. [Google Scholar] [CrossRef]
- Havaux, M.; Gruszecki, W.I. heat-and light-induced chlorophyll a fluorescence changes in potato leaves containing high or low levels of the carotenoid zeaxanthin: Indications of a regulatory effect of zeaxanthin on thylakoid membrane fluidity. Photochem. Photobiol. 1993, 58, 607–614. [Google Scholar] [CrossRef]
- Gould, K.S. Nature’s Swiss Army Knife: The Diverse Protective Roles of Anthocyanins in Leaves. J. Biomed. Biotechnol. 2004, 2004, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Margaria, P.; Ferrandino, A.; Caciagli, P.; Kedrina, O.; Schubert, A.; Palmano, S. Metabolic and Transcript Analysis of the Flavonoid Pathway in Diseased and Recovered Nebbiolo and Barbera Grapevines (Vitis vinifera L.) Following Infection by Flavescence Dorée Phytoplasma. Plant Cell Environ. 2014, 37, 2183–2200. [Google Scholar] [CrossRef]
- Ferrandino, A.; Lovisolo, C. Abiotic Stress Effects on Grapevine (Vitis vinifera L.): Focus on Abscisic Acid-Mediated Consequences on Secondary Metabolism and Berry Quality. Environ. Exp. Bot. 2014, 103, 138–147. [Google Scholar] [CrossRef]
- Bogs, J.; Ebadi, A.; McDavid, D.; Robinson, S.P. Identification of the Flavonoid Hydroxylases from Grapevine and Their Regulation during Fruit Development. Plant Physiol. 2006, 140, 279–291. [Google Scholar] [CrossRef]
- Gollop, R.; Even, S.; Colova-Tsolova, V.; Perl, A. Expression of the Grape Dihydroflavonol Reductase Gene and Analysis of Its Promoter Region1. J. Exp. Bot. 2002, 53, 1397–1409. [Google Scholar] [CrossRef]
- Solfanelli, C.; Poggi, A.; Loreti, E.; Alpi, A.; Perata, P. Sucrose-Specific Induction of the Anthocyanin Biosynthetic Pathway in Arabidopsis. Plant Physiol. 2006, 140, 637–646. [Google Scholar] [CrossRef]
- Coombe, B.G. Growth Stages of the Grapevine: Adoption of a System for Identifying Grapevine Growth Stages. Aust. J. Grape Wine Res. 1995, 1, 104–110. [Google Scholar] [CrossRef]
- Wang, C.; Lv, S.; Wu, Y.; Lian, M.; Gao, X.; Meng, Q. Study of Aroma Formation and Transformation during the Manufacturing Process of Biluochun Green Tea in Yunnan Province by HS-SPME and GC-MS. J. Sci. Food Agric. 2016, 96, 4492–4498. [Google Scholar] [CrossRef]
- Fan, X.D.; Dong, Y.F.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Zhu, H.J. Molecular identification and gene sequence analysis of Grapevine virus E. Acta Phytopathol. Sin. 2014, 44, 455–460. (In Chinese) [Google Scholar]
- Zhang, M.Y.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Fan, X.D.; Dong, Y.F. Establishment and Application of a Real-time Fluorescent Quantitative RT-PCR for Detection of Grapevine fabavirus. Acta Hortic. Sin. 2020, 47, 187. [Google Scholar]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An Optimized Grapevine RNA Isolation Procedure and Statistical Determination of Reference Genes for Real-Time RT-PCR during Berry Development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraga, C.G.; Clowers, B.H.; Moore, R.J.; Zink, E.M. Signature-Discovery Approach for Sample Matching of a Nerve-Agent Precursor Using Liquid Chromatography-Mass Spectrometry, XCMS, and Chemometrics. Anal. Chem. 2010, 82, 4165–4173. [Google Scholar] [CrossRef]






| Data | 2018 | 2019 | ||||
|---|---|---|---|---|---|---|
| GFabV-Free | GFabV-Infected | p-Value | GFabV-Free | GFabV-Infected | p-Value | |
| Berry weight (g) | 10.38 ± 0.08 | 9.14 ± 0.33 | ** | 7.67 ± 0.98 | 5.33 ± 1.03 | NS |
| Berry length (cm) | 27.6 ± 0.13 | 26.64 ± 1.41 | * | 16.24 ± 0.34 | 14.60 ± 0.14 | ** |
| Berry width (cm) | 27.53 ± 0.23 | 26.92 ± 1.16 | ** | 16.63 ± 0.86 | 15.23 ± 0.95 | NS |
| Soluble solids (°Brix) | 18.43 ± 0.06 | 18.4 ± 0.26 | NS | 18.60 ± 0.41 | 17.63 ± 1.05 | NS |
| Titratable acidity (g/L) | 0.31 ± 0.001 | 0.34 ± 0.01 | * | 0.43 ± 0.08 | 0.48 ± 0.05 | NS |
| Total cyanine glycosides (mg/100 g) | 114.9 ± 6.38 | 191.2 ± 2.38 | ** | 112.93 ± 2.82 | 191.79 ± 1.87 | ** |
| Bunch weight (g) | 253 ± 8.89 | 290 ± 13.5 | NS | 506.7 ± 56.62 | 440 ± 56.57 | NS |
| Bunch length (cm) | 11.33 ± 0.58 | 9.33 ± 2.52 | NS | 14 ± 1.63 | 12.33 ± 0.47 | NS |
| Bunch width (cm) | 0.47 ± 0.06 | 0.6 ± 0.10 | NS | 1 ± 0.08 | 0.73 ± 0.12 | NS |
| Unique ID | Functional Annotation | Log2(Fold Change) | ||
|---|---|---|---|---|
| Leaf | Berry | |||
| EL15 | EL31 | EL38 | ||
| Photosynthesis and energy metabolism | ||||
| VIT_18s0001g11470 | Cytochrome P450 82A3 | −0.86 | ||
| VIT_00s0505g00040 | Photosystem II reaction center protein H | −1.45 | ||
| VIT_07s0141g00890 | Cytochrome P450 94A1 | 0.85 | ||
| VIT_13s0019g02630 | Photosystem II protein D1 | 0.82 | −1.92 | |
| VIT_18s0122g00460 | Calvin cycle protein CP12–3, chloroplastic | 0.64 | ||
| VIT_08s0007g07530 | NADPH-dependentaldehyde reductase-like protein, chloroplastic | −1.09 | ||
| Carbohydrate metabolism | ||||
| VIT_06s0061g00360 | UDP-glycosyltransferase 86A1 | −0.67 | ||
| VIT_05s0062g01280 | UDP-glycosyltransferase 92A1 | −1.15 | ||
| VIT_11s0052g01190 | Xyloglucanendotransglucosylase/hydrolase protein 23 | 1.14 | ||
| VIT_03s0038g04570 | Glucose-1-phosphate adenylyltransferase large subunit 1 (Fragment) | −1.58 | ||
| VIT_05s0062g00240 | Xyloglucan endotransglucosylase/hydrolase 2 | 0.62 | ||
| VIT_00s0233g00030 | Trehalose-phosphate phosphatase F | 0.69 | ||
| VIT_05s0062g00250 | Xyloglucan endotransglucosylase/hydrolase 2 | 1.81 | ||
| VIT_05s0102g00710 | Fructose-2 | 0.71 | ||
| VIT_14s0108g00890 | Beta-1,3-galactosyltransferase GALT1 | −0.66 | ||
| Nucleic acid metabolism and transcriptional regulation | ||||
| VIT_08s0058g00690 | Probable WRKY transcription factor 33 | −0.61 | ||
| VIT_08s0105g00370 | MYB family transcription factor PHL7 | 1.13 | ||
| VIT_08s0007g05030 | Transcription factor MYB36 | 0.71 | ||
| VIT_06s0004g04140 | Transcription factor MYB59 | −0.64 | ||
| VIT_18s0001g10030 | WRKY transcription factor 7 | 0.63 | ||
| VIT_05s0124g00240 | Transcription factor bHLH128 | 1.14 | ||
| VIT_01s0011g03070 | AP2/ERF and B3 domain-containing transcription repressor TEM1 | 0.62 | ||
| Lipid metabolism | ||||
| VIT_11s0016g00120 | Omega-6fattyaciddesaturase, chloroplastic | 0.62 | ||
| VIT_07s0141g00340 | Linoleate 9S-lipoxygenase 6 (Fragment) | 0.63 | ||
| VIT_00s0583g00030 | Sphingolipiddelta (4)-desaturaseDES1-like | −0.72 | ||
| VIT_11s0052g01090 | 4-coumarate—CoA ligase 1 | 1.03 | ||
| VIT_02s0109g00250 | 4-coumarate—CoA ligase-like 6 | −0.63 | ||
| VIT_14s0171g00110 | Triacylglycerol lipase 2 | 0.81 | ||
| Secondary metabolism | ||||
| VIT_11s0016g01520 | Phenylalanine ammonia-lyase | −0.61 | ||
| VIT_06s0009g03050 | Flavonoid 3′,5′-hydroxylase 2 | 0.91 | ||
| VIT_11s0065g00130 | Beta-amyrin 28-oxidase | −0.69 | ||
| VIT_06s0004g06130 | Phosphomethylpyrimidinesynthase, chloroplastic | −0.73 | ||
| VIT_16s0100g01200 | Stilbene synthase 6 | 2.19 | ||
| VIT_02s0025g04880 | Geraniol 8-hydroxylase | 0.59 | ||
| VIT_06s0009g02910 | Flavonoid 3′,5′-hydroxylase 2 | 0.66 | ||
| VIT_18s0001g03470 | Flavonolsynthase/flavanone 3-hydroxylase | −1.41 | ||
| VIT_03s0038g04620 | Isoflavone reductase homolog PCBER | −0.78 | ||
| Response to hormone stimulus and hormone biosynthesis | ||||
| VIT_16s0013g01000 | Ethylene-responsive transcription factor 5 | −0.66 | ||
| VIT_06s0004g06790 | Gibberellin 2-beta-dioxygenase 8 | −0.59 | ||
| VIT_05s0020g04680 | Auxin-induced protein 22D | −0.80 | ||
| VIT_17s0000g02420 | Auxin efflux carrier component 1 | 1.07 | ||
| VIT_00s2086g00010 | 1-aminocyclopropane-1-carboxylate oxidase | −0.67 | 0.59 | |
| VIT_17s0000g06210 | Gibberellin-regulated protein 6 | 1.39 | ||
| VIT_10s0116g00410 | Gibberellin 2-beta-dioxygenase 8 | 0.65 | ||
| Response to endogenous stimulus and signal transduction | ||||
| VIT_10s0003g02050 | Cyclic phosphodiesterase | −0.70 | ||
| VIT_09s0002g07720 | Receptor-like protein 1 | 0.66 | ||
| VIT_19s0014g00520 | Leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840 | 1.02 | ||
| VIT_16s0050g01530 | Receptor-like protein EIX1 | −0.74 | ||
| VIT_12s0034g01300 | TMV resistance protein N | −0.74 | ||
| VIT_14s0108g01000 | Calcium-binding protein CML45 | −0.90 | ||
| VIT_00s0125g00090 | LRR receptor-like serine/threonine-protein kinase | |||
| At1g56130 | −0.62 | 0.70 | ||
| Response to disease and abiotic stress | ||||
| VIT_13s0019g02840 | 18.2 kDa class I heat shock protein | −1.16 | ||
| VIT_04s0008g00150 | NAC domain-containing protein 22 | −0.79 | ||
| VIT_06s0004g07750 | Cationic peroxidase 1 | 0.69 | ||
| VIT_09s0096g00200 | Disease resistance protein RPS5 | −1.03 | ||
| VIT_03s0091g00560 | Leucine-rich repeat protein 2 | −0.67 | ||
| VIT_17s0000g02950 | Glutathione S-transferase | −0.89 | ||
| VIT_06s0004g03120 | MLO-like protein 3 | 0.62 | ||
| VIT_04s0023g02570 | Peroxidase 72 | −1.01 | ||
| VIT_02s0025g00280 | Heat shock protein 83 | 0.70 | ||
| VIT_02s0012g01040 | NAC domain-containing protein 71 | −0.76 | ||
| VIT_15s0046g01570 | Acidic endochitinase | −1.25 | ||
| VIT_06s0004g01180 | Peroxidase 15 | 1.01 | ||
| VIT_13s0067g03220 | ProteinENHANCEDDISEASE RESISTANCE 4 | −0.66 | ||
| Transport | ||||
| VIT_05s0020g00890 | ABC transporter B family member 11 | −0.66 | ||
| VIT_14s0066g01000 | Phosphoenolpyruvate/phosphate translocator 2, chloroplastic | 0.60 | ||
| VIT_17s0000g02500 | Ras-related protein RABA6a | −0.91 | ||
| VIT_01s0127g00070 | High affinity nitrate transporter 2.5 | 1.02 | ||
| VIT_03s0063g02250 | Polyol transporter 5 | 2.15 | ||
| VIT_03s0017g02170 | Zinc transporter 5 | −0.63 | ||
| VIT_04s0008g04180 | Silicon efflux transporter LSI2 | −0.61 | ||
| Cell cycle, morphogenesis, and homeostasis | ||||
| VIT_00s0414g00010 | Cellulose synthase-like protein E6 | −0.63 | ||
| VIT_11s0016g01530 | Ankyrin repeat-containing protein BDA1 | −0.68 | ||
| VIT_15s0021g02680 | Cyclin-U2-1 | 1.71 | ||
| VIT_06s0009g02020 | Tubulin beta-6 chain | −0.63 | ||
| VIT_02s0025g01850 | Cellulose synthase-like protein G3 | 0.60 | ||
| VIT_03s0017g01010 | Metacaspase-1 | 0.74 | ||
| VIT_10s0003g05480 | Patellin-4 | 2.02 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.; Zhang, M.; Jia, X.; Hu, G.; Ren, F.; Fan, X.; Dong, Y. Integrated Transcriptome and Metabolome Dissecting Interaction between Vitis vinifera L. and Grapevine Fabavirus. Int. J. Mol. Sci. 2023, 24, 3247. https://doi.org/10.3390/ijms24043247
Zhang B, Zhang M, Jia X, Hu G, Ren F, Fan X, Dong Y. Integrated Transcriptome and Metabolome Dissecting Interaction between Vitis vinifera L. and Grapevine Fabavirus. International Journal of Molecular Sciences. 2023; 24(4):3247. https://doi.org/10.3390/ijms24043247
Chicago/Turabian StyleZhang, Baodong, Mengyan Zhang, Xiaojun Jia, Guojun Hu, Fang Ren, Xudong Fan, and Yafeng Dong. 2023. "Integrated Transcriptome and Metabolome Dissecting Interaction between Vitis vinifera L. and Grapevine Fabavirus" International Journal of Molecular Sciences 24, no. 4: 3247. https://doi.org/10.3390/ijms24043247






