Apalutamide Prevents SARS-CoV-2 Infection in Lung Epithelial Cells and in Human Nasal Epithelial Cells
Abstract
1. Introduction
2. Results
2.1. TMPRSS2 and AR Genes Are Expressed in Lung Cells
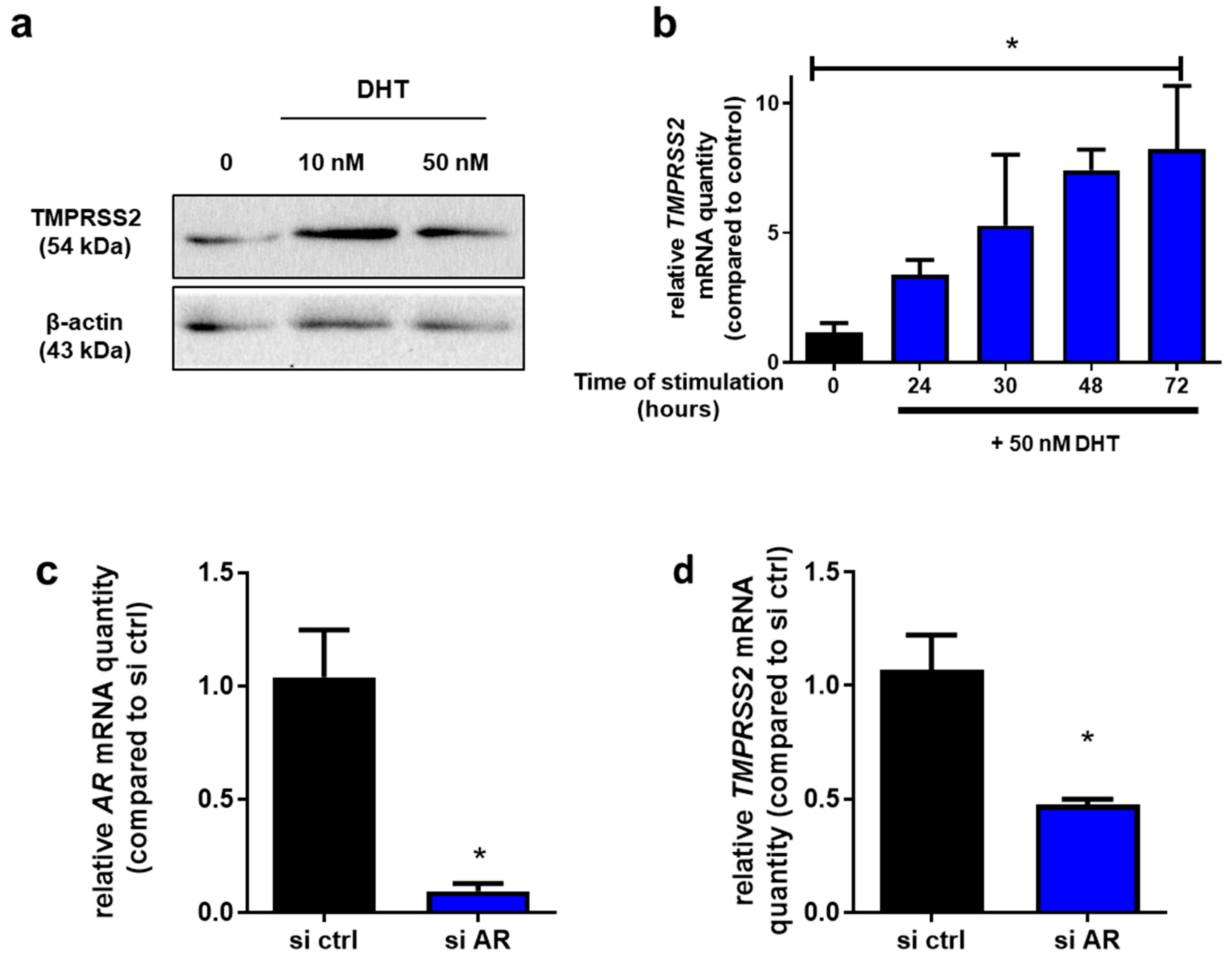
2.2. TMPRSS2 Is an Androgen-Responsive Gene in Calu-3 Cells
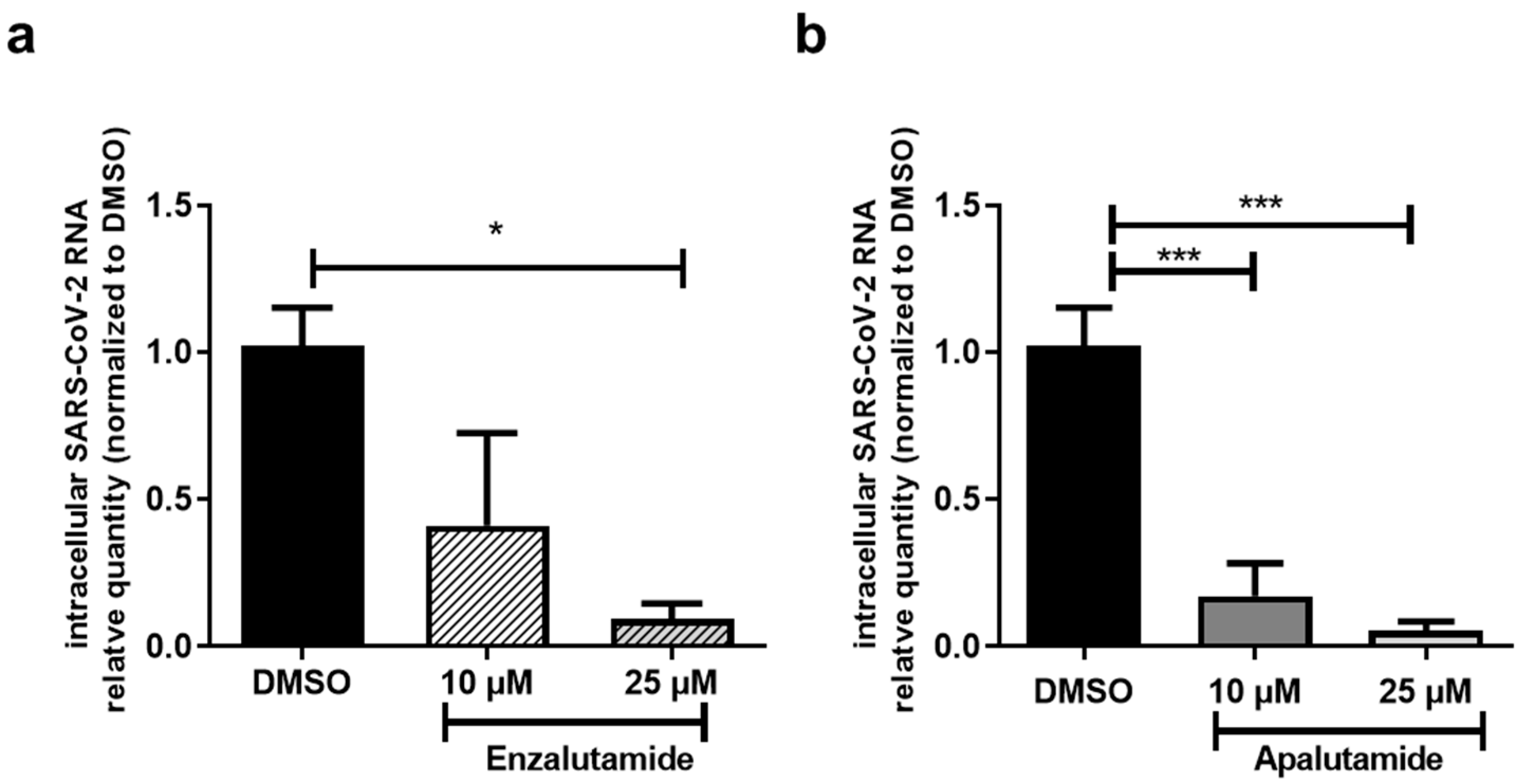
2.3. AR Antagonists Impact SARS-CoV-2 Infection in Calu-3 and HNECs Cultures
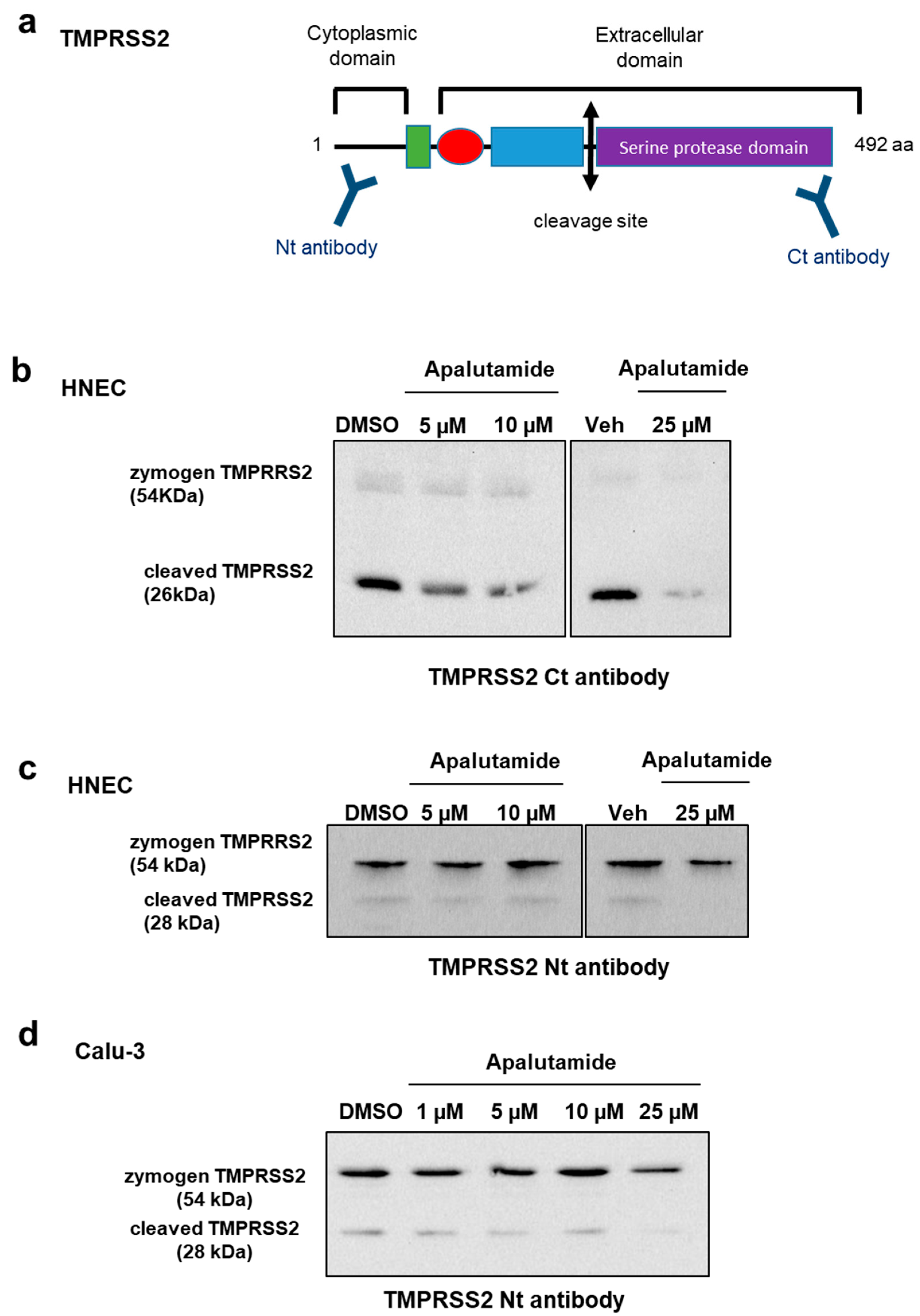
2.4. Apalutamide Blocks TMPRSS2 Catalytic Cleavage in Lung Cells and HNECs Cultures
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Preparation of Human Nasal Epithelial Cells (HNECs)
4.3. Viruses
4.4. SARS-CoV-2 Infection
4.5. Extraction and Quantification of Viral RNA by RT-qPCR
4.6. Cellular RNA Extraction and Quantification by RT-qPCR
4.7. Western Blot
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; McGoogan, J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef]
- Al-Jarallah, M.; Rajan, R.; Dashti, R.; Al Saber, A.; Brady, P.A.; Abdelnaby, H.; Alroomi, M.; Aboelhassan, W.; Abdullah, M.; AlNasrallah, N.; et al. In-Hospital Mortality in SARS-CoV-2 Stratified by Sex Diffrences: A Retrospective Cross-Sectional Cohort Study. Ann. Med. Surg. 2022, 79, 104026. [Google Scholar] [CrossRef]
- Haitao, T.; Vermunt, J.V.; Abeykoon, J.; Ghamrawi, R.; Gunaratne, M.; Jayachandran, M.; Narang, K.; Parashuram, S.; Suvakov, S.; Garovic, V.D. COVID-19 and Sex Differences. Mayo. Clin. Proc. 2020, 95, 2189–2203. [Google Scholar] [CrossRef] [PubMed]
- Scully, E.P.; Haverfield, J.; Ursin, R.L.; Tannenbaum, C.; Klein, S.L. Considering How Biological Sex Impacts Immune Responses and COVID-19 Outcomes. Nat. Rev. Immunol. 2020, 20, 442–447. [Google Scholar] [CrossRef]
- Vahidy, F.S.; Pan, A.P.; Ahnstedt, H.; Munshi, Y.; Choi, H.A.; Tiruneh, Y.; Nasir, K.; Kash, B.A.; Andrieni, J.D.; McCullough, L.D. Sex Differences in Susceptibility, Severity, and Outcomes of Coronavirus Disease 2019: Cross-Sectional Analysis from a Diverse US Metropolitan Area. PLoS ONE 2021, 16, e0245556. [Google Scholar] [CrossRef]
- Karlberg, J.; Chong, D.S.Y.; Lai, W.Y.Y. Do Men Have a Higher Case Fatality Rate of Severe Acute Respiratory Syndrome than Women Do? Am. J. Epidemiol. 2004, 159, 229–231. [Google Scholar] [CrossRef]
- Chen, X.; Chughtai, A.A.; Dyda, A.; MacIntyre, C.R. Comparative Epidemiology of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in Saudi Arabia and South Korea. Emerg. Microbes Infect. 2017, 6, e51. [Google Scholar] [CrossRef]
- Mousavizadeh, L.; Ghasemi, S. Genotype and Phenotype of COVID-19: Their Roles in Pathogenesis. J. Microbiol. Immunol. Infect. 2020, 54, 159–163. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Lin, B.; Ferguson, C.; White, J.T.; Wang, S.; Vessella, R.; True, L.D.; Hood, L.; Nelson, P.S. Prostate-Localized and Androgen-Regulated Expression of the Membrane-Bound Serine Protease TMPRSS2. Cancer Res. 1999, 59, 4180–4184. [Google Scholar]
- Afar, D.E.; Vivanco, I.; Hubert, R.S.; Kuo, J.; Chen, E.; Saffran, D.C.; Raitano, A.B.; Jakobovits, A. Catalytic Cleavage of the Androgen-Regulated TMPRSS2 Protease Results in Its Secretion by Prostate and Prostate Cancer Epithelia. Cancer Res. 2001, 61, 1686–1692. [Google Scholar]
- Lucas, J.M.; True, L.; Hawley, S.; Matsumura, M.; Morrissey, C.; Vessella, R.; Nelson, P.S. The Androgen-Regulated Type II Serine Protease TMPRSS2 Is Differentially Expressed and Mislocalized in Prostate Adenocarcinoma. J. Pathol. 2008, 215, 118–125. [Google Scholar] [CrossRef]
- Lucas, J.M.; Heinlein, C.; Kim, T.; Hernandez, S.A.; Malik, M.S.; True, L.D.; Morrissey, C.; Corey, E.; Montgomery, B.; Mostaghel, E.; et al. The Androgen-Regulated Protease TMPRSS2 Activates a Proteolytic Cascade Involving Components of the Tumor Microenvironment and Promotes Prostate Cancer Metastasis. Cancer Discov. 2014, 4, 1310–1325. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Rasool, R.U.; Russell, R.M.; Natesan, R.; Asangani, I.A. Targeting Androgen Regulation of TMPRSS2 and ACE2 as a Therapeutic Strategy to Combat COVID-19. iScience 2021, 24, 102254. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Wang, X.-M.; Mannan, R.; Pitchiaya, S.; Zhang, Y.; Wotring, J.W.; Xiao, L.; Robinson, D.R.; Wu, Y.-M.; Tien, J.C.-Y.; et al. Targeting Transcriptional Regulation of SARS-CoV-2 Entry Factors ACE2 and TMPRSS2. Proc. Natl. Acad. Sci. USA 2021, 118, e2021450118. [Google Scholar] [CrossRef] [PubMed]
- Leach, D.A.; Mohr, A.; Giotis, E.S.; Cil, E.; Isac, A.M.; Yates, L.L.; Barclay, W.S.; Zwacka, R.M.; Bevan, C.L.; Brooke, G.N. The Antiandrogen Enzalutamide Downregulates TMPRSS2 and Reduces Cellular Entry of SARS-CoV-2 in Human Lung Cells. Nat. Commun. 2021, 12, 4068. [Google Scholar] [CrossRef]
- Guo, W.; Porter, L.M.; Crozier, T.W.; Coates, M.; Jha, A.; McKie, M.; Nathan, J.A.; Lehner, P.J.; Greenwood, E.J.; McCaughan, F. Topical TMPRSS2 Inhibition Prevents SARS-CoV-2 Infection in Differentiated Human Airway Cultures. Life Sci. Alliance 2022, 5, e202101116. [Google Scholar] [CrossRef]
- Baratchian, M.; McManus, J.M.; Berk, M.P.; Nakamura, F.; Mukhopadhyay, S.; Xu, W.; Erzurum, S.; Drazba, J.; Peterson, J.; Klein, E.A.; et al. Androgen Regulation of Pulmonary AR, TMPRSS2 and ACE2 with Implications for Sex-Discordant COVID-19 Outcomes. Sci. Rep. 2021, 11, 11130. [Google Scholar] [CrossRef]
- Li, F.; Han, M.; Dai, P.; Xu, W.; He, J.; Tao, X.; Wu, Y.; Tong, X.; Xia, X.; Guo, W.; et al. Distinct Mechanisms for TMPRSS2 Expression Explain Organ-Specific Inhibition of SARS-CoV-2 Infection by Enzalutamide. Nat. Commun. 2021, 12, 866. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Feng, F.; Hu, G.; Wang, Y.; Yu, Y.; Zhu, Y.; Xu, W.; Cai, X.; Sun, Z.; Han, W.; et al. A Genome-Wide CRISPR Screen Identifies Host Factors That Regulate SARS-CoV-2 Entry. Nat. Commun. 2021, 12, 961. [Google Scholar] [CrossRef]
- Koch, J.; Uckeley, Z.M.; Doldan, P.; Stanifer, M.; Boulant, S.; Lozach, P.-Y. TMPRSS2 Expression Dictates the Entry Route Used by SARS-CoV-2 to Infect Host Cells. EMBO J. 2021, 40, e107821. [Google Scholar] [CrossRef]
- Mori, K.; Mostafaei, H.; Pradere, B.; Motlagh, R.S.; Quhal, F.; Laukhtina, E.; Schuettfort, V.M.; Abufaraj, M.; Karakiewicz, P.I.; Kimura, T.; et al. Apalutamide, Enzalutamide, and Darolutamide for Non-Metastatic Castration-Resistant Prostate Cancer: A Systematic Review and Network Meta-Analysis. Int. J. Clin. Oncol. 2020, 25, 1892–1900. [Google Scholar] [CrossRef] [PubMed]
- Coste, A.; Brugel, L.; Maître, B.; Boussat, S.; Papon, J.F.; Wingerstmann, L.; Peynègre, R.; Escudier, E. Inflammatory Cells as Well as Epithelial Cells in Nasal Polyps Express Vascular Endothelial Growth Factor. Eur. Respir. J. 2000, 15, 367–372. [Google Scholar] [CrossRef]
- Papon, J.-F.; Coste, A.; Gendron, M.-C.; Cordonnier, C.; Wingerstmann, L.; Peynègre, R.; Escudier, E. HLA-DR and ICAM-1 Expression and Modulation in Epithelial Cells from Nasal Polyps. Laryngoscope 2002, 112, 2067–2075. [Google Scholar] [CrossRef]
- Bequignon, E.; Dhommée, C.; Angely, C.; Thomas, L.; Bottier, M.; Escudier, E.; Isabey, D.; Coste, A.; Louis, B.; Papon, J.-F.; et al. FcRn-Dependent Transcytosis of Monoclonal Antibody in Human Nasal Epithelial Cells In Vitro: A Prerequisite for a New Delivery Route for Therapy? Int. J. Mol. Sci. 2019, 20, 1379. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.; Brighton, L.E.; Carson, J.L.; Fischer, W.A.; Jaspers, I. Culturing of Human Nasal Epithelial Cells at the Air Liquid Interface. J. Vis. Exp. 2013, 80, e50646. [Google Scholar] [CrossRef]
- Treppiedi, D.; Marra, G.; Di Muro, G.; Catalano, R.; Mangili, F.; Esposito, E.; Barbieri, A.M.; Arosio, M.; Mantovani, G.; Peverelli, E. TMPRSS2 Expression and Activity Modulation by Sex-Related Hormones in Lung Calu-3 Cells: Impact on Gender-Specific SARS-CoV-2 Infection. Front. Endocrinol. 2022, 13, 862789. [Google Scholar] [CrossRef]
- Global Health 50/50. The COVID-19 Sex-Disaggregated Data Tracker. 2022. Available online: https://globalhealth5050.org/the-sex-gender-and-covid-19-project/the-data-tracker/ (accessed on 8 November 2022).
- Peckham, H.; de Gruijter, N.M.; Raine, C.; Radziszewska, A.; Ciurtin, C.; Wedderburn, L.R.; Rosser, E.C.; Webb, K.; Deakin, C.T. Male Sex Identified by Global COVID-19 Meta-Analysis as a Risk Factor for Death and ITU Admission. Nat. Commun. 2020, 11, 6317. [Google Scholar] [CrossRef]
- Mikkonen, L.; Pihlajamaa, P.; Sahu, B.; Zhang, F.-P.; Jänne, O.A. Androgen Receptor and Androgen-Dependent Gene Expression in Lung. Mol. Cell. Endocrinol. 2010, 317, 14–24. [Google Scholar] [CrossRef]
- Baczenas, J.J.; Andersen, H.; Rashid, S.; Yarmosh, D.; Puthuveetil, N.; Parker, M.; Bradford, R.; Florence, C.; Stemple, K.J.; Lewis, M.G.; et al. Propagation of SARS-CoV-2 in Calu-3 Cells to Eliminate Mutations in the Furin Cleavage Site of Spike. Viruses 2021, 13, 2434. [Google Scholar] [CrossRef]
- Recchia, A.G.; Musti, A.M.; Lanzino, M.; Panno, M.L.; Turano, E.; Zumpano, R.; Belfiore, A.; Andò, S.; Maggiolini, M. A Cross-Talk between the Androgen Receptor and the Epidermal Growth Factor Receptor Leads to P38MAPK-Dependent Activation of MTOR and CyclinD1 Expression in Prostate and Lung Cancer Cells. Int. J. Biochem. Cell Biol. 2009, 41, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Ujike, M.; Morikawa, S.; Tashiro, M.; Taguchi, F. Protease-Mediated Enhancement of Severe Acute Respiratory Syndrome Coronavirus Infection. Proc. Natl. Acad. Sci. USA 2005, 102, 12543–12547. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Nagata, N.; Shirato, K.; Kawase, M.; Takeda, M.; Taguchi, F. Efficient Activation of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein by the Transmembrane Protease TMPRSS2. J. Virol. 2010, 84, 12658–12664. [Google Scholar] [CrossRef] [PubMed]
- Bestle, D.; Heindl, M.R.; Limburg, H.; Van Lam, T.; Pilgram, O.; Moulton, H.; Stein, D.A.; Hardes, K.; Eickmann, M.; Dolnik, O.; et al. TMPRSS2 and Furin Are Both Essential for Proteolytic Activation of SARS-CoV-2 in Human Airway Cells. Life Sci. Alliance 2020, 3, e202000786. [Google Scholar] [CrossRef]
- Montopoli, M.; Zumerle, S.; Vettor, R.; Rugge, M.; Zorzi, M.; Catapano, C.V.; Carbone, G.M.; Cavalli, A.; Pagano, F.; Ragazzi, E.; et al. Androgen-Deprivation Therapies for Prostate Cancer and Risk of Infection by SARS-CoV-2: A Population-Based Study (N=4532). Ann. Oncol. 2020, 31, 1040–1045. [Google Scholar] [CrossRef]
- Patel, V.G.; Zhong, X.; Liaw, B.; Tremblay, D.; Tsao, C.-K.; Galsky, M.D.; Oh, W.K. Does Androgen Deprivation Therapy Protect against Severe Complications from COVID-19? Ann. Oncol. 2020, 31, 1419–1420. [Google Scholar] [CrossRef]
- Chakravarty, D.; Nair, S.S.; Hammouda, N.; Ratnani, P.; Gharib, Y.; Wagaskar, V.; Mohamed, N.; Lundon, D.; Dovey, Z.; Kyprianou, N.; et al. Sex Differences in SARS-CoV-2 Infection Rates and the Potential Link to Prostate Cancer. Commun. Biol. 2020, 3, 374. [Google Scholar] [CrossRef]
- Koskinen, M.; Carpen, O.; Honkanen, V.; Seppänen, M.R.J.; Miettinen, P.J.; Tuominen, J.A.; Raivio, T. Androgen Deprivation and SARS-CoV-2 in Men with Prostate Cancer. Ann. Oncol. 2020, 31, 1417–1418. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.L.; Tucker, M.D.; Bakouny, Z.; Labaki, C.; Hsu, C.-Y.; Shyr, Y.; Armstrong, A.J.; Beer, T.M.; Bijjula, R.R.; Bilen, M.A.; et al. Association Between Androgen Deprivation Therapy and Mortality Among Patients With Prostate Cancer and COVID-19. JAMA Netw. Open 2021, 4, e2134330. [Google Scholar] [CrossRef] [PubMed]
- Gedeborg, R.; Loeb, S.; Styrke, J.; Kiiski-Berggren, R.; Garmo, H.; Stattin, P. Susceptibility to SARS-Cov-2 Infection and Risk for Severe COVID-19 in Patients with Prostate Cancer on Androgen Deprivation Therapy. Int. J. Cancer 2022, 151, 1925–1934. [Google Scholar] [CrossRef]
- Klein, E.A.; Li, J.; Milinovich, A.; Schold, J.D.; Sharifi, N.; Kattan, M.W.; Jehi, L. Androgen Deprivation Therapy in Men with Prostate Cancer Does Not Affect Risk of Infection with SARS-CoV-2. J. Urol. 2021, 205, 441–443. [Google Scholar] [CrossRef]
- Karimi, A.; Nowroozi, A.; Alilou, S.; Amini, E. Effects of Androgen Deprivation Therapy on COVID-19 in Patients with Prostate Cancer: A Systematic Review and Meta-Analysis. Urol. J. 2021, 18, 577–584. [Google Scholar] [CrossRef]
- Shah, N.J.; Patel, V.G.; Zhong, X.; Pina, L.; Hawley, J.E.; Lin, E.; Gartrell, B.A.; Febles, V.A.; Wise, D.R.; Qin, Q.; et al. The Impact of Androgen Deprivation Therapy on COVID-19 Illness in Men With Prostate Cancer. JNCI Cancer Spectr. 2022, 6, pkac035. [Google Scholar] [CrossRef]
- Samuel, R.M.; Majd, H.; Richter, M.N.; Ghazizadeh, Z.; Zekavat, S.M.; Navickas, A.; Ramirez, J.T.; Asgharian, H.; Simoneau, C.R.; Bonser, L.R.; et al. Androgen Signaling Regulates SARS-CoV-2 Receptor Levels and Is Associated with Severe COVID-19 Symptoms in Men. Cell Stem Cell 2020, 27, 876–889.e12. [Google Scholar] [CrossRef]
- Welén, K.; Rosendal, E.; Gisslén, M.; Lenman, A.; Freyhult, E.; Fonseca-Rodríguez, O.; Bremell, D.; Stranne, J.; Balkhed, Å.Ö.; Niward, K.; et al. A Phase 2 Trial of the Effect of Antiandrogen Therapy on COVID-19 Outcome: No Evidence of Benefit, Supported by Epidemiology and In Vitro Data. Eur. Urol. 2022, 81, 285–293. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majidipur, A.; Morin-Dewaele, M.; Gaspar Lopes, J.; Berry, F.; Fouchet, J.; Bartier, S.; Dufros Duval, A.; Soyeux, P.; Huet, E.; Louis, B.; et al. Apalutamide Prevents SARS-CoV-2 Infection in Lung Epithelial Cells and in Human Nasal Epithelial Cells. Int. J. Mol. Sci. 2023, 24, 3288. https://doi.org/10.3390/ijms24043288
Majidipur A, Morin-Dewaele M, Gaspar Lopes J, Berry F, Fouchet J, Bartier S, Dufros Duval A, Soyeux P, Huet E, Louis B, et al. Apalutamide Prevents SARS-CoV-2 Infection in Lung Epithelial Cells and in Human Nasal Epithelial Cells. International Journal of Molecular Sciences. 2023; 24(4):3288. https://doi.org/10.3390/ijms24043288
Chicago/Turabian StyleMajidipur, Amene, Margot Morin-Dewaele, Jeanne Gaspar Lopes, Francois Berry, Julien Fouchet, Sophie Bartier, Anais Dufros Duval, Pascale Soyeux, Eric Huet, Bruno Louis, and et al. 2023. "Apalutamide Prevents SARS-CoV-2 Infection in Lung Epithelial Cells and in Human Nasal Epithelial Cells" International Journal of Molecular Sciences 24, no. 4: 3288. https://doi.org/10.3390/ijms24043288
APA StyleMajidipur, A., Morin-Dewaele, M., Gaspar Lopes, J., Berry, F., Fouchet, J., Bartier, S., Dufros Duval, A., Soyeux, P., Huet, E., Louis, B., Coste, A., Béquignon, É., Saldana, C., Le Corvoisier, P., Destouches, D., Pawlotsky, J.-M., de la Taille, A., Vacherot, F., Bruscella, P., & Firlej, V. (2023). Apalutamide Prevents SARS-CoV-2 Infection in Lung Epithelial Cells and in Human Nasal Epithelial Cells. International Journal of Molecular Sciences, 24(4), 3288. https://doi.org/10.3390/ijms24043288






