Genome-Wide Analysis of WOX Multigene Family in Sunflower (Helianthus annuus L.)
Abstract
1. Introduction
2. Results
2.1. Identification of HaWOX in Sunflower Genome
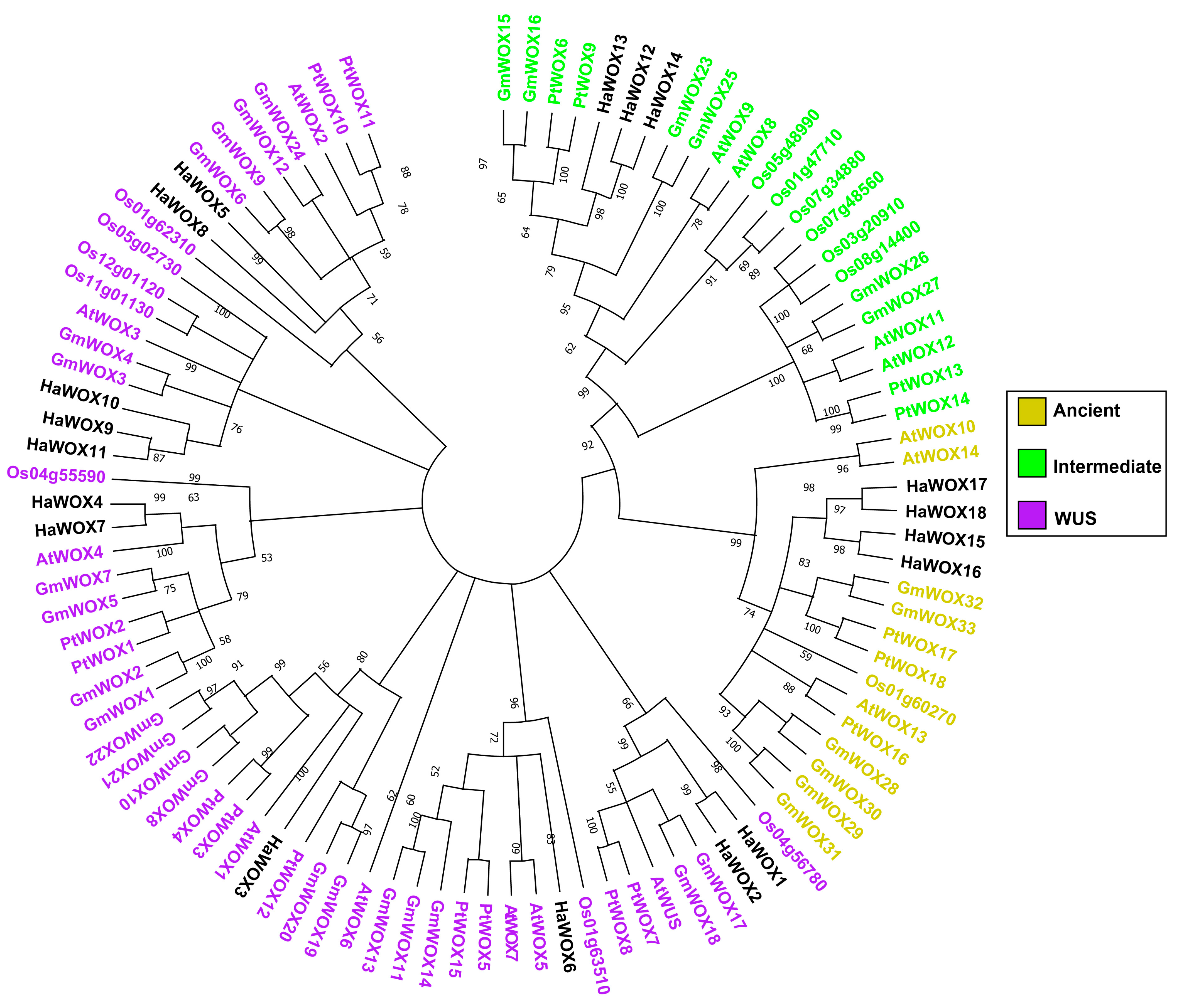
2.2. HaWOX Phylogenetic Analysis and Clade Subdivision
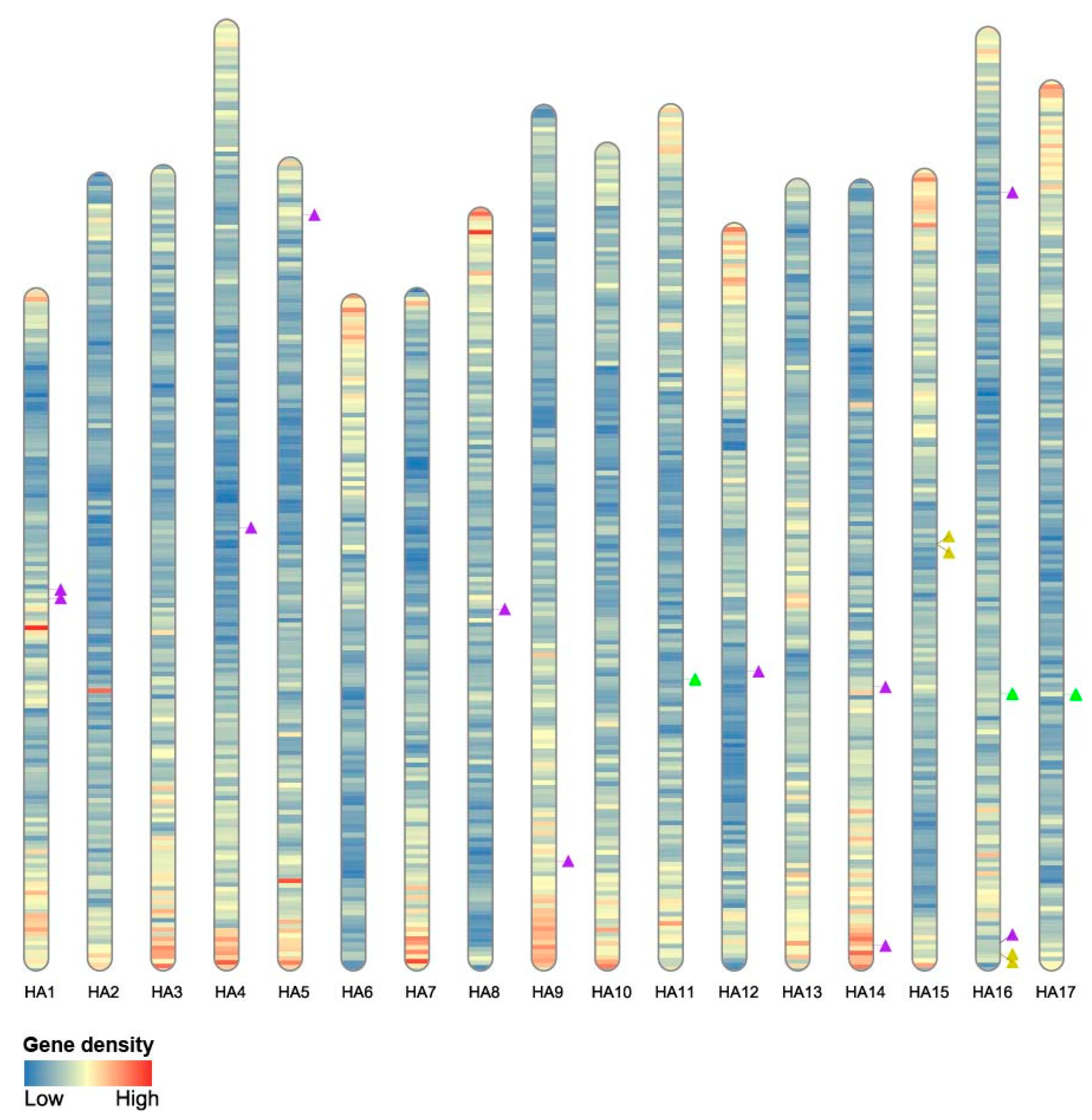
2.3. HaWOX Distribution in Sunflower Genome
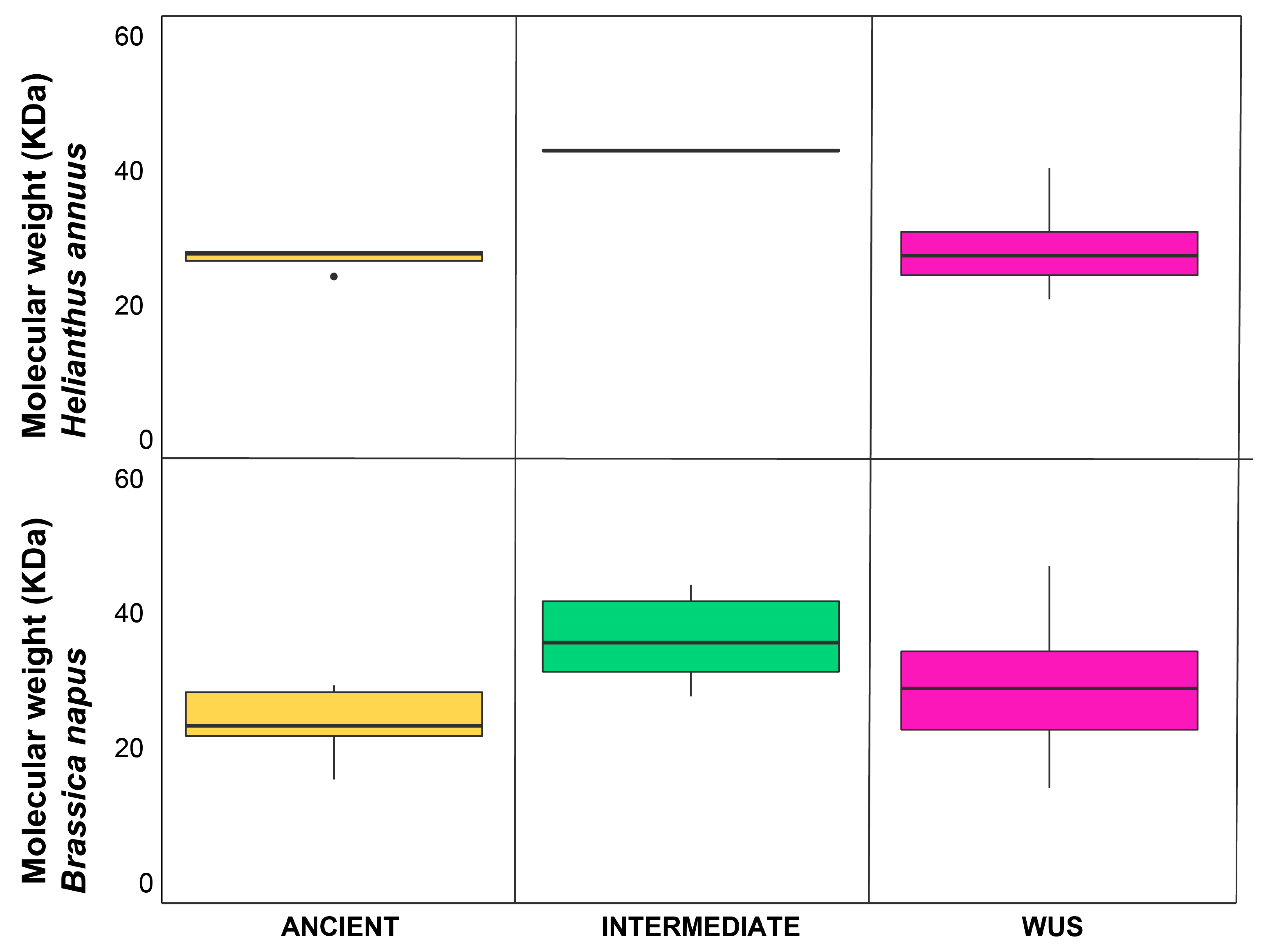
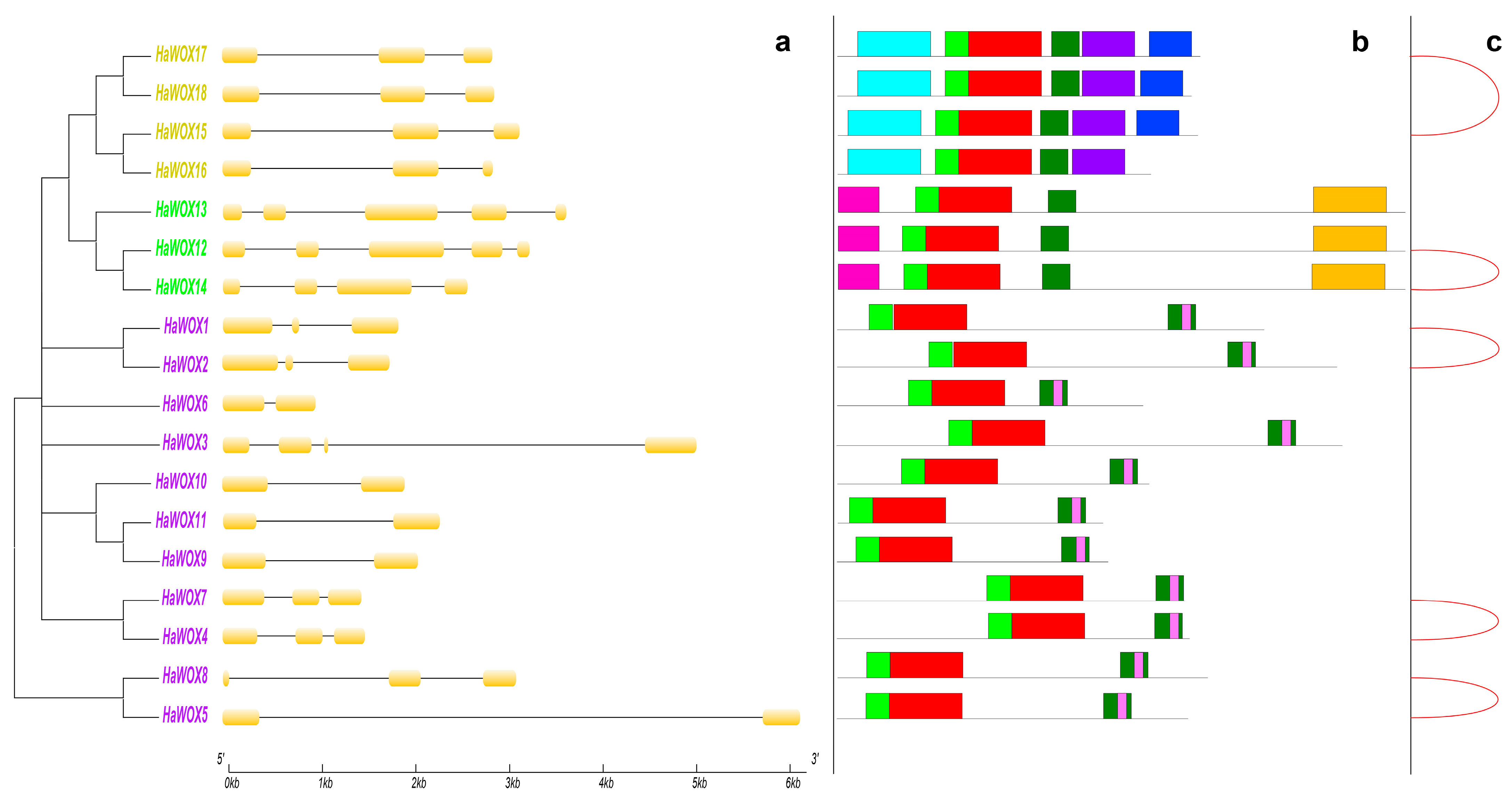
2.4. Biochemical, Structural, and Genomic Characterization of Sunflower WOX
2.5. HaWOX Genes Duplication Events in Sunflower Genome
2.6. HaWOX Gene Expression
3. Discussion
3.1. Structural Conservation of the Sunflower WOX Gene Family
3.2. Gene Expression Analysis
4. Materials and Methods
4.1. WOX Identification in Sunflower Genome
4.2. Alignments and Phylogenetic Analysis for HaWOX
4.3. Structural and Genomic Characterization of HaWOX Sequences
4.4. HaWOX Chromosomal Distribution and Gene Duplication Events
4.5. Gene Expression Analysis by Quantitative Real-Time Quantitative PCR (qRT–PCR)
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| AG | AGAMOUS |
| CLV | CLAVATA |
| DAP | Days after pollination |
| E | Embryo |
| EAR | ERF-associated amphiphilic repression |
| IM | Inflorescence meristem |
| HaL1L | LEAFY COTYLEDON1-Like |
| HB | WOX homeodomain |
| HLHTH | Helix–loop–helix–turn–helix |
| Kn-1 | Knotted-1 |
| OC | Organizing center |
| O | Ovule |
| qRT–PCR | Quantitative real-time polymerase chain reaction |
| RAM | Root apical meristem |
| SAM | Shoot apical meristem |
| TF | Transcription factor |
| TPL | TOPLESS |
| WOX | WUSCHEL-related homeobox genes |
| WUS | WUSCHEL |
References
- Howard-Ashby, M.; Materna, S.C.; Brown, C.T.; Chen, L.; Cameron, R.A.; Davidson, E.H. Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus and their expression in embryonic development. Dev. Biol. 2006, 300, 74–89. [Google Scholar] [CrossRef]
- Takatori, N.; Butts, T.; Candiani, S.; Pestarino, M.; Ferrier, D.E.; Saiga, H.; Holland, P.W. Comprehensive survey and classification of homeobox genes in the genome of amphioxus, Branchiostoma floridae. Dev. Genes Evol. 2008, 218, 579–590. [Google Scholar] [CrossRef]
- De Mendoza, A.; Sebe-Pedros, A.; Sestak, M.S.; Matejcic, M.; Torruella, G.; Domazet-Loso, T.; Ruiz-Trillo, I. Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc. Natl. Acad. Sci. USA 2013, 110, E4858–E4866. [Google Scholar] [CrossRef]
- Holland, P.W. Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 31–45. [Google Scholar] [CrossRef]
- Morino, Y.; Okada, K.; Niikura, M.; Honda, M.; Satoh, N.; Wada, H. A genome-wide survey of genes encoding transcription factors in the Japanese pearl oyster, Pinctada fucata: I. homeobox genes. Zool. Sci. 2013, 30, 851–857. [Google Scholar] [CrossRef]
- Simakov, O.; Marletaz, F.; Cho, S.J.; Edsinger-Gonzales, E.; Havlak, P.; Hellsten, U.; Kuo, D.-H.; Larsson, T.; Lv, J.; Rokhsar, D.S.; et al. Insights into bilaterian evolution from three spiralian genomes. Nature 2013, 493, 526–531. [Google Scholar] [CrossRef]
- Hench, J.; Henriksson, J.; Abou-Zied, A.M.; Luppert, M.; Dethlefsen, J.; Mukherjee, K.; Tong, Y.G.; Tang, L.; Gangishetti, U.; Baillie, D.L.; et al. The homeobox genes of Caenorhabditis elegans and insights into their spatio-temporal expression dynamics during embryogenesis. PLoS ONE 2015, 10, e01226947. [Google Scholar] [CrossRef]
- Bürglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef]
- Spencer, V.; Nemec Venza, Z.; Harrison, C.J. What can lycophytes teach us about plant evolution and development? Modern perspectives on an ancient lineage. Evol. Dev. 2021, 23, 174–196. [Google Scholar] [CrossRef]
- Gehring, W.J.; Hiromi, Y. Homeotic genes and the homeobox. Annu. Rev. Genet. 1986, 20, 147–173. [Google Scholar] [CrossRef]
- Maeda, R.K.; Karch, F. Cis-regulation in the Drosophila Bithorax complex. Adv. Exp. Med. Biol. 2010, 89, 17–40. [Google Scholar]
- McGinnis, W.; Levine, M.S.; Hafen, E.; Kuroiwa, A.; Gehring, W.J. A conserved DNA sequence in homeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 1984, 308, 428–433. [Google Scholar] [CrossRef]
- Scott, M.P.; Weiner, A.J. Structural relationships among genes that control development: Sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc. Nat. Acad. Sci. USA 1984, 81, 4115–4119. [Google Scholar] [CrossRef]
- Vollbrecht, E.; Veit, B.; Sinha, N.; Hake, S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 1991, 350, 241–243. [Google Scholar] [CrossRef]
- Bryan, A.A.; Sass, J.E. Heritable characters in maize. J. Hered. 1941, 32, 343–346. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Weigel, D.; Jürgens, G. Stem cells that make stems. Nature 2002, 415, 751–754. [Google Scholar] [CrossRef]
- Reiser, L.; Sanchez-Baracaldo, P.; Hake, S. Knots in the family three: Evolutionary relationships and function of knox homeobox genes. Plant. Mol. Biol. 2000, 42, 151–166. [Google Scholar] [CrossRef]
- Basile, A.; Fambrini, M.; Pugliesi, C. The vascular plants: Open system of growth. Dev. Genes Evol. 2017, 227, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Coen, E.S.; Meyerowitz, E. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Fambrini, M.; Pugliesi, C. CYCLOIDEA 2 clade genes: Key players in the of floral symmetry, inflorescence architecture, and reproductive organ development. Plant Mol. Biol. Rep. 2017, 35, 20–36. [Google Scholar] [CrossRef]
- Brandt, R.; Cabedo, M.; Xie, Y.; Wenkel, S. Homeodomain in leucine-zipper proteins and their role in synchronizing growth and development with the environment. J. Integr. Plant Biol. 2014, 56, 518–526. [Google Scholar] [CrossRef]
- Costanzo, E.; Trehin, C.; Vandenbussche, M. The role of WOX genes in flower development. Ann. Bot. 2014, 114, 1545–1553. [Google Scholar] [CrossRef]
- Laux, T.; Mayer, K.F.X.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef]
- Jha, P.; Ochatt, S.J.; Kumar, V. WUSCHEL: A master regulator in plant growth signaling. Plant Cell Rep. 2020, 39, 431–444. [Google Scholar] [CrossRef]
- van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Wang, M.-M.; Liu, M.-M.; Ran, F.; Guo, P.-C.; Ke, Y.-Z.; Wu, Y.-W.; Wen, J.; Li, P.-F.; Li, J.-N.; Du, H. Global analysis of WOX transcription factor gene family in Brassica napus reveals their stress- and hormone-responsive patterns. Int. J. Mol. Sci. 2018, 19, 3470. [Google Scholar] [CrossRef]
- Gu, R.; Song, X.; Liu, X.; Yan, L.; Zhou, Z.; Zhang, X. Genome-wide analysis of CsWOX transcription factor gene family in cucumber (Cucumis sativus L.). Sci. Rep. 2020, 10, 6216. [Google Scholar] [CrossRef]
- Paponov, I.A.; Teale, W.; Lang, D.; Paponov, M.; Reski, R.; Rensing, S.A.; Palme, K. The evolution of nuclear auxin signalling. BMC Evol. Biol. 2009, 9, 126. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 2009, 21, 3493–3505. [Google Scholar] [CrossRef] [PubMed]
- Long, J.A.; Ohno, C.; Smith, Z.R.; Meyerowitz, E.M. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 2006, 312, 1520–1523. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hu, Y.; Dai, M.; Huang, L.; Zhou, D.X. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 2009, 21, 736–748. [Google Scholar] [CrossRef]
- Dai, M.; Hu, Y.; Zhao, Y.; Liu, H.; Zhou, D.X. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 2007, 144, 380–390. [Google Scholar] [CrossRef]
- Sarkar, A.K.; Luijten, M.; Miyashima, S.; Lenhard, M.; Hashimoto, T.; Nakajima, K.; Sheres, B.; Heidstra, R.; Laux, T. Conserved factors regulate signaling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007, 446, 811–814. [Google Scholar] [CrossRef]
- Ditengou, F.A.; Teale, W.D.; Kochersperger, P.; Flittner, K.A.; Kneuper, I.; van der Graaff, E.; Nziengi, K.; Pinosa, F.; Li, X.; Nitschke, R.; et al. Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 18818–18823. [Google Scholar] [CrossRef]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef]
- Kieffer, M.; Stern, Y.; Cook, H.; Clerici, E.; Maulbetsch, C.; Laux, T.; Davies, B. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 2006, 18, 560–573. [Google Scholar] [CrossRef] [PubMed]
- Nardmann, J.; Werr, W. The invention of WUS-like stem cell-promoting functions in plants predates leptosporangiate ferns. Plant Mol. Biol. 2012, 78, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Deyhle, F.; Sarkar, A.K.; Tucker, E.J.; Laux, T. WUSCHEL regulates cell differentiation during anther development. Dev. Biol. 2007, 302, 154–159. [Google Scholar] [CrossRef]
- Lenhard, M.; Bohnert, A.; Jurgens, G.; Laux, T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 2001, 105, 805–814. [Google Scholar] [CrossRef]
- Salvini, M.; Fambrini, M.; Giorgetti, L.; Pugliesi, C. Molecular aspects of zygotic embryogenesis in sunflower (Helianthus annuus L.): Correlation of positive histone marks with HaWUS expression and putative link HaWUS/HaL1L. Planta 2016, 243, 199–215. [Google Scholar] [CrossRef]
- Salvini, M.; Sani, E.; Fambrini, M.; Pistelli, L.; Pucciariello, C.; Pugliesi, C. Molecular analysis of a sunflower gene encoding an homologous of the B subunit of a CAAT binding factor. Mol. Biol. Rep. 2012, 39, 6449–6465. [Google Scholar] [CrossRef]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Briere, C.; Owens, G.L.; Carrere, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef]
- Tang, Y.; Li, H.; Guan, Y.; Li, S.; Xun, C.; Dong, Y.; Huo, R.; Guo, Y.; Bao, X.; Pei, E.; et al. Genome-wide identification of the physic nut WUSCHEL-related homeobox gene family and functional analysis of the abiotic stress responsive gene JcWOX5. Front. Genet. 2020, 11, 670. [Google Scholar] [CrossRef]
- Zhang, X.; Zong, J.; Liu, J.; Yin, J.; Zhang, D. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef]
- Vandenbussche, M.; Horstman, A.; Zethof, J.; Koes, R.; Rijpkema, A.S.; Gerats, T. Differential recruitment of WOX transcription factors for lateral development and organ fusion in petunia and Arabidopsis. Plant Cell 2009, 21, 2269–2283. [Google Scholar] [CrossRef]
- Lin, H.; Niu, L.; McHale, N.A.; Ohme-Takagi, M.; Mysore, K.S.; Tadege, M. Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Plant Signal. Behav. 2013, 110, 366–371. [Google Scholar] [CrossRef]
- Li, M.; Wang, R.; Liu, Z.; Wu, X.; Wang, J. Genome-wide identification and analysis of the WUSCHEL-related homeobox (WOX) gene family in allotetraploid Brassica napus reveals changes in WOX genes during polyploidization. BMC Genomics 2019, 20, 317. [Google Scholar] [CrossRef]
- Wang, Z.; Cai, Q.; Xia, H.; Han, B.; Li, M.; Wang, Y.; Zhu, M.; Jiao, C.; Wang, D.; Zhu, J.; et al. Genome-wide identification and comparative analysis of WOX genes in four Euphorbiaceae species and their expression patterns in Jatropha curcas. Front. Genet. 2022, 13, 878554. [Google Scholar] [CrossRef]
- Yang, Z.; Gong, Q.; Qin, W.; Yang, Z.; Cheng, Y.; Lu, L.; Ge, X.; Zhang, C.; Wu, Z.; Li, F. Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 2017, 17, 113. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hamyat, M.; Liu, C.; Salman, A.; Gao, X.; Guo, C.; Wang, Y.; Guo, Y. Identification and characterization of the WOX family genes in five Solanaceae species reveal their conserved roles in peptide signaling. Genes 2018, 9, 260. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Guo, Y.; Zheng, Y.; Sun, Y.; Yang, J.; Ye, N. Genome-wide identification of WOX genes and their expression patterns under different hormone and abiotic stress treatments in tea plant (Camellia sinensis). Trees 2019, 33, 1129–1142. [Google Scholar] [CrossRef]
- Hao, Q.; Zhang, L.; Yang, Y.; Shan, Z.; Zhou, X.-a. Genome-wide analysis of the WOX gene family and function exploration of GmWOX18 in soybean. Plants 2019, 8, 215. [Google Scholar] [CrossRef]
- Alvarez, J.M.; Bueno, N.; Cañas, R.A.; Avila, C.; Cánovas, F.; Ordás, R.J. Analysis of the WUSCHEL-RELATED HOMEOBOX gene family in Pinus pinaster: New insights into the gene family evolution. Plant Physiol. Biochem. 2018, 123, 304–318. [Google Scholar] [CrossRef]
- Gambino, G.; Minuto, M.; Boccacci, P.; Perrone, I.; Vallania, R.; Gribuaudo, I. Characterization of expression dynamics of WOX homeodomain transcription factors during somatic embryogenesis in Vitis vinifera. J. Exp. Bot. 2011, 62, 1089–1101. [Google Scholar] [CrossRef]
- Palovaara, J.; Hakman, I. Conifer WOX-related homeodomain transcription factors, developmental consideration and expression dynamic of WOX2 during Picea abies somatic embryogenesis. Plant Mol. Biol. 2008, 66, 533–549. [Google Scholar] [CrossRef]
- Wang, X.; Bi, C.; Wang, C.; Ye, Q.; Yin, T.; Ye, N. Genome-wide identification and characterization of WUSCHEL-Related Homeobox (WOX) genes in Salix Suchowensis. J. For. Res. 2018, 30, 1811–1822. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Meng, D.; Li, G.; Li, D.; Abdullah, M.; Jin, Q.; Lin, Y.; Cai, Y. Genome-wide analysis suggests the relaxed purifying selection affect the evolution of WOX genes in Pyrus bretschneideri, Prunus persica, Prunus mume, and Fragaria vesca. Front. Genet. 2017, 8, 78. [Google Scholar] [CrossRef]
- Lian, G.; Ding, Z.; Wang, Q.; Zhang, D.; Xu, J. Origins and Evolution of WUSCHEL-related homeobox protein family in plant kingdom. Sci. World J. 2014, 2014, 534140. [Google Scholar] [CrossRef]
- Deveaux, Y.; Toffano-Nioche, C.; Claisse, G.; Thareau, V.; Morin, H.; Laufs, P.; Moreau, H.; Kreis, M.; Lecharny, A. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol. Biol. 2008, 8, 291. [Google Scholar] [CrossRef]
- Almoguera, C.; Rojas, A.; Díaz-Martín, J.; Prieto-Dapena, P.; Carranco, R.; Juan Jordano, J. A seed-specific heat-shock transcription factor involved in developmental regulation during embryogenesis in sunflower. J. Biol. Chem. 2002, 277, 43866–43872. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, Y.; Li, G.; Tang, Y.; Kramer, E.M.; Tadege, M. STENOFOLIA recruits TOPLESS to repress ASYMMETRIC LEAVES2 at the leaf margin and promote leaf blade outgrowth in Medicago truncatula. Plant Cell 2014, 26, 650–664. [Google Scholar] [CrossRef]
- Pi, L.; Aichinger, E.; van der Graaff, E.; Llavata-Peris, C.I.; Weijers, D.; Hennig, L.; Groot, E.; Laux, T. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell 2015, 33, 576–588. [Google Scholar] [CrossRef]
- Dolzblasz, A.; Nardmann, J.; Clerici, E.; Causier, B.; van der Graaff, E.; Chen, J.; Davies, B.; Werr, W.; Laux, T. Stem cell regulation by Arabidopsis WOX genes. Mol. Plant 2016, 9, 1028–1039. [Google Scholar] [CrossRef]
- Ohta, M.; Matsui, K.; Hiratsu, K.; Shinshi, H.; Ohme-Takagi, M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 2001, 13, 1959–1968. [Google Scholar] [CrossRef]
- Ma, J.; Ptashne, M. A new class of yeast transcriptional activators. Cell 1987, 51, 113–119. [Google Scholar] [CrossRef]
- Romera-Branchat, M.; Ripoll, J.J.; Yanofsky, M.F.; Pelaz, S. The WOX13 homeobox gene promotes replum formation in the Arabidopsis thaliana fruit. Plant J. 2012, 73, 37–49. [Google Scholar] [CrossRef]
- Tang, F.; Chen, N.; Zhao, M.; Wang, Y.; He, R.; Peng, X.; Shen, S. Identification and functional divergence analysis of WOX gene family in paper mulberry. Int. J. Mol. Sci. 2017, 18, 1782. [Google Scholar] [CrossRef]
- Lieber, D.; Lora, J.; Schrempp, S.; Lenhard, M.; Laux, T. Arabidopsis WIH1 and WIH2 genes act in the transition from somatic to reproductive cell fate. Curr. Biol. 2011, 21, 1009–1017. [Google Scholar] [CrossRef]
- Le Page-Degivry, M.T.; Garello, G. In situ abscisic acid synthesis: A requirement for induction of embryo dormancy in Helianthus annuus. Plant Physiol. 1992, 98, 1386–1390. [Google Scholar] [CrossRef] [PubMed]
- Pugliesi, C.; Fambrini, M.; Vernieri, P.; Baroncelli, S. Characterization of a wilty sunflower (Helianthus annuus L.) mutant I. Abscisic acid content, light-dark changes in the stomatal conductance and genetic analysis. J. Exp. Bot. 1994, 45, 533–538. [Google Scholar] [CrossRef]
- Giordani, T.; Natali, L.; D’Ercole, A.; Pugliesi, C.; Fambrini, M.; Vernieri, P.; Vitagliano, C.; Cavallini, A. Expression of a dehydrin gene during embryo development and drought stress in ABA-deficient mutants of sunflower (Helianthus annuus L.). Plant Mol. Biol. 1999, 39, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Mantese, A.I.; Medan, D.; Hall, A.J. Achene structure, development and lipid accumulation in sunflower cultivars differing in oil content at maturity. Ann. Bot. 2006, 97, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Haecker, A.; Gross-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef]
- Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Birney, E.; Biswas, M.; Bucher, P.; Cerutti, L.; Corpet, F.; Croning, M.D.R. The InterPro database, an integrated documentation resource for protein families, domain and functional sites. Nucleic Acid Res. 2001, 29, 37–40. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bailey, T.L.; Bodén, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Hao, Z.; Lv, D.; Ge, Y.; Shi, J.; Weijers, D.; Yu, G.; Chen, J. RIdeogram: Drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput. Sci. 2020, 6, 251. [Google Scholar] [CrossRef]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609–W612. [Google Scholar] [CrossRef]
- Fambrini, M.; Bellanca, M.; Costa Muñoz, M.; Usai, G.; Cavallini, A.; Pugliesi, C. Ligulate inflorescence of Helianthus × multiflorus, cv. Soleil d’Or, correlates with a mis-regulation of a CYCLOIDEA gene characterised by insertion of a transposable element. Plant Biol. 2018, 20, 956–967. [Google Scholar] [CrossRef]
- Tähtiharju, S.; Rijpkema, A.S.; Vetterli, A.; Albert, V.A.; Teeri, T.H.; Elomaa, P. Evolution and diversification of the CYC/TB1 gene family in Asteraceae—A comparative study in gerbera (Mutisieae) and sunflower (Heliantheae). Mol. Biol. Evol. 2012, 29, 1155–1166. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2-∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riccucci, E.; Vanni, C.; Vangelisti, A.; Fambrini, M.; Giordani, T.; Cavallini, A.; Mascagni, F.; Pugliesi, C. Genome-Wide Analysis of WOX Multigene Family in Sunflower (Helianthus annuus L.). Int. J. Mol. Sci. 2023, 24, 3352. https://doi.org/10.3390/ijms24043352
Riccucci E, Vanni C, Vangelisti A, Fambrini M, Giordani T, Cavallini A, Mascagni F, Pugliesi C. Genome-Wide Analysis of WOX Multigene Family in Sunflower (Helianthus annuus L.). International Journal of Molecular Sciences. 2023; 24(4):3352. https://doi.org/10.3390/ijms24043352
Chicago/Turabian StyleRiccucci, Ettore, Cosimo Vanni, Alberto Vangelisti, Marco Fambrini, Tommaso Giordani, Andrea Cavallini, Flavia Mascagni, and Claudio Pugliesi. 2023. "Genome-Wide Analysis of WOX Multigene Family in Sunflower (Helianthus annuus L.)" International Journal of Molecular Sciences 24, no. 4: 3352. https://doi.org/10.3390/ijms24043352
APA StyleRiccucci, E., Vanni, C., Vangelisti, A., Fambrini, M., Giordani, T., Cavallini, A., Mascagni, F., & Pugliesi, C. (2023). Genome-Wide Analysis of WOX Multigene Family in Sunflower (Helianthus annuus L.). International Journal of Molecular Sciences, 24(4), 3352. https://doi.org/10.3390/ijms24043352





