Nanoscale Iron-Based Metal–Organic Frameworks: Incorporation of Functionalized Drugs and Degradation in Biological Media
Abstract
:1. Introduction
2. Results and Discussion
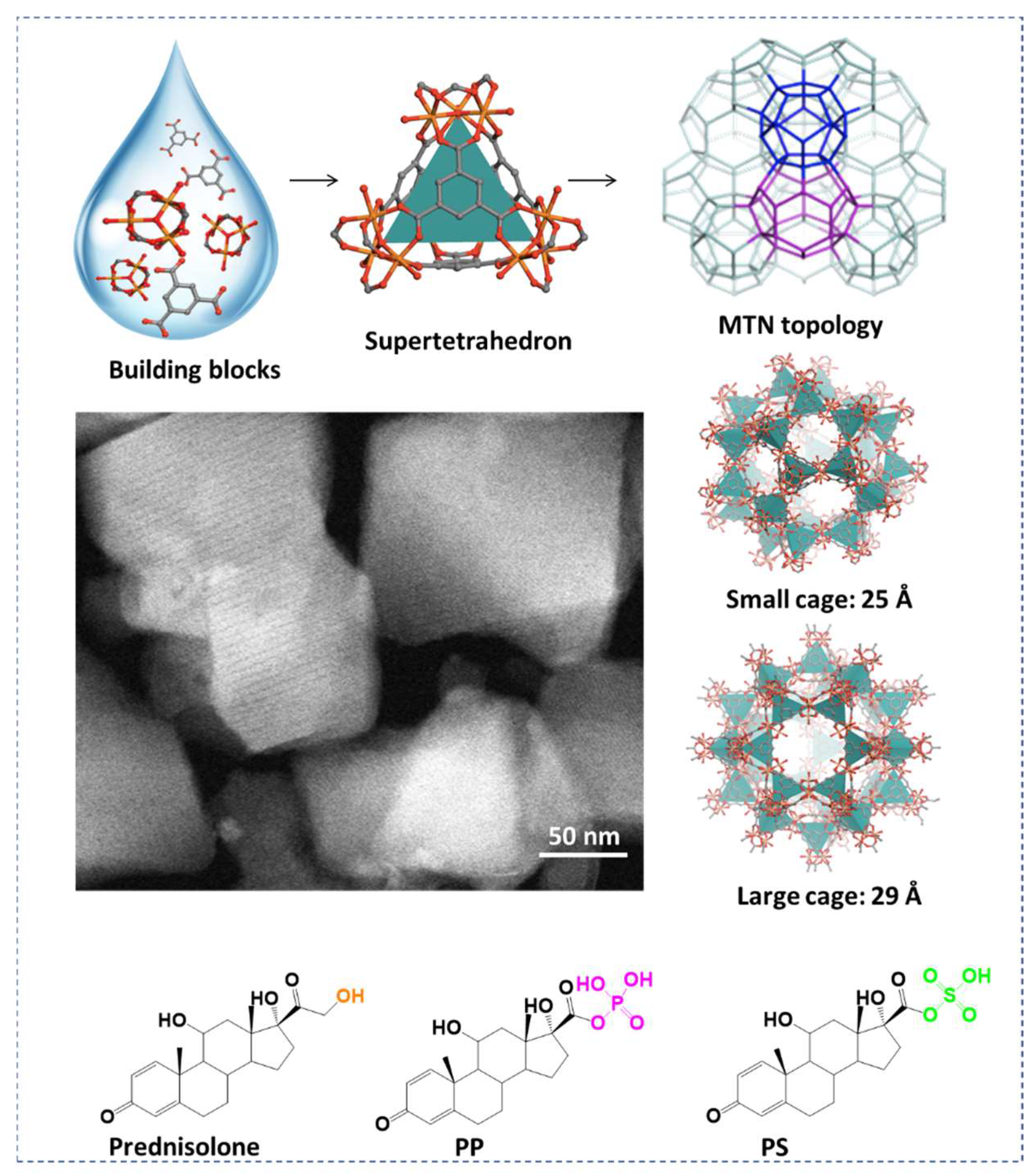
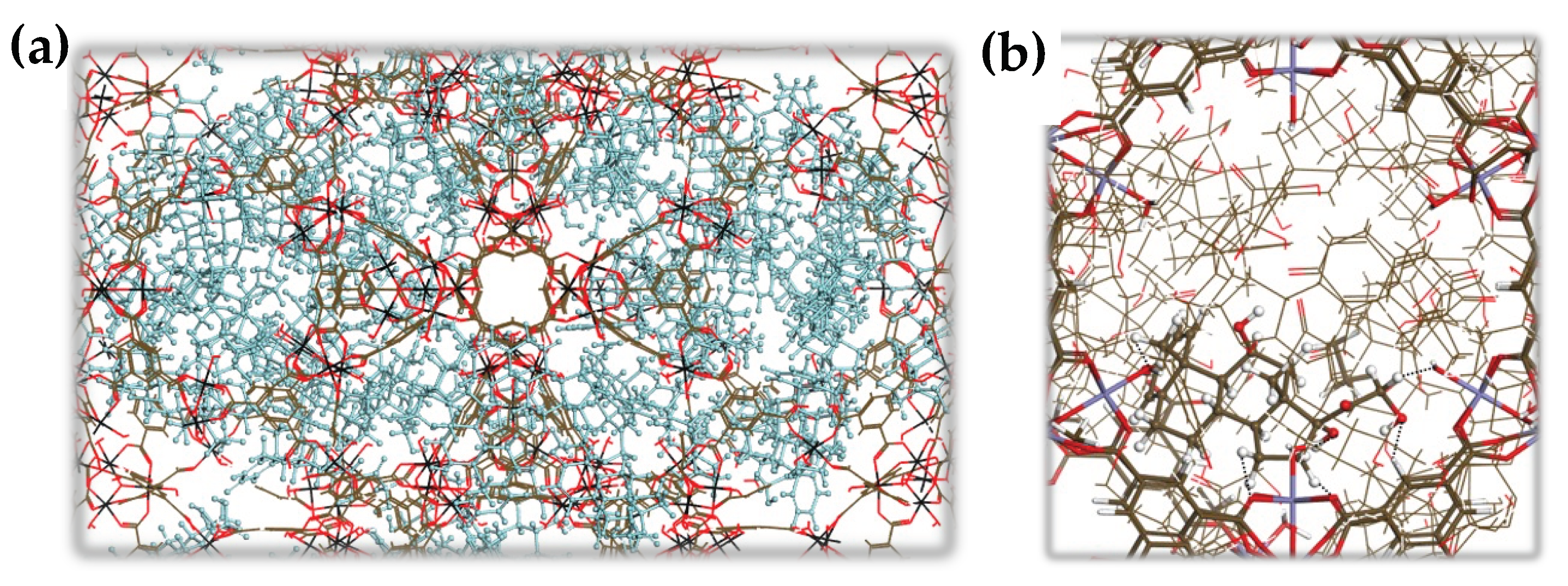
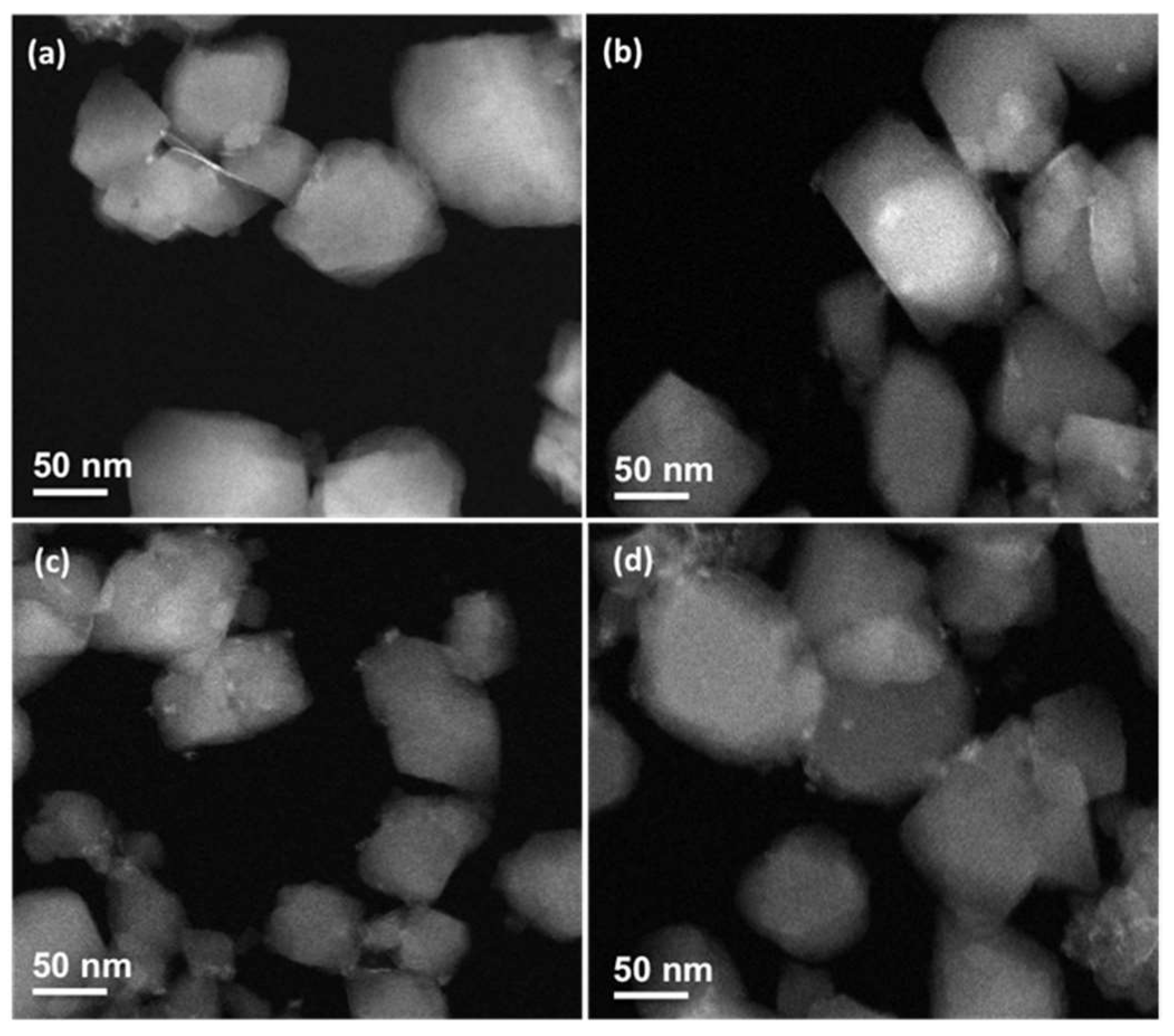
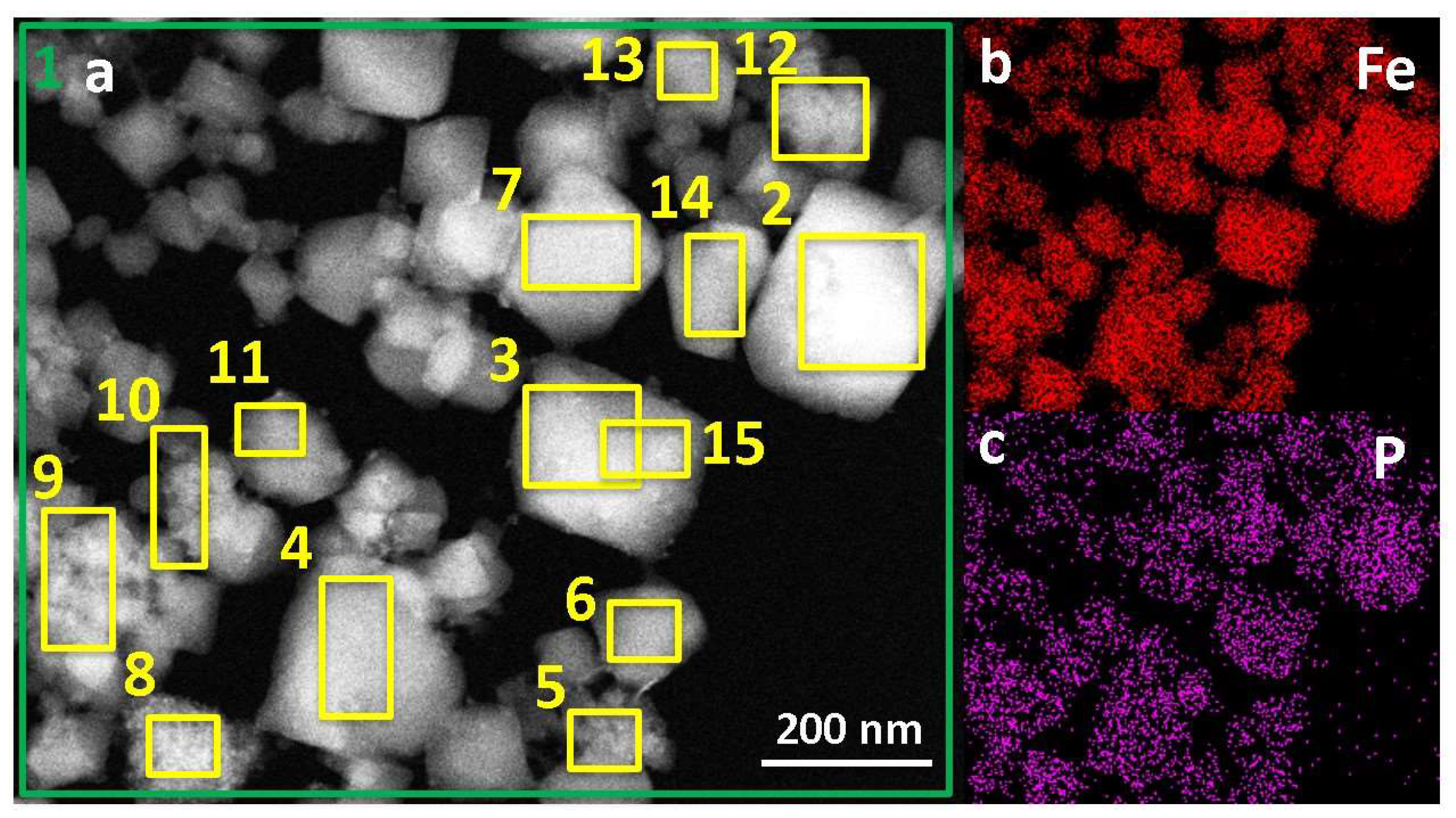
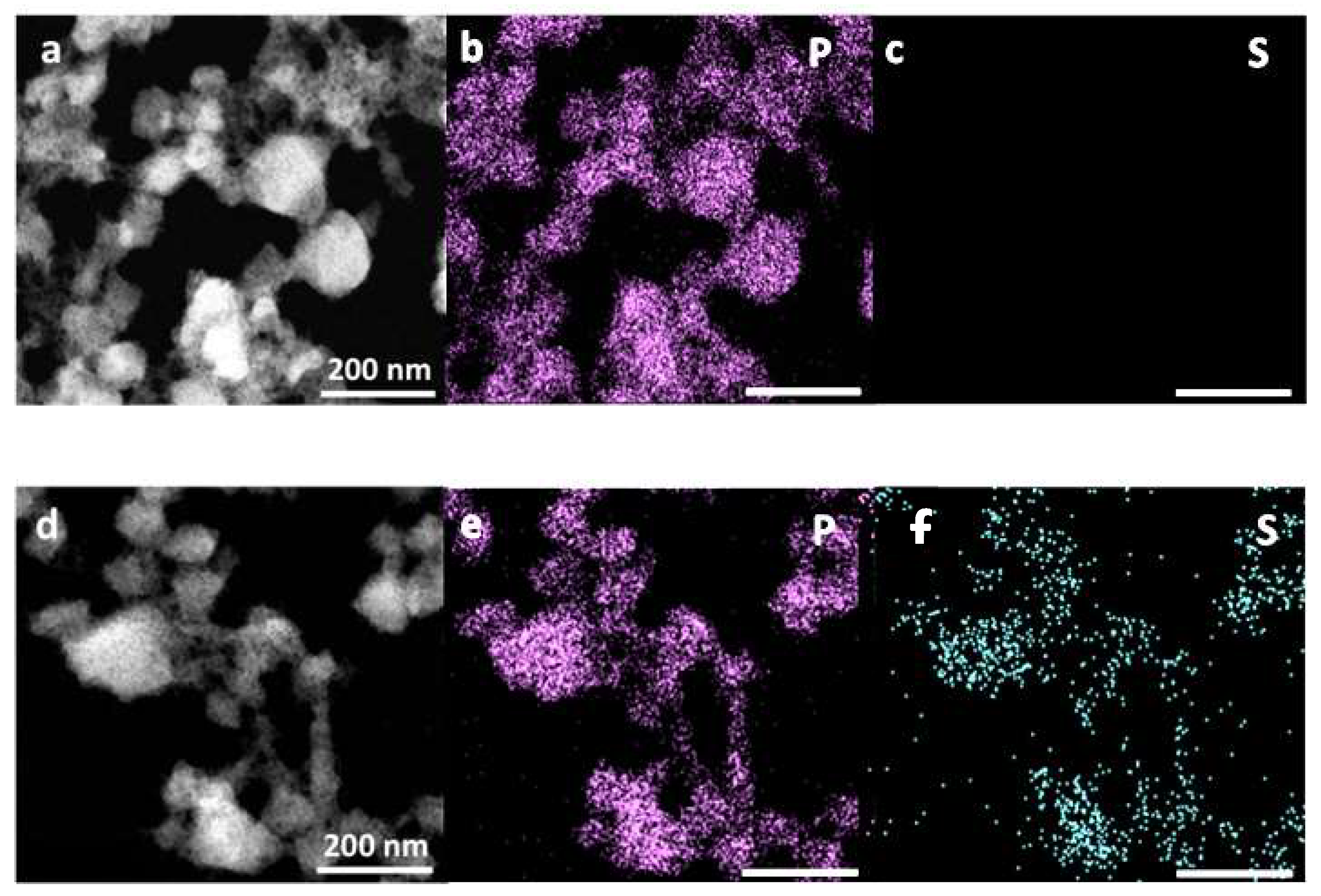
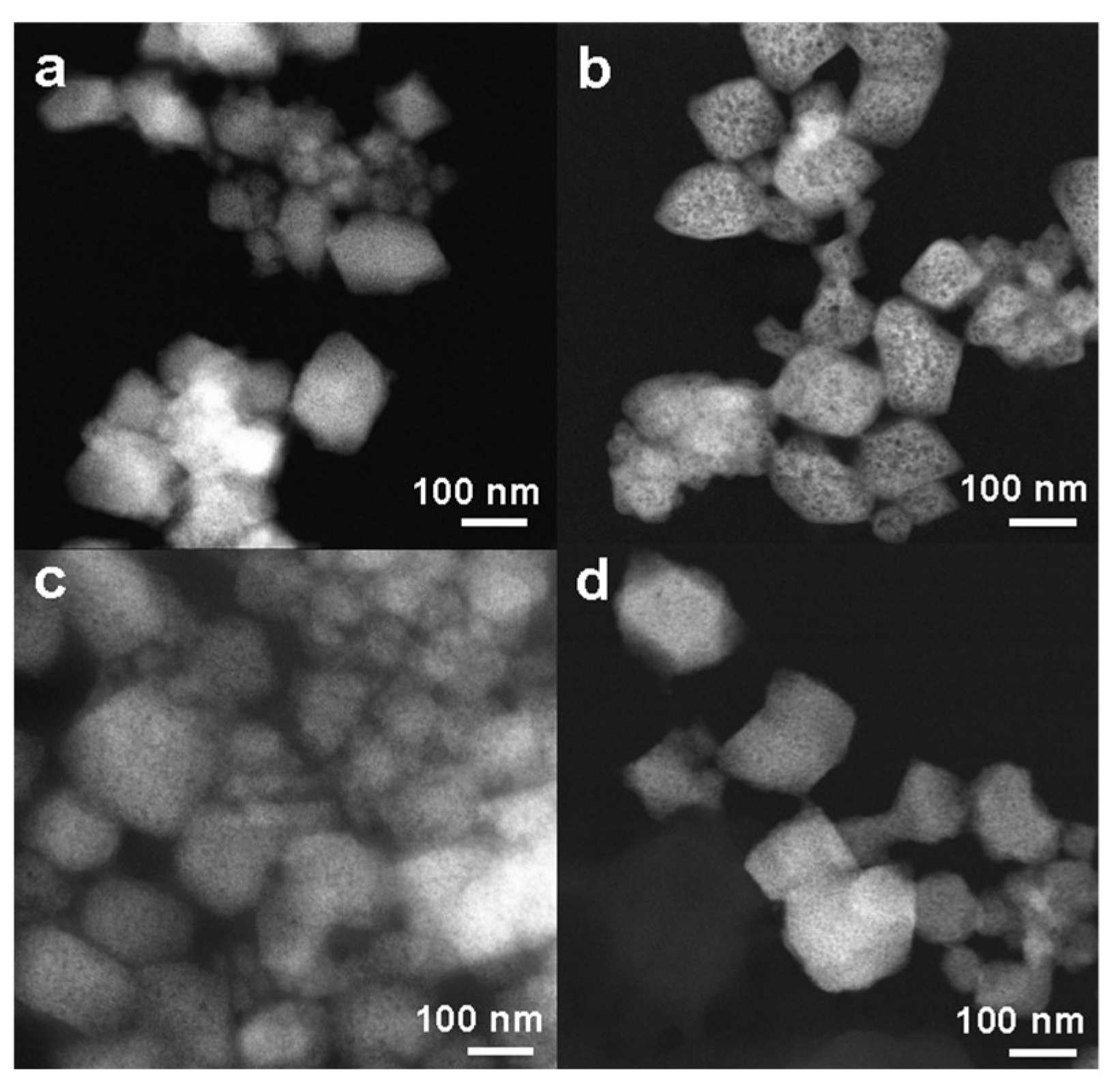
2.1. Synthesis and Characterization
2.2. Degradation Study of Empty nanoMOFs
2.3. Comparative Study of Release Kinetics in Different Media
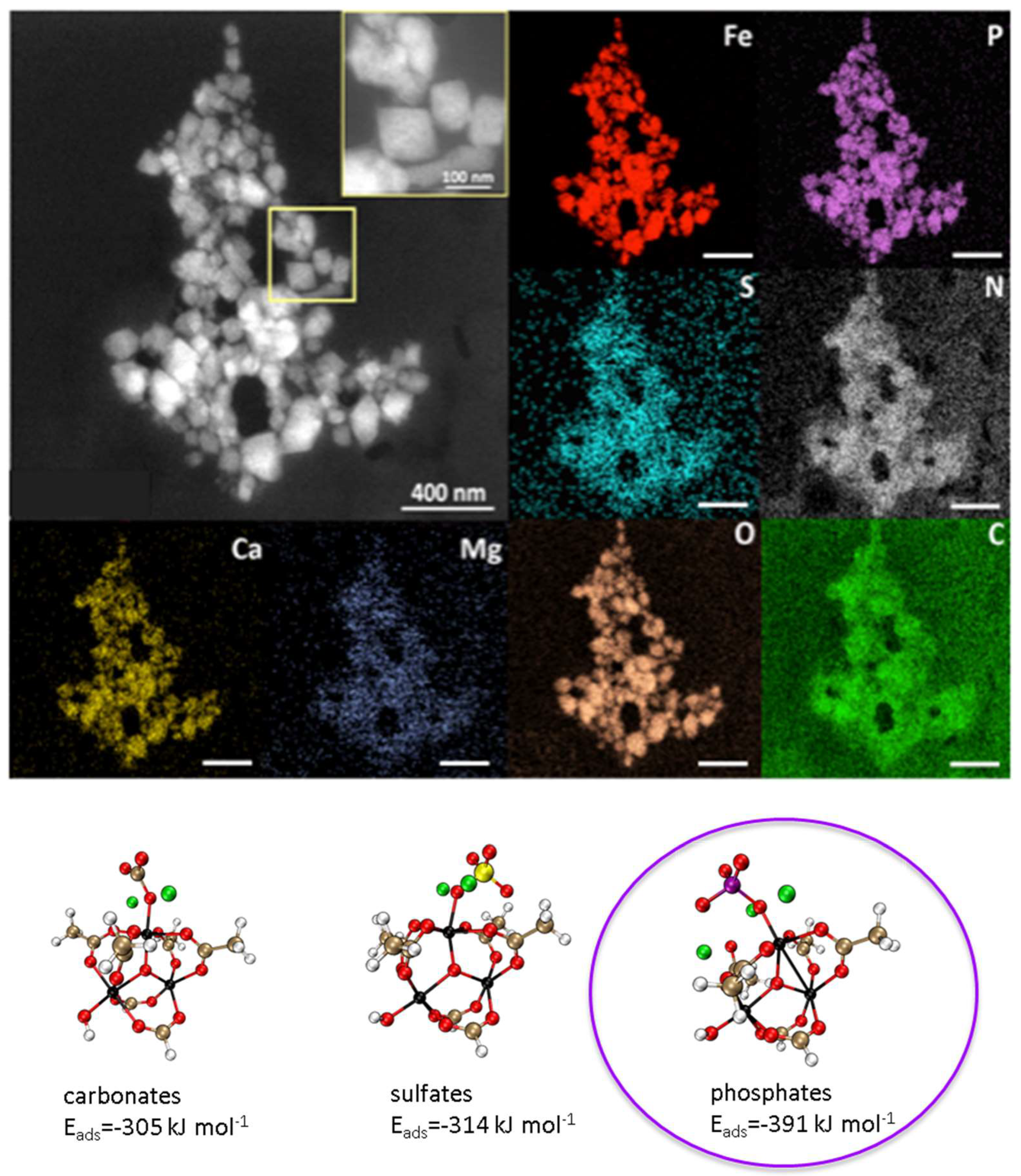
2.4. Degradation Study in Blood and Serum
3. Materials and Methods
3.1. Materials
3.2. Synthesis of nanoMOFs
3.3. Characterization of nanoMOFs
3.4. Drug Encapsulation
3.5. Drug and Ligand Release
3.6. HPLC Measurement Conditions
3.6.1. Quantification of Prednisolone
3.6.2. Quantification of PP
3.6.3. Quantification of PS
3.6.4. Quantification of Trimesate Ligand Release
3.7. Computational Section
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering Precision Nanoparticles for Drug Delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar]
- Bangham, A.D.; Glover, J.C.; Hollingshead, S.; Pethica, B.A. The Surface Properties of Some Neoplastic Cells. Biochem. J. 1962, 84, 513–517. [Google Scholar]
- Horcajada, P.; Chalati, T.; Serre, C.; Gillet, B.; Sebrie, C.; Baati, T.; Eubank, J.F.; Heurtaux, D.; Clayette, P.; Kreuz, C.; et al. Porous Metal-Organic-Framework Nanoscale Carriers as a Potential Platform for Drug Deliveryand Imaging. Nat. Mater. 2010, 9, 172–178. [Google Scholar] [PubMed]
- Li, S.; Tan, L.; Meng, X. Nanoscale Metal-Organic Frameworks: Synthesis, Biocompatibility, Imaging Applications, and Thermal and Dynamic Therapy of Tumors. Adv. Funct. Mater. 2020, 30, 1908924. [Google Scholar] [CrossRef]
- Rodriguez-Ruiz, V.; Maksimenko, A.; Anand, R.; Monti, S.; Agostoni, V.; Couvreur, P.; Lampropoulou, M.; Yannakopoulou, K.; Gref, R. Efficient “Green” Encapsulation of a Highly Hydrophilic Anticancer Drug in Metal-Organic Framework Nanoparticles. J. Drug Target. 2015, 23, 759–767. [Google Scholar] [PubMed]
- Di Nunzio, M.R.; Agostoni, V.; Cohen, B.; Gref, R.; Douhal, A. A “Ship in a Bottle” Strategy to Load a Hydrophilic Anticancer Drug in Porous Metal Organic Framework Nanoparticles: Efficient Encapsulation, Matrix Stabilization, and Photodelivery. J. Med. Chem. 2014, 57, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Agostoni, V.; Horcajada, P.; Rodriguez-Ruiz, V.; Willaime, H.; Couvreur, P.; Serre, C.; Gref, R. ‘Green’ Fluorine-Free Mesoporous Iron(III) Trimesate Nanoparticles for Drug Delivery. Green Mater. 2013, 1, 209–217. [Google Scholar] [CrossRef]
- Anand, R.; Borghi, F.; Manoli, F.; Manet, I.; Agostoni, V.; Reschiglian, P.; Gref, R.; Monti, S. Host-Guest Interactions in Fe(III)-Trimesate MOF Nanoparticles Loaded with Doxorubicin. J. Phys. Chem. B 2014, 118, 8532–8539. [Google Scholar] [CrossRef]
- Ding, M.; Liu, W.; Gref, R. Nanoscale MOFs: From Synthesis to Drug Delivery and Theranostics Applications. Adv. Drug Deliv. Rev. 2022, 190, 114496. [Google Scholar] [PubMed]
- Grall, R.; Hidalgo, T.; Delic, J.; Garcia-Marquez, A.; Chevillard, S.; Horcajada, P. In Vitro Biocompatibility of Mesoporous Metal (III; Fe, Al, Cr) Trimesate MOF Nanocarriers. J. Mater. Chem. B 2015, 3, 8279–8292. [Google Scholar]
- Wuttke, S.; Zimpel, A.; Bein, T.; Braig, S.; Stoiber, K.; Vollmar, A.; Müller, D.; Haastert-Talini, K.; Schaeske, J.; Stiesch, M.; et al. Validating Metal-Organic Framework Nanoparticles for Their Nanosafety in Diverse Biomedical Applications. Adv. Healthc. Mater. 2017, 6, 1600818. [Google Scholar]
- Ruyra, A.; Yazdi, A.; Espín, J.; Carné-Sánchez, A.; Roher, N.; Lorenzo, J.; Imaz, I.; Maspoch, D. Synthesis, Culture Medium Stability, and in Vitro and in Vivo Zebrafish Embryo Toxicity of Metal-Organic Framework Nanoparticles. Chem.-A Eur. J. 2015, 21, 2508–2518. [Google Scholar] [CrossRef] [PubMed]
- Simon-Yarza, M.T.; Baati, T.; Paci, A.; Lesueur, L.L.; Seck, A.; Chiper, M.; Gref, R.; Serre, C.; Couvreur, P.; Horcajada, P. Antineoplastic Busulfan Encapsulated in a Metal Organic Framework Nanocarrier: First in Vivo Results. J. Mater. Chem. B 2016, 4, 585–588. [Google Scholar] [PubMed]
- Baati, T.; Njim, L.; Neffati, F.; Kerkeni, A.; Bouttemi, M.; Gref, R.; Najjar, M.F.; Zakhama, A.; Couvreur, P.; Serre, C.; et al. In Depth Analysis of the in Vivo Toxicity of Nanoparticles of Porous Iron(Iii) Metal–Organic Frameworks. Chem. Sci. 2013, 4, 1597–1607. [Google Scholar] [CrossRef]
- Simon-Yarza, T.; Giménez-Marqués, M.; Mrimi, R.; Mielcarek, A.; Gref, R.; Horcajada, P.; Serre, C.; Couvreur, P. A Smart Metal–Organic Framework Nanomaterial for Lung Targeting. Angew. Chem.-Int. Ed. 2017, 56, 15565–15569. [Google Scholar]
- Tamames-Tabar, C.; Cunha, D.; Imbuluzqueta, E.; Ragon, F.; Serre, C.; Blanco-Prieto, M.J.; Horcajada, P. Cytotoxicity of Nanoscaled Metal-Organic Frameworks. J. Mater. Chem. B 2014, 2, 262–271. [Google Scholar] [CrossRef]
- Ettlinger, R.; Lä, U.; Gref, R.; Horcajada, P.; Lammers, T.; Serre, C.; Couvreur, P.; Morris, R.E.; Wuttke, S. Toxicity of Metal-Organic Framework Nanoparticles: From Essential Analyses to Potential Applications. Chem. Soc. Rev. 2022, 464, 464. [Google Scholar] [CrossRef]
- Li, X.; Lachmanski, L.; Safi, S.; Sene, S.; Serre, C.; Grenèche, J.M.; Zhang, J.; Gref, R. New Insights into the Degradation Mechanism of Metal-Organic Frameworks Drug Carriers. Sci. Rep. 2017, 7, 13142. [Google Scholar] [CrossRef]
- Bellido, E.; Guillevic, M.; Hidalgo, T.; Santander-Ortega, M.J.; Serre, C.; Horcajada, P. Understanding the Colloidal Stability of the Mesoporous MIL-100(Fe) Nanoparticles in Physiological Media. Langmuir 2014, 30, 5911–5920. [Google Scholar] [CrossRef]
- Christodoulou, I.; Serre, C.; Gref, R. Metal-organic frameworks for drug delivery: Degradation mechanism and in vivo fate. In Metal-Organic Frameworks for Biomedical Applications, 1st ed.; Woodhead Publishing: Sawston, UK, 2020; ISBN 978-01-2816-985-8. [Google Scholar]
- Agostoni, V.; Chalati, T.; Horcajada, P.; Willaime, H.; Anand, R.; Semiramoth, N.; Baati, T.; Hall, S.; Maurin, G.; Chacun, H.; et al. Towards an Improved Anti-HIV Activity of NRTI via Metal-Organic Frameworks Nanoparticles. Adv. Healthc. Mater. 2013, 2, 1630–1637. [Google Scholar]
- Agostoni, V.; Anand, R.; Monti, S.; Hall, S.; Maurin, G.; Horcajada, P.; Serre, C.; Bouchemal, K.; Gref, R. Impact of Phosphorylation on the Encapsulation of Nucleoside Analogues within Porous Iron(Iii) Metal-Organic Framework MIL-100(Fe) Nanoparticles. J. Mater. Chem. B 2013, 1, 4231–4242. [Google Scholar] [CrossRef] [PubMed]
- Roda, B.; Marassi, V.; Zattoni, A.; Borghi, F.; Anand, R.; Agostoni, V.; Gref, R.; Reschiglian, P.; Monti, S. Flow Field-Flow Fractionation and Multi-Angle Light Scattering as a Powerful Tool for the Characterization and Stability Evaluation of Drug-Loaded Metal–Organic Framework Nanoparticles. Anal. Bioanal. Chem. 2018, 410, 5245–5253. [Google Scholar] [CrossRef] [PubMed]
- Granner, D.K.; Wang, J.C.; Yamamoto, K.R. Regulatory Actions of Glucocorticoid Hormones: From Organisms to Mechanisms. Adv. Exp. Med. Biol. 2015, 872, 3–31. [Google Scholar] [PubMed]
- Hua, C.; Buttgereit, F.; Combe, B. Glucocorticoids in Rheumatoid Arthritis: Current Status and Future Studies. RMD Open 2020, 6, e000536. [Google Scholar] [PubMed]
- Kearns, N.; Maijers, I.; Harper, J.; Beasley, R.; Weatherall, M. Inhaled Corticosteroids in Acute Asthma: A Systemic Review and Meta-Analysis. J. Allergy Clin. Immunol. Pract. 2020, 8, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Fizazi, K.; Tran, N.; Fein, L.; Matsubara, N.; Rodriguez-Antolin, A.; Alekseev, B.Y.; Özgüroğlu, M.; Ye, D.; Feyerabend, S.; Protheroe, A.; et al. Abiraterone plus Prednisone in Metastatic, Castration-Sensitive Prostate Cancer. N. Engl. J. Med. 2017, 377, 352–360. [Google Scholar]
- Fulmali, A.; Bharate, S.S. Phosphate Moiety in FDA-Approved Pharmaceutical Salts and Prodrugs. Drug Dev. Res. 2022, 83, 1059–1074. [Google Scholar]
- Shinde, G.; Shiyani, S.; Shelke, S.; Chouthe, R.; Kulkarni, D.; Marvaniya, K. Enhanced brain targeting efficiency using 5-FU (fluorouracil) lipid–drug conjugated nanoparticles in brain cancer therapy. Prog. Biomater. 2020, 9, 259–275. [Google Scholar] [PubMed]
- Pagar, R.R.; Musale, S.R.; Pawar, G.; Kulkarni, D.; Giram, P.S. Comprehensive Review on the Degradation Chemistry and Toxicity Studies of Functional Materials. ACS Biomater. Sci. Eng. 2022, 8, 2161–2195. [Google Scholar] [CrossRef]
- Férey, G.; Serre, C.; Mellot-Draznieks, C.; Millange, F.; Surblé, S.; Dutour, J.; Margiolaki, I. A Hybrid Solid with Giant Pores Prepared by a Combination of Targeted Chemistry, Simulation, and Powder Diffraction. Angew. Chem.-Int. Ed. 2004, 43, 6296–6301. [Google Scholar]
- Horcajada, P.; Surblé, S.; Serre, C.; Hong, D.Y.; Seo, Y.K.; Chang, J.S.; Grenèche, J.M.; Margiolaki, I.; Férey, G. Synthesis and Catalytic Properties of MIL-100(Fe), an Iron(III) Carboxylate with Large Pores. Chem. Commun. 2007, 100, 2820–2822. [Google Scholar]
- Horcajada, P.; Serre, C.; Vallet-Regí, M.; Sebban, M.; Taulelle, F.; Férey, G. Metal-Organic Frameworks as Efficient Materials for Drug Delivery. Angew. Chem.-Int. Ed. 2006, 45, 5974–5978. [Google Scholar] [CrossRef] [PubMed]
- Sene, S.; Marcos-Almaraz, M.T.; Menguy, N.; Scola, J.; Volatron, J.; Rouland, R.; Grenèche, J.M.; Miraux, S.; Menet, C.; Guillou, N.; et al. Maghemite-NanoMIL-100(Fe) Bimodal Nanovector as a Platform for Image-Guided Therapy. Chem 2017, 3, 303–322. [Google Scholar]
- García Márquez, A.; Demessence, A.; Platero-Prats, A.E.; Heurtaux, D.; Horcajada, P.; Serre, C.; Chang, J.S.; Férey, G.; De La Peña-O’Shea, V.A.; Boissière, C.; et al. Green Microwave Synthesis of MIL-100(Al, Cr, Fe) Nanoparticles for Thin-Film Elaboration. Eur. J. Inorg. Chem. 2012, 100, 5165–5174. [Google Scholar]
- Li, X.; Porcel, E.; Menendez-Miranda, M.; Qiu, J.; Yang, X.; Serre, C.; Pastor, A.; Desmaële, D.; Lacombe, S.; Gref, R. Highly Porous Hybrid Metal–Organic Nanoparticles Loaded with Gemcitabine Monophosphate: A Multimodal Approach to Improve Chemo- and Radiotherapy. ChemMedChem 2020, 15, 274–283. [Google Scholar]
- Inês Severino, M.; Al Mohtar, A.; Vieira Soares, C.; Freitas, C.; Sadovnik, N.; Nandi, S.; Mouchaham, G.; Pimenta, V.; Nouar, F.; Daturi, M.; et al. Metal-Organic Frameworks Hot Paper MOFs with Open Metal(III) Sites for the Environmental Capture of Polar Volatile Organic Compounds. Angew. Chem. Int. Ed. 2022, 62, e202211583. [Google Scholar]
- Bayard, F.J.C.; Thielemans, W.; Pritchard, D.I.; Paine, S.W.; Young, S.S.; Bäckman, P.; Ewing, P.; Bosquillon, C. Polyethylene Glycol-Drug Ester Conjugates for Prolonged Retention of Small Inhaled Drugs in the Lung. J. Control. Release 2013, 171, 234–240. [Google Scholar] [CrossRef]
- Huanbutta, K.; Sangnim, T.; Limmatvapirat, S.; Nunthanid, J.; Sriamornsak, P. Design and Characterization of Prednisolone-Loaded Nanoparticles Fabricated by Electrohydrodynamic Atomization Technique. Chem. Eng. Res. Des. 2016, 109, 816–823. [Google Scholar]
- Rojas, S.; Colinet, I.; Cunha, D.; Hidalgo, T.; Salles, F.; Serre, C.; Guillou, N.; Horcajada, P. Toward Understanding Drug Incorporation and Delivery from Biocompatible Metal-Organic Frameworks in View of Cutaneous Administration. ACS Omega 2018, 3, 2994–3003. [Google Scholar]
- Cunha, D.; Ben Yahia, M.; Hall, S.; Miller, S.R.; Chevreau, H.; Elkaïm, E.; Maurin, G.; Horcajada, P.; Serre, C. Rationale of Drug Encapsulation and Release from Biocompatible Porous Metal-Organic Frameworks. Chem. Mater. 2013, 25, 2767–2776. [Google Scholar] [CrossRef]
- Khoe, G.H.; Robins, R.G. The complexation of iron(III) with sulphate, phosphate, or arsenate ion in sodium nitrate medium at 25 °C. J. Chem. Soc. Dalton Trans. 1988, 8, 2015–2021. [Google Scholar]
- Nezafati, N.; Moztarzadeh, F.; Hesaraki, S. Surface Reactivity and in Vitro Biological Evaluation of Sol Gel Derived Silver/Calcium Silicophosphate Bioactive Glass. Biotechnol. Bioprocess Eng. 2012, 17, 746–754. [Google Scholar] [CrossRef]
- Anderson, N.L.; Anderson, N.G. The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Mol. Cell. Proteom. MCP 2002, 1, 845–867. [Google Scholar] [PubMed]
- Simon-Yarza, T.; Baati, T.; Neffati, F.; Njim, L.; Couvreur, P.; Serre, C.; Gref, R.; Najjar, M.F.; Zakhama, A.; Horcajada, P. In Vivo Behavior of MIL-100 Nanoparticles at Early Times after Intravenous Administration. Int. J. Pharm. 2016, 511, 1042–1047. [Google Scholar] [PubMed]
- Li, X.; Salzano, G.; Qiu, J.; Menard, M.; Berg, K.; Theodossiou, T.; Ladavière, C.; Gref, R. Drug-Loaded Lipid-Coated Hybrid Organic-Inorganic “Stealth” Nanoparticles for Cancer Therapy. Front. Bioeng. Biotechnol. 2020, 8, 1027. [Google Scholar]
- Cutrone, G.; Li, X.; Casas-Solvas, J.M.; Menendez-Miranda, M.; Qiu, J.; Benkovics, G.; Constantin, D.; Malanga, M.; Moreira-Alvarez, B.; Costa-Fernandez, J.M.; et al. Design of Engineered Cyclodextrin Derivatives for Spontaneous Coating of Highly Porous Metal-Organic Framework Nanoparticles in Aqueous Media. Nanomaterials 2019, 9, 1103. [Google Scholar]
- Finšgar, M.; Perva-Uzunalić, A.; Behr, H.; Ledinek, N.; Knez, Ž.; Novak, Z. An Improved Reversed-Phase High-Performance Liquid Chromatography Method for the Analysis of Related Substances of Prednisolone in Active Ingredient. ACS Omega 2020, 5, 7987–8000. [Google Scholar] [CrossRef]
- Abdullah, N.; Karamat, F.; Qamar, S.; Abbass, M.; Khan, A.M.; Ullah, N. Development and Validation of RP-HPLC Method for Simultaneous Quantification of Sulfacetamide Sodium and Prednisolone Sodium Phosphate. Acta Pol. Pharm.-Drug Res. 2019, 76, 37–47. [Google Scholar]
- Schäfer, A.; Huber, C.; Ahlrichs, R. Fully Optimized Contracted Gaussian Basis Sets of Triple Zeta Valence Quality for Atoms Li to Kr. J. Chem. Phys. 1994, 100, 5829–5835. [Google Scholar]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A Consistent and Accurate Ab Initio Parametrization of Density Functional Dispersion Correction (DFT-D) for the 94 Elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar]
- Rappé, A.K.; Casewit, C.J.; Colwell, K.S.; Goddard, W.A.; Skiff, W.M. UFF, a Full Periodic Table Force Field for Molecular Mechanics and Molecular Dynamics Simulations. J. Am. Chem. Soc. 1992, 114, 10024–10035. [Google Scholar] [CrossRef]
- Mayo, S.L.; Olafson, B.D.; Goddard, W.A. DREIDING: A Generic Force Field for Molecular Simulations. J. Phys. Chem. 1990, 94, 8897–8909. [Google Scholar]
- Loschwitz, J.; Jäckering, A.; Keutmann, M.; Olagunju, M.; Olubiyi, O.O.; Strodel, B. Dataset of AMBER Force Field Parameters of Drugs, Natural Products and Steroids for Simulations Using GROMACS. Data Brief 2021, 35, 106948. [Google Scholar] [CrossRef] [PubMed]
- Heinz, H.; Suter, U.W. Atomic Charges for Classical Simulations of Polar Systems. J. Phys. Chem. B 2004, 108, 18341–18352. [Google Scholar] [CrossRef]
- Martin, F.; Zipse, H. Charge Distribution in the Water Molecule -A Comparison of Methods. J. Comput. Chem. 2005, 26, 97–105. [Google Scholar]
- Lin, Z.S.; Chen, D.; Nie, H.Y.; Wong, Y.T.A.; Huang, Y. Investigations of the Formation of Zeolite ZSM-39 (MTN). Can. J. Chem. 2019, 97, 840–847. [Google Scholar] [CrossRef] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Christodoulou, I.; Lyu, P.; Soares, C.V.; Patriarche, G.; Serre, C.; Maurin, G.; Gref, R. Nanoscale Iron-Based Metal–Organic Frameworks: Incorporation of Functionalized Drugs and Degradation in Biological Media. Int. J. Mol. Sci. 2023, 24, 3362. https://doi.org/10.3390/ijms24043362
Christodoulou I, Lyu P, Soares CV, Patriarche G, Serre C, Maurin G, Gref R. Nanoscale Iron-Based Metal–Organic Frameworks: Incorporation of Functionalized Drugs and Degradation in Biological Media. International Journal of Molecular Sciences. 2023; 24(4):3362. https://doi.org/10.3390/ijms24043362
Chicago/Turabian StyleChristodoulou, Ioanna, Pengbo Lyu, Carla Vieira Soares, Gilles Patriarche, Christian Serre, Guillaume Maurin, and Ruxandra Gref. 2023. "Nanoscale Iron-Based Metal–Organic Frameworks: Incorporation of Functionalized Drugs and Degradation in Biological Media" International Journal of Molecular Sciences 24, no. 4: 3362. https://doi.org/10.3390/ijms24043362




