Eukaryotic Ribosomal Protein S5 of the 40S Subunit: Structure and Function
Abstract
:1. Introduction
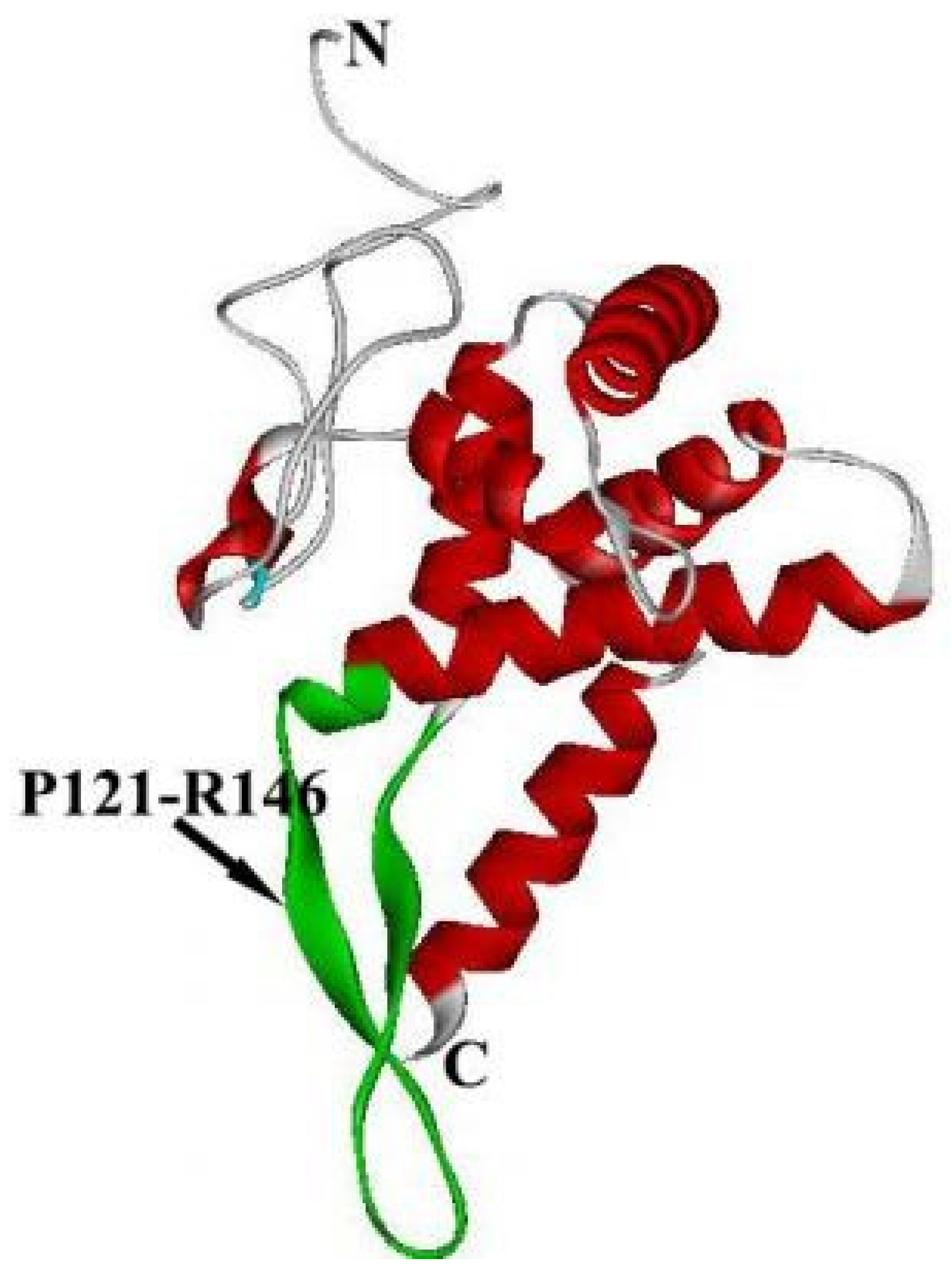
2. The Structure of RPS5
3. Role of RPS5 in Translation
4. Potential rRNA Binding Sites on the N-Terminus of RPS5
5. RPS5 as a Potential Target for Liver Disease and Cancer
6. The Non-Ribosomal Functions of the RPS5 N-Terminus
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bowman, J.C.; Petrov, A.S.; Frenkel-Pinter, M.; Penev, P.I.; Williams, L.D. Root of the Tree: The Significance, Evolution, and Origins of the Ribosome. Chem. Rev. 2020, 120, 4848–4878. [Google Scholar] [CrossRef] [PubMed]
- Moraleva, A.A.; Deryabin, A.S.; Rubtsov, Y.P.; Rubtsova, M.P.; Dontsova, O.A. Eukaryotic Ribosome Biogenesis: The 40S Subunit. Acta Nat. 2022, 14, 14–30. [Google Scholar] [CrossRef] [PubMed]
- Jenner, L.; Melnikov, S.; Garreau, D.L.N.; Ben-Shem, A.; Iskakova, M.; Urzhumtsev, A.; Meskauskas, A.; Dinman, J.; Yusupova, G.; Yusupov, M. Crystal structure of the 80S yeast ribosome. Curr. Opin. Struct. Biol. 2012, 22, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shem, A.; Garreau, D.L.N.; Melnikov, S.; Jenner, L.; Yusupova, G.; Yusupov, M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 2011, 334, 1524–1529. [Google Scholar] [CrossRef]
- Wang, X.; Ren, Y.; Gong, C.; Chen, Y.F.; Ge, X.P.; Kong, J.P.; Sun, W.J.; Du, X.Y. 40S ribosomal protein S18 is a novel maternal peptidoglycan-binding protein that protects embryos of zebrafish from bacterial infections. Dev. Comp. Immunol. 2021, 125, 104212. [Google Scholar] [CrossRef]
- Linnemann, J.; Pöll, G.; Jakob, S.; Ferreira-Cerca, S.; Griesenbeck, J.; Tschochner, H.; Milkereit, P. Impact of two neighbouring ribosomal protein clusters on biogenesis factor binding and assembly of yeast late small ribosomal subunit precursors. PLoS ONE 2019, 14, e203415. [Google Scholar] [CrossRef]
- Kuwano, Y.; Olvera, J.; Wool, I.G. The primary structure of rat ribosomal protein S5. A ribosomal protein present in the rat genome in a single copy. J. Biol. Chem. 1993, 267, 25304–25308. [Google Scholar] [CrossRef]
- Frigerio, J.M.; Dagorn, J.C.; Iovanna, J.L. Cloning, sequencing and expression of the L5, L21, L27a, L28, S5, S9, S10 and S29 human ribosomal protein mRNAs. Biochim. Biophys. Acta 1995, 1262, 64–68. [Google Scholar] [CrossRef]
- Ignatovich, O.; Cooper, M.; Kulesza, H.M.; Beggs, J.D. Cloning and characterisation of the gene encoding the ribosomal protein S5 (also known as rp14, S2, YS8) of Saccharomyces cerevisiae. Nucleic Acids Res. 1995, 23, 4616–4619. [Google Scholar] [CrossRef]
- Pelava, A.; Schneider, C.; Watkins, N.J. The importance of ribosome production, and the 5S RNP–MDM2 pathway, in health and disease. Biochem. Soc. Trans. 2016, 44, 1086–1090. [Google Scholar] [CrossRef] [Green Version]
- Yusupov, M.M.; Yusupova, G.Z.; Baucom, A.; Lieberman, K.; Earnest, T.N.; Cate, J.H.; Noller, H.F. Crystal structure of the ribosome at 5.5 A resolution. Science 2001, 292, 883–896. [Google Scholar] [CrossRef]
- Spahn, C.M.; Gomez-Lorenzo, M.G.; Grassucci, R.A.; Jørgensen, R.; Andersen, G.R.; Beckmann, R.; Penczek, P.A.; Ballesta, J.P.; Frank, J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004, 23, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Galkin, O.; Bentley, A.A.; Gupta, S.; Compton, B.A.; Mazumder, B.; Kinzy, T.G.; Merrick, W.C.; Hatzoglou, M.; Pestova, T.V.; Hellen, C.U.; et al. Roles of the negatively charged N-terminal extension of Saccharomyces cerevisiae ribosomal protein S5 revealed by characterization of a yeast strain containing human ribosomal protein S5. RNA 2007, 13, 2116–2128. [Google Scholar] [CrossRef] [PubMed]
- Jakob, S.; Ohmayer, U.; Neueder, A.; Hierlmeier, T.; Perez-Fernandez, J.; Hochmuth, E.; Deutzmann, R.; Griesenbeck, J.; Tschochner, H.; Milkereit, P. Interrelationships between yeast ribosomal protein assembly events and transient ribosome biogenesis factors interactions in early pre-ribosomes. PLoS ONE 2012, 7, e32552. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Jindal, S.; Bentley, A.A.; Hinnebusch, A.G.; Komar, A.A. Rps5-Rps16 communication is essential for efficient translation initiation in yeast S. cerevisiae. Nucleic Acids Res. 2014, 42, 8537–8555. [Google Scholar] [CrossRef]
- Visweswaraiah, J.; Hinnebusch, A.G. Interface between 40S exit channel protein uS7/Rps5 and eIF2α modulates start codon recognition in vivo. eLife 2017, 6, e22572. [Google Scholar] [CrossRef]
- Dong, J.; Hinnebusch, A.G. uS5/Rps2 residues at the 40S ribosome entry channel enhance initiation at suboptimal start codons in vivo. Genetics 2022, 220, iyab176. [Google Scholar] [CrossRef]
- Matragkou, C.N.; Papachristou, E.T.; Tezias, S.S.; Tsiftsoglou, A.S.; Choli-Papadopoulou, T.; Vizirianakis, I.S. The potential role of ribosomal protein S5 on cell cycle arrest and initiation of murine erythroleukemia cell differentiation. J. Cell. Biochem. 2008, 104, 1477–1490. [Google Scholar] [CrossRef]
- Vizirianakis, I.S.; Papachristou, E.T.; Andreadis, P.; Zopounidou, E.; Matragkou, C.N.; Tsiftsoglou, A.S. Genetic manipulation of RPS5 gene expression modulates the initiation of commitment of MEL cells to erythroid maturation: Implications in understanding ribosomopathies. Int. J. Oncol. 2015, 47, 303–314. [Google Scholar] [CrossRef]
- Xu, W.H.; Hu, H.G.; Tian, Y.; Wang, S.Z.; Li, J.; Li, J.Z.; Deng, X.; Qian, H.; Qiu, L.; Hu, Z.L.; et al. Bioactive compound reveals a novel function for ribosomal protein S5 in hepatic stellate cell activation and hepatic fibrosis. Hepatology 2014, 60, 648–660. [Google Scholar] [CrossRef]
- Yang, H.L.; Tu, Z.L.; Yang, D.; Hu, M.T.; Zhou, L.L.; Li, Q.H.; Yu, B.; Hou, S.X. Exosomes from hypoxic pre-treated ADSCs attenuateacute ischemic stroke-induced brain injury via delivery of circ-Rps5 and promote M2 microglia/macrophage polarization. Neurosci. Lett. 2022, 769, 136389. [Google Scholar] [CrossRef]
- Lumsden, T.; Bentley, A.A.; Beutler, W.; Ghosh, A.; Galkin, O.; Komar, A.A. Yeast strains with N-terminally truncated ribosomal protein S5: Implications for the evolution, structure and function of the Rps5/Rps7 proteins. Nucleic Acids Res. 2010, 38, 1261–1272. [Google Scholar] [CrossRef]
- Davies, C.; Bussiere, D.E.; Golden, B.L.; Porter, S.J.; Ramakrishnan, V.; White, S.W. Ribosomal proteins S5 and L6: High-resolution crystal structures and roles in protein synthesis and antibiotic resistance. J. Mol. Biol. 1998, 279, 873–888. [Google Scholar] [CrossRef]
- Malygin, A.; Baranovskaya, O.; Ivanov, A.; Karpova, G. Expression and purification of human ribosomal proteins S3, S5, S10, S19, and S26. Protein Expr. Purif. 2003, 28, 57–62. [Google Scholar] [CrossRef]
- Harada, N.; Sano, K.; Kimura, M.; Hosaka, H.; Nakagawa, A.; Tanaka, I. Crystallization and preliminary X-ray crystallographic study of the ribosomal protein S7 from Bacillus stearothermophilus. J. Struct. Biol. 1997, 120, 112–114. [Google Scholar] [CrossRef]
- Hosaka, H.; Yao, M.; Kimura, M.; Tanaka, I. The structure of the archaebacterial ribosomal protein S7 and its possible interaction with 16S rRNA. J. Biochem. 2001, 130, 695–701. [Google Scholar] [CrossRef]
- Li, Z.; Wu, D.; Zhan, B.; Hu, X.; Gan, J.; Ji, C.; Li, J. Structural insights into the complex of trigger factor chaperone and ribosomal protein S7 from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2019, 512, 838–844. [Google Scholar] [CrossRef]
- Miyamoto, A.; Usui, M.; Yamasaki, N.; Yamada, N.; Kuwano, E.; Tanaka, I.; Kimura, M. Role of the N-terminal region of ribosomal protein S7 in its interaction with 16S rRNA which binds to the concavity formed by the beta-ribbon arm and the alpha-helix. Eur. J. Biochem. 1999, 266, 591–598. [Google Scholar] [CrossRef]
- Djumagulov, M.; Demeshkina, N.; Jenner, L.; Rozov, A.; Yusupov, M.; Yusupova, G. Accuracy mechanism of eukaryotic ribosome translocation. Nature 2021, 600, 543–546. [Google Scholar] [CrossRef]
- Rabl, J.; Leibundgut, M.; Ataide, S.F.; Haag, A.; Ban, N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 2011, 331, 730–736. [Google Scholar] [CrossRef] [Green Version]
- Yusupova, G.; Yusupov, M. High-resolution structure of the eukaryotic 80S ribosome. Annu. Rev. Biochem. 2014, 83, 467–486. [Google Scholar] [CrossRef] [PubMed]
- Neueder, A.; Jakob, S.; Pöll, G.; Linnemann, J.; Deutzmann, R.; Tschochner, H.; Milkereit, P. A local role for the small ribosomal subunit primary binder rpS5 in final 18S rRNA processing in yeast. PLoS ONE 2010, 5, e10194. [Google Scholar] [CrossRef]
- Bheemireddy, S.; Sandhya, S.; Srinivasan, N. Comparative Analysis of Structural and Dynamical Features of Ribosome Upon Association With mRNA Reveals Potential Role of Ribosomal Proteins. Front. Mol. Biosci. 2021, 8, 654164. [Google Scholar] [CrossRef] [PubMed]
- Robert, F.; Brakier-Gingras, L. Ribosomal protein S7 from Escherichia coli uses the same determinants to bind 16S ribosomal RNA and its messenger RNA. Nucleic Acids Res. 2001, 29, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Duss, O.; Stepanyuk, G.A.; Puglisi, J.D.; Williamson, J.R. Transient Protein-RNA Interactions Guide Nascent Ribosomal RNA Folding. Cell 2019, 179, 1357–1369. [Google Scholar] [CrossRef]
- Fredrick, K.; Dunny, G.M.; Noller, H.F. Tagging ribosomal protein S7 allows rapid identification of mutants defective in assembly and function of 30 S subunits. J. Mol. Biol. 2000, 298, 379–394. [Google Scholar] [CrossRef]
- Noller, H.F.; Hoang, L.; Fredrick, K. The 30S ribosomal P site: A function of 16S rRNA. FEBS Lett. 2005, 579, 855–858. [Google Scholar] [CrossRef]
- Greuer, B.; Thiede, B.; Brimacombe, R. The cross-link from the upstream region of mRNA to ribosomal protein S7 is located in the C-terminal peptide: Experimental verification of a prediction from modeling studies. RNA 1999, 5, 1521–1525. [Google Scholar] [CrossRef]
- Saito, K.; Mattheakis, L.C.; Nomura, M. Post-transcriptional regulation of the str operon in Escherichia coli. Ribosomal protein S7 inhibits coupled translation of S7 but not its independent translation. J. Mol. Biol. 1994, 235, 111–124. [Google Scholar] [CrossRef]
- Robert, F.; Brakier-Gingras, L. A functional interaction between ribosomal proteins S7 and S11 within the bacterial ribosome. J. Biol. Chem. 2003, 278, 44913–44920. [Google Scholar] [CrossRef] [Green Version]
- Ian’Shina, D.D.; Malygin, A.A.; Karpova, G.G. The mutual effect of human ribosomal proteins S5 and S16 on their binding with 18S rRNA fragment 1203–1236/1521–1698. Mol. Biol. 2009, 43, 643–651. [Google Scholar] [CrossRef]
- Malygin, A.A.; Yanshina, D.D.; Karpova, G.G. Interactions of human ribosomal proteins S16 and S5 with an 18S rRNA fragment containing their binding sites. Biochimie 2009, 91, 1180–1186. [Google Scholar] [CrossRef] [PubMed]
- Le, S.N.; Brown, C.R.; Harvey, S.; Boege, R.H.; Elmlund, H.; Elmlund, D. The TAFs of TFIID Bind and Rearrange the Topology of the TATA-Less RPS5 Promoter. Int. J. Mol. Sci. 2019, 20, 3290. [Google Scholar] [CrossRef] [PubMed]
- Bischof, O.; Kruft, V.; Wittmann-Liebold, B. Analysis of the puromycin binding site in the 70 S ribosome of Escherichia coli at the peptide level. J. Biol. Chem. 1994, 269, 18315–18319. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, H.; Nakagawa, A.; Tanaka, I.; Harada, N.; Sano, K.; Kimura, M.; Yao, M.; Wakatsuki, S. Ribosomal protein S7: A new RNA-binding motif with structural similarities to a DNA architectural factor. Structure 1997, 5, 1199–1208. [Google Scholar] [CrossRef]
- Golovin, A.V.; Khayrullina, G.A.; Kraal, B.; Kopylov, C.A. Identification of Novel RNA-Protein Contact in Complex of Ribosomal Protein S7 and 3′-Terminal Fragment of 16S rRNA in E. coli. Acta Nat. 2012, 4, 65–72. [Google Scholar] [CrossRef]
- Urlaub, H.; Kruft, V.; Bischof, O.; Müller, E.C.; Wittmann-Liebold, B. Protein-rRNA binding features and their structural and functional implications in ribosomes as determined by cross-linking studies. EMBO J. 1995, 14, 4578–4588. [Google Scholar] [CrossRef]
- Urlaub, H.; Thiede, B.; Müller, E.C.; Brimacombe, R.; Wittmann-Liebold, B. Identification and sequence analysis of contact sites between ribosomal proteins and rRNA in Escherichia coli 30 S subunits by a new approach using matrix-assisted laser desorption/ionization-mass spectrometry combined with N-terminal microsequencing. J. Biol. Chem. 1997, 272, 14547–14555. [Google Scholar] [CrossRef]
- Robert, F.; Gagnon, M.; Sans, D.; Michnick, S.; Brakier-Gingras, L. Mapping of the RNA recognition site of Escherichia coli ribosomal protein S7. RNA 2000, 6, 1649–1659. [Google Scholar] [CrossRef]
- Spahn, C.M.; Beckmann, R.; Eswar, N.; Penczek, P.A.; Sali, A.; Blobel, G.; Frank, J. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell 2001, 107, 373–386. [Google Scholar] [CrossRef] [Green Version]
- Sharifulin, D.; Babaylova, E.; Kossinova, O.; Bartuli, Y.; Graifer, D.; Karpova, G. Ribosomal protein S5e is implicated in translation initiation through its interaction with the N-terminal domain of initiation factor eIF2α. Chembiochem 2013, 14, 2136–2143. [Google Scholar] [CrossRef]
- Spahn, C.M.; Jan, E.; Mulder, A.; Grassucci, R.A.; Sarnow, P.; Frank, J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell 2004, 118, 465–475. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. Structural Insights into the Mechanism of Scanning and Start Codon Recognition in Eukaryotic Translation Initiation. Trends Biochem. Sci. 2017, 42, 589–611. [Google Scholar] [CrossRef]
- Pisarev, A.V.; Kolupaeva, V.G.; Pisareva, V.P.; Merrick, W.C.; Hellen, C.U.; Pestova, T.V. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Gene Dev. 2006, 20, 624–636. [Google Scholar] [CrossRef]
- Tsukihashi, Y.; Kawaichi, M.; Kokubo, T. Requirement for yeast TAF145 function in transcriptional activation of the RPS5 promoter that depends on both core promoter structure and upstream activating sequences. J. Biol. Chem. 2001, 276, 25715–25726. [Google Scholar] [CrossRef]
- Matragkou, C.; Papachristou, H.; Karetsou, Z.; Papadopoulos, G.; Papamarcaki, T.; Vizirianakis, I.S.; Tsiftsoglou, A.S.; Choli-Papadopoulou, T. On the intracellular trafficking of mouse S5 ribosomal protein from cytoplasm to nucleoli. J. Mol. Biol. 2009, 392, 1192–1204. [Google Scholar] [CrossRef]
- Visweswaraiah, J.; Pittman, Y.; Dever, T.E.; Hinnebusch, A.G. The β -hairpin of 40S exit channel protein Rps5/uS7 promotes efficient and accurate translation initiation in vivo. eLife 2015, 4, e7939. [Google Scholar] [CrossRef]
- Stainslas, P.; Sylvie, L. The remarkable history of the hepatitis C virus. Microbes Infect. 2019, 21, 263–270. [Google Scholar] [CrossRef]
- Bhat, P.; Shwetha, S.; Sharma, D.K.; Joseph, A.P.; Srinivasan, N.; Das, S. The beta hairpin structure within ribosomal protein S5 mediates interplay between domains II and IV and regulates HCV IRES function. Nucleic Acids Res. 2015, 43, 2888–2901. [Google Scholar] [CrossRef]
- Brown, Z.P.; Abaeva, I.S.; De, S.; Hellen, C.; Pestova, T.V.; Frank, J. Molecular architecture of 40S translation initiation complexes on IRES. EMBO J. 2022, 41, e110581. [Google Scholar] [CrossRef]
- Babaylova, E.S.; Graifer, D.M.; Malygin, A.A.; Karpova, G.G. Arrangements ofnucleotides flanking the start codon in the IRES of the hepatitis C virus in the IRES binary complex with the human 40S ribosomal subunit. Biochimie 2018, 148, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Quade, N.; Boehringer, D.; Leibundgut, M.; van den Heuvel, J.; Ban, N. Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution. Nat. Commun. 2015, 6, 7646. [Google Scholar] [CrossRef] [PubMed]
- Malygin, A.A.; Kossinova, O.A.; Shatsky, I.N.; Karpova, G.G. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res. 2013, 41, 8706–8714. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.G.; Grosely, R.; Petrov, A.N.; Puglisi, J.D. Dynamics of IRES-mediated translation. Philos. Trans. R. Soc. B 2017, 372, 20160177. [Google Scholar] [CrossRef] [PubMed]
- Joseph, A.P.; Bhat, P.; Das, S.; Srinivasan, N. Re-analysis of cryoEM data on HCV IRES bound to 40S subunit of human ribosome integrated with recent structural information suggests new contact regions between ribosomal proteins and HCV RNA. RNA Biol. 2014, 11, 891–905. [Google Scholar] [CrossRef]
- Dong, J.; Aitken, C.E.; Thakur, A.; Shin, B.S.; Lorsch, J.R. Rps3/uS3 promotes mRNA binding at the 40S ribosome entry channel and stabilizes preinitiation complexes at start codons. Proc. Natl. Acad. Sci. USA 2017, 114, E2126–E2135. [Google Scholar] [CrossRef]
- Guo, H.Y.; Zhu, J.; Miao, Q.H.; Qi, R.B.; Liu, C.C.; Yang, H.Z.; Yuan, L.G.; Liu, G.Q. RPS5 interacts with the rabbit hemorrhagic disease virus 3′ extremities region and plays a role in virus replication. Vet. Microbiol. 2020, 249, 108858. [Google Scholar] [CrossRef]
- Tomioka, M.; Shimobayashi, M.; Kitabatake, M.; Ohno, M.; Kozutsumi, Y.; Oka, S.; Takematsu, H. Ribosomal protein uS7/Rps5 serine-223 in protein kinase-mediated phosphorylation and ribosomal small subunit maturation. Sci. Rep. 2018, 8, 1244. [Google Scholar] [CrossRef]
- Hernandez-Gea, V.; Friedman, S.L. Pathogenesis of liver fibrosis. Annu. Rev. Pathol.-Mech. 2011, 6, 425–456. [Google Scholar] [CrossRef]
- Paez, J.; Sellers, W.R. PI3K/PTEN/AKT pathway: A critical mediator of oncogenic signaling. Cancer Treat Res. 2004, 115, 145–167. [Google Scholar] [CrossRef]
- Vizirianakis, I.S.; Pappas, I.S.; Gougoumas, D.; Tsiftsoqlou, A.S. Expression of ribosomal protein S5 cloned gene during differentiation and apoptosis in murine erythroleukemia (MEL) cells. Oncol. Res. 1999, 11, 409–419. [Google Scholar]
- Su, Z.X.; Gu, Y.Q. Identification of key genes and pathways involved in abdominal aortic aneurysm initiation and progression. Vascular 2021, 30, 639–664. [Google Scholar] [CrossRef]
- Pan, R.; Yu, C.; Shao, Y.; Hong, H.; Sun, J.; Zhang, Z.; Li, P.; Zheng, M. Identification of Key Genes and Pathways Involved in Circulating Tumor Cells in Colorectal Cancer. Anal. Cell. Pathol. 2022, 2022, 9943571. [Google Scholar] [CrossRef]
- Weijers, D.; Franke-Van, D.M.; Vencken, R.J.; Quint, A.; Hooykaas, P.; Offringa, R. An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 2001, 128, 4289–4299. [Google Scholar] [CrossRef]
- Zhang, J.; Yuan, H.; Yang, Y.; Fish, T.; Lyi, S.M.; Thannhauser, T.W.; Zhang, L.; Li, L. Plastid ribosomal protein S5 is involved in photosynthesis, plant development, and cold stress tolerance in Arabidopsis. J. Exp. Bot. 2016, 67, 2731–2744. [Google Scholar] [CrossRef]
- Francis, T.A.; Roberts, S. Influence of the state of ribosome association on the phosphorylation of ribosomal proteins in isolated ribosome--protein kinase systems from rat cerebral cortex. Biochem. J. 1982, 208, 289–300. [Google Scholar] [CrossRef]
- Madjar, J.J.; Traut, R.R. Differences in electrophoretic behaviour of eight ribosomal proteins from rat and rabbit tissues and evidence for proteolytic action on liver proteins. Mol. Gen. Genet. 1980, 179, 89–101. [Google Scholar] [CrossRef]
- Tian, Z.C.; Gao, S.D.; Du, J.Z.; Liu, G.Y.; Luo, J.X.; Yin, H. Comparative evaluation of two informative markers in the phylogeny of Babesia and Theileria parasites. Exp. Parasitol. 2020, 212, 107870. [Google Scholar] [CrossRef]
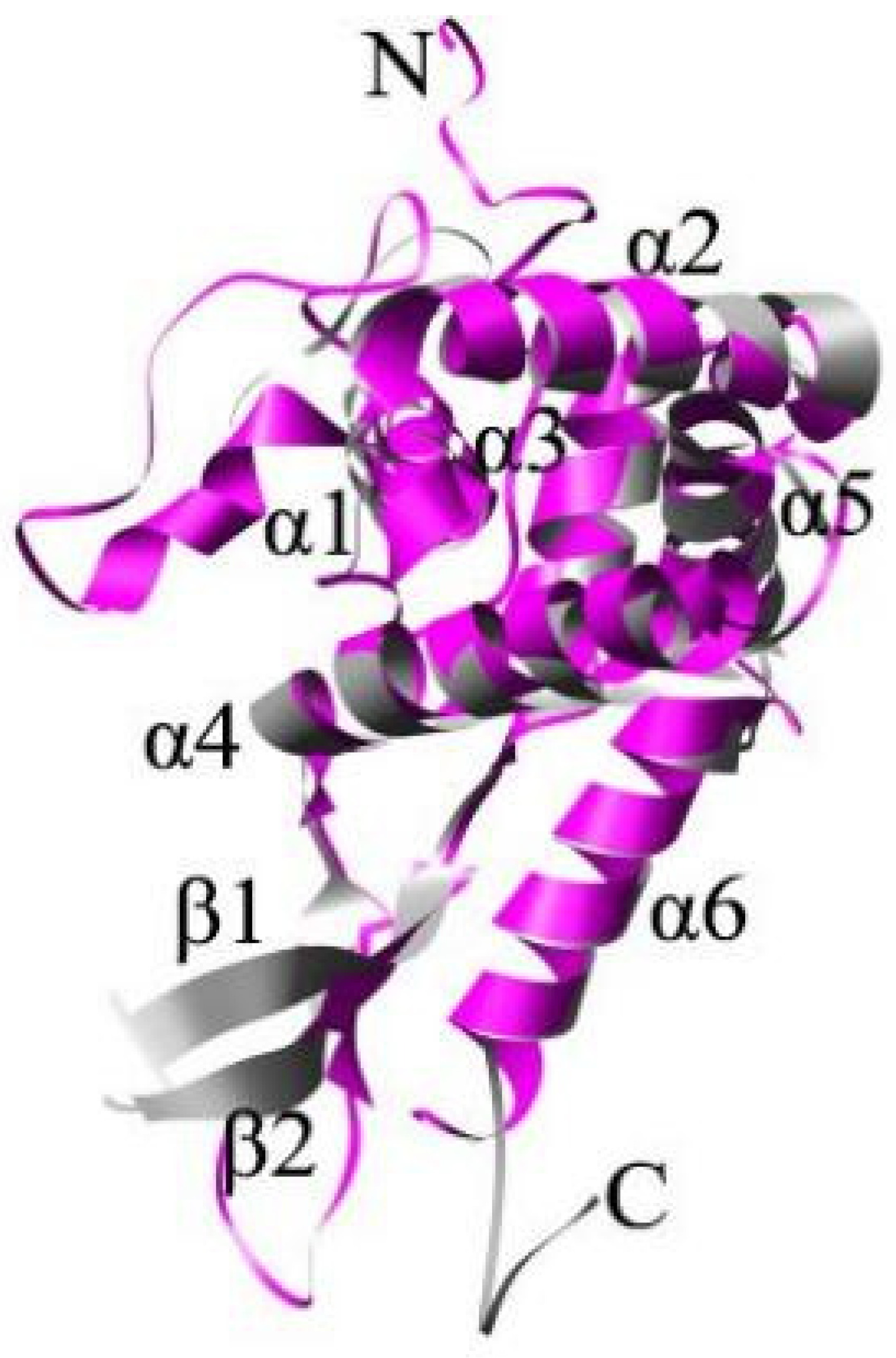
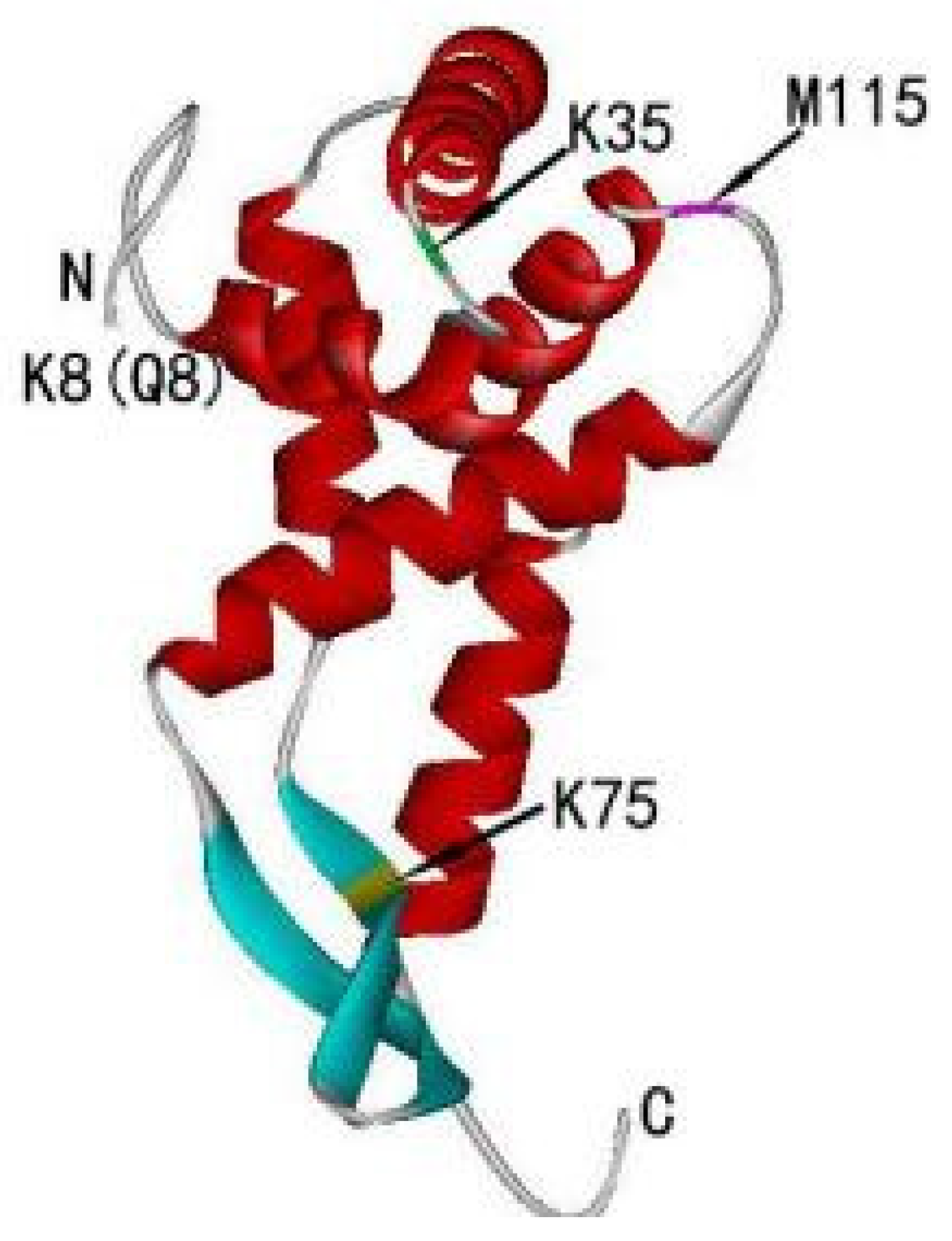
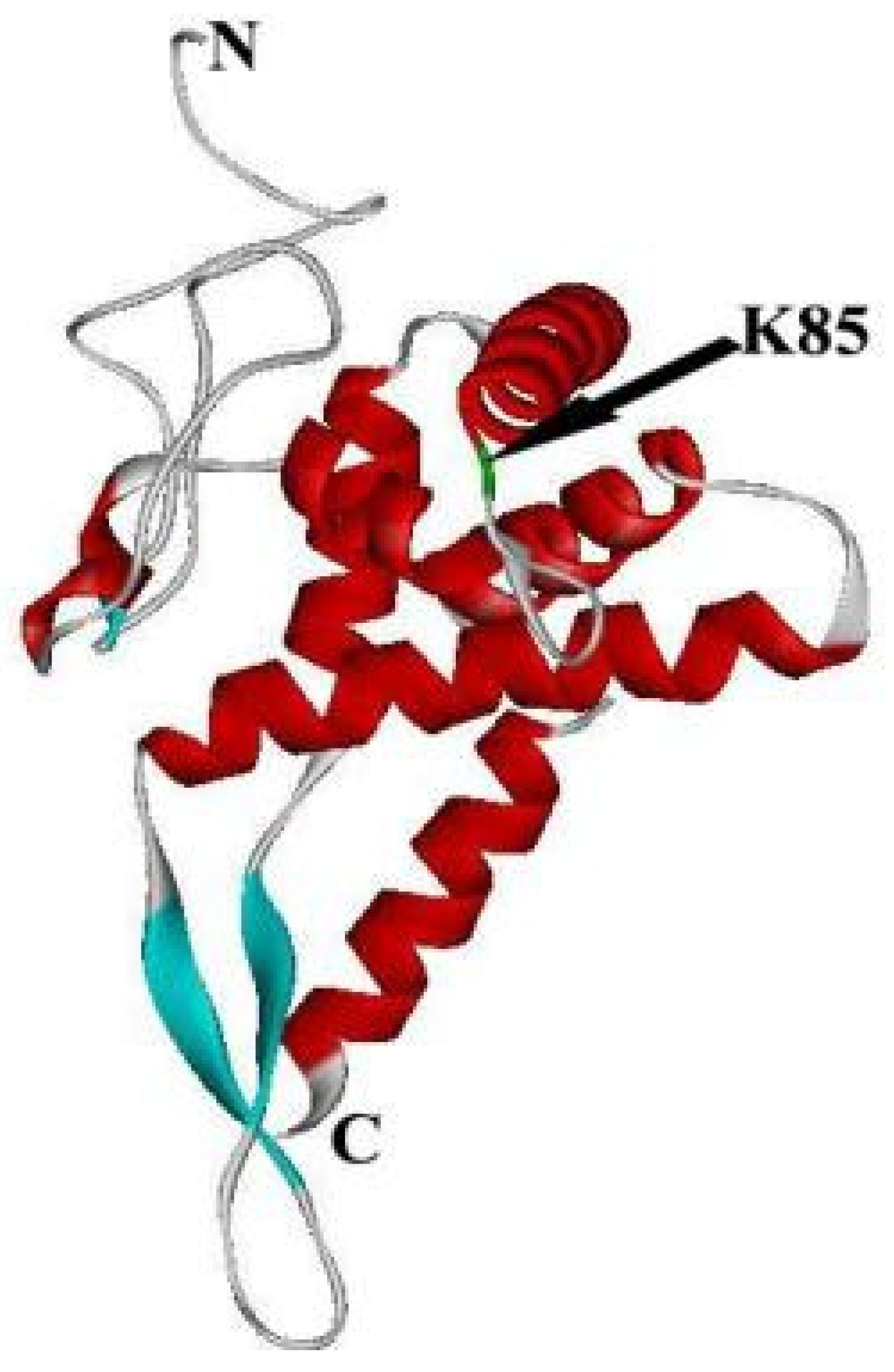
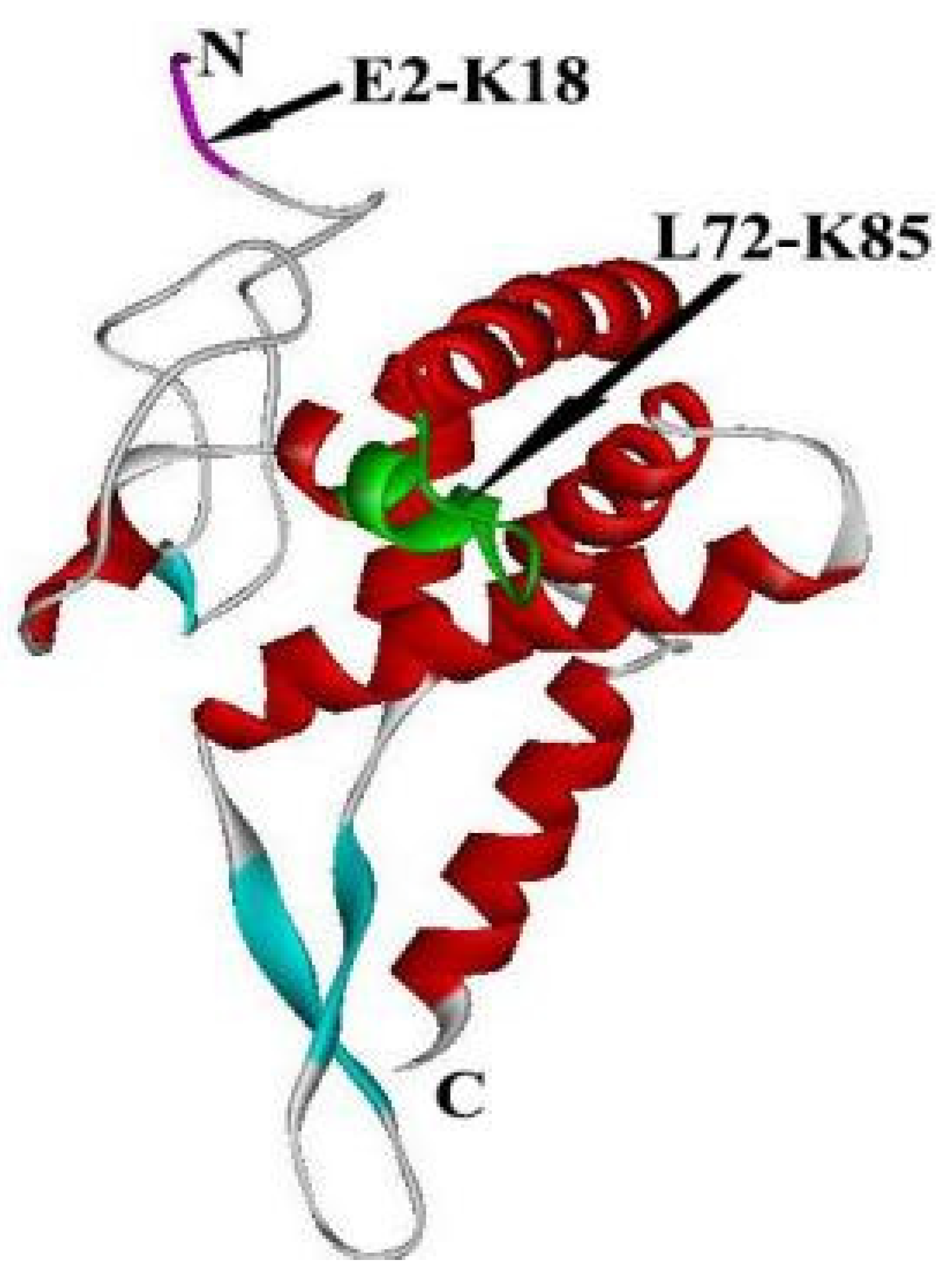

| Homologous | Gene Copy | Nucleotides | Amino Acids | Amino Acid Differences | Highly Conserved C-Terminal Structure and Variable N-Terminal Structure | |
|---|---|---|---|---|---|---|
| eukaryotic | RPS5 | 3~6 | 701 | 204 | A6 A60 A181 | More basic amino acids yes |
| rat | RPS5 | single | 701 | 204 (98% identity) | T6 G60 R181 | More basic amino acids yes |
| mouse | RPS5 | 3~6 | 701 | 204 | yes | |
| yeast | RPS5 | single | 908 | 204 + 69 | 4truncated variants (lacking 13, 24, 30, and 46 N-terminal amino acids) | |
| Escherichia coli | RPS7 | 557 | 204 − 48 | no | ||
| 30% identity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, L.; Chao, W.; Zhong, S.; Ren, A.-J. Eukaryotic Ribosomal Protein S5 of the 40S Subunit: Structure and Function. Int. J. Mol. Sci. 2023, 24, 3386. https://doi.org/10.3390/ijms24043386
Qiu L, Chao W, Zhong S, Ren A-J. Eukaryotic Ribosomal Protein S5 of the 40S Subunit: Structure and Function. International Journal of Molecular Sciences. 2023; 24(4):3386. https://doi.org/10.3390/ijms24043386
Chicago/Turabian StyleQiu, Lijuan, Wen Chao, Shan Zhong, and An-Jing Ren. 2023. "Eukaryotic Ribosomal Protein S5 of the 40S Subunit: Structure and Function" International Journal of Molecular Sciences 24, no. 4: 3386. https://doi.org/10.3390/ijms24043386
APA StyleQiu, L., Chao, W., Zhong, S., & Ren, A.-J. (2023). Eukaryotic Ribosomal Protein S5 of the 40S Subunit: Structure and Function. International Journal of Molecular Sciences, 24(4), 3386. https://doi.org/10.3390/ijms24043386





