Extracellular Vesicle Treatment Alleviates Neurodevelopmental and Neurodegenerative Pathology in Cortical Spheroid Model of Down Syndrome
Abstract
1. Introduction
2. Results
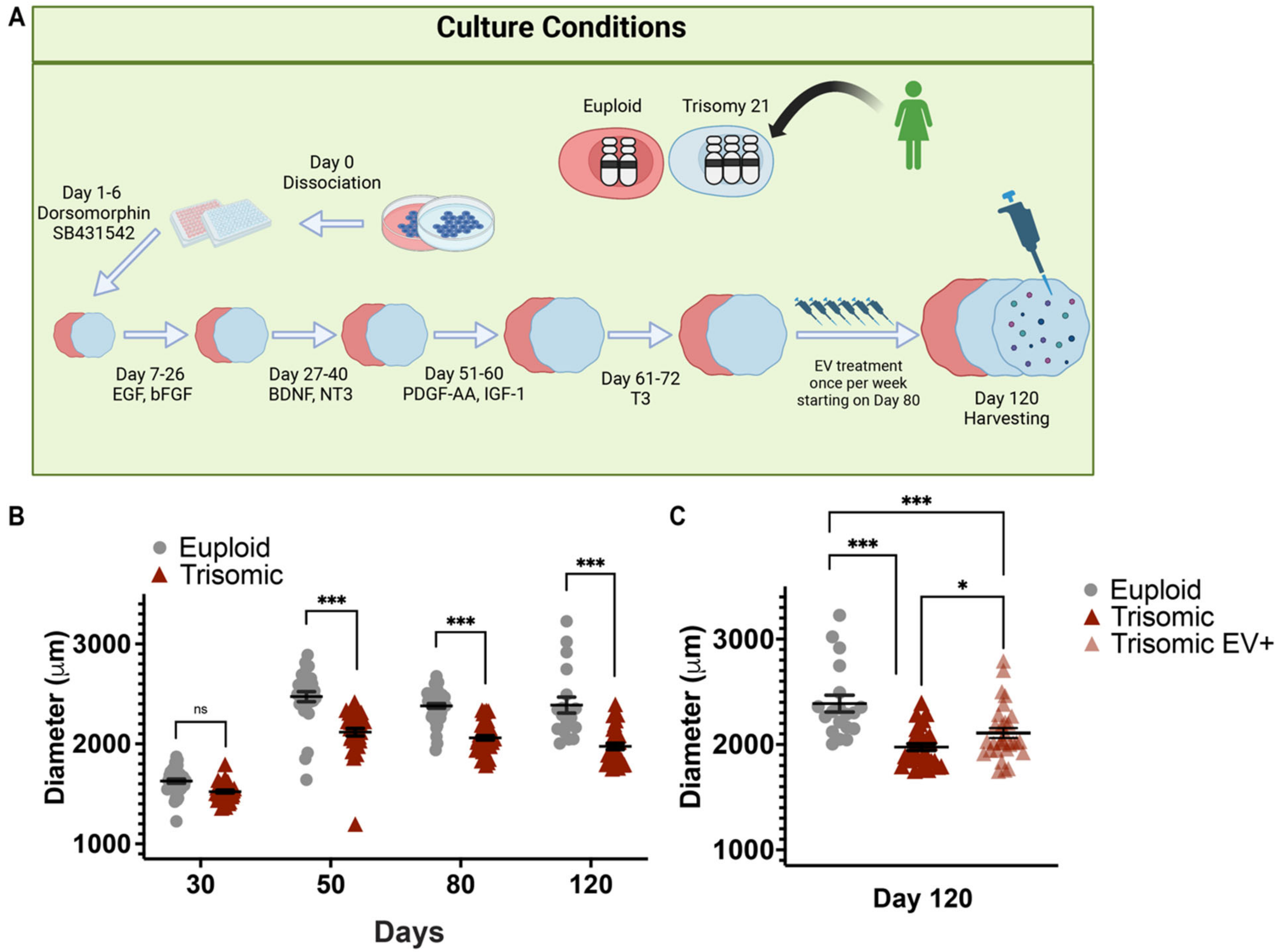
2.1. EV Treatment Preserved the Size of Trisomic CS
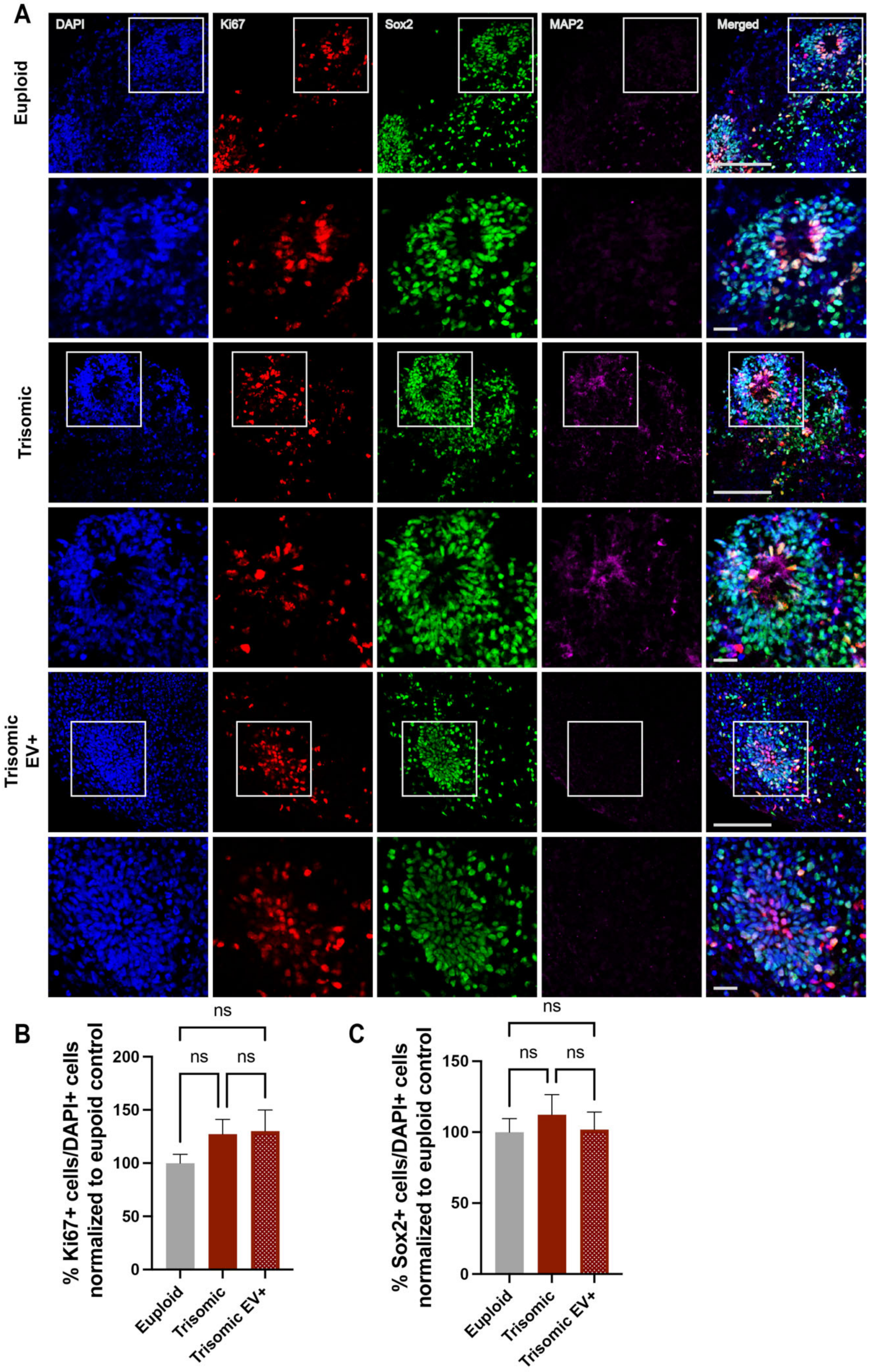
2.2. EV Treatment Did Not Affect the Proliferation of Neural Progenitors (NPCs) within Trisomic CS
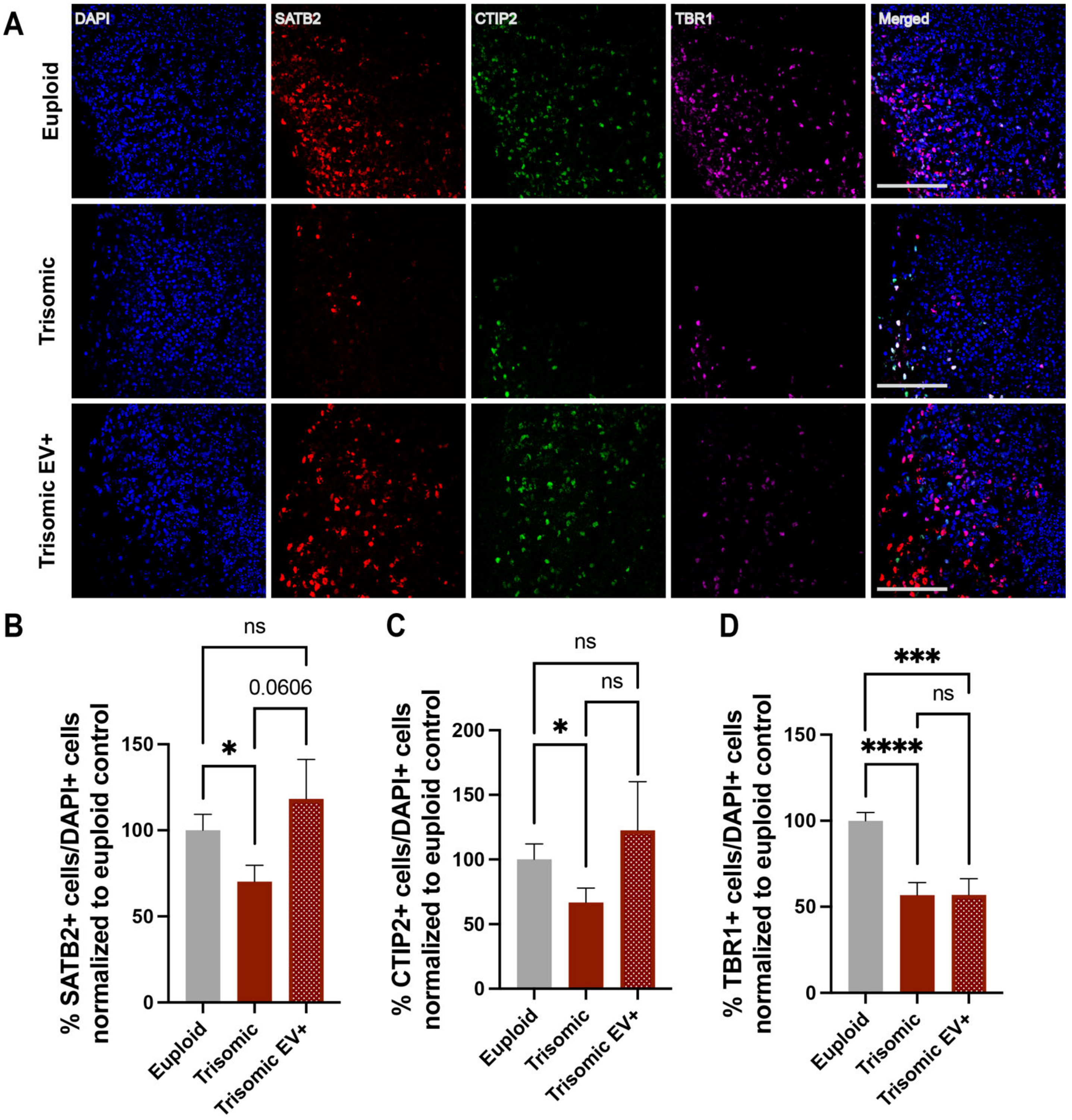
2.3. Treatment with EVs Can Ameliorate Aberrant Neurogenesis of Specific Neuronal Subpopulations in Trisomic CS
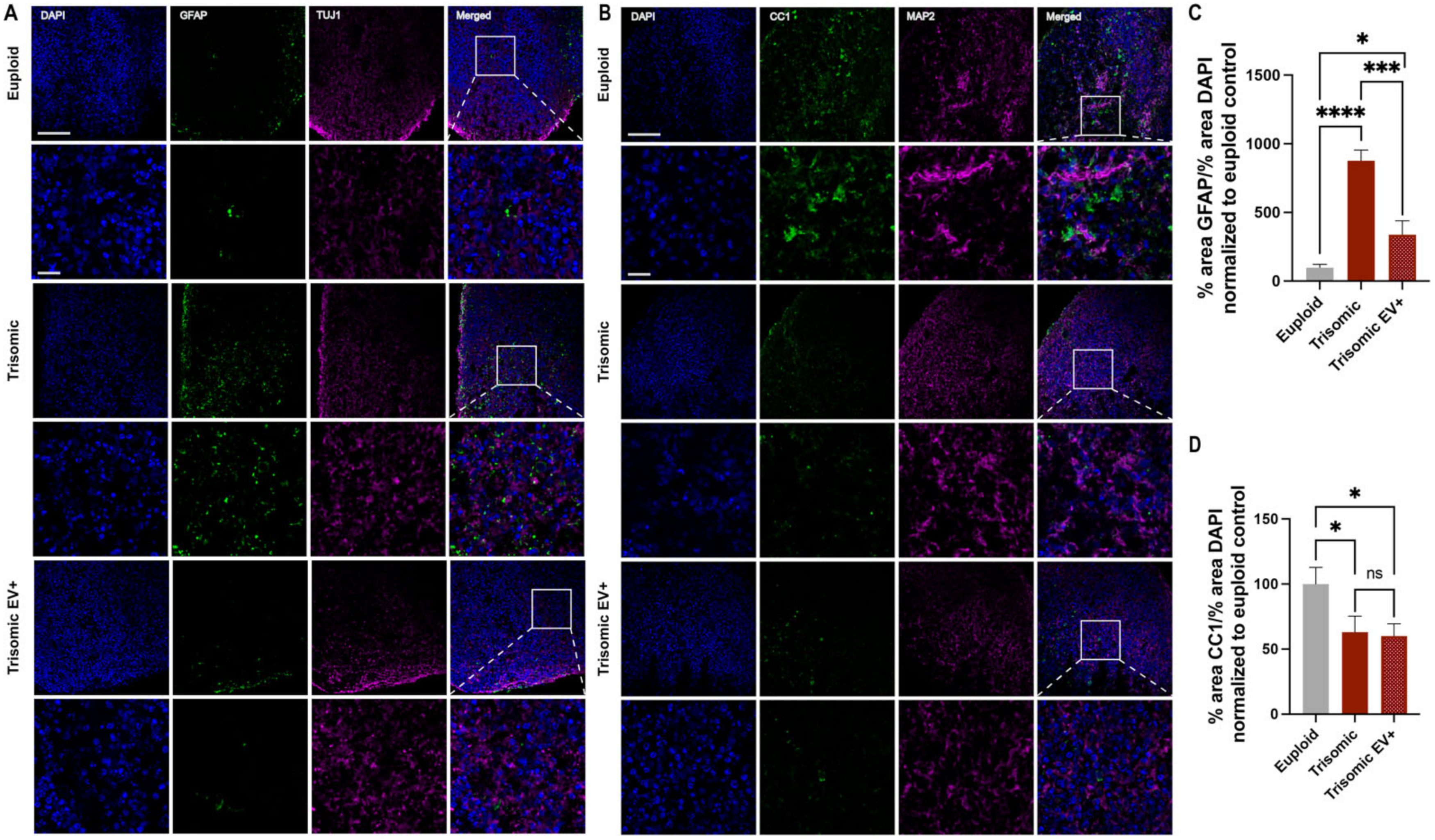
2.4. EV Treatment Modified GFAP and CC1 Expression in Trisomic CS
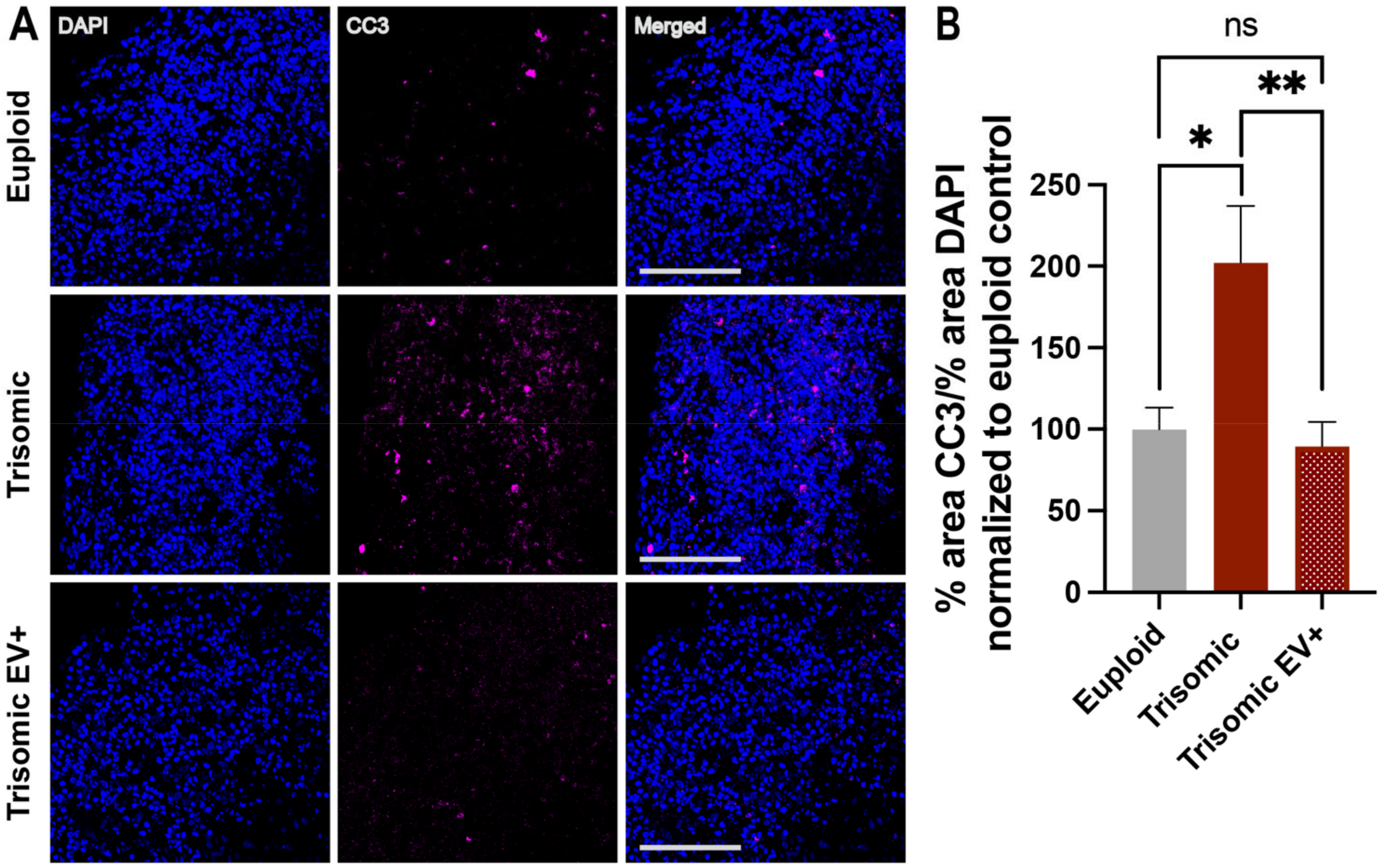
2.5. EV Treatment Decreased Cell Death in Trisomic CS
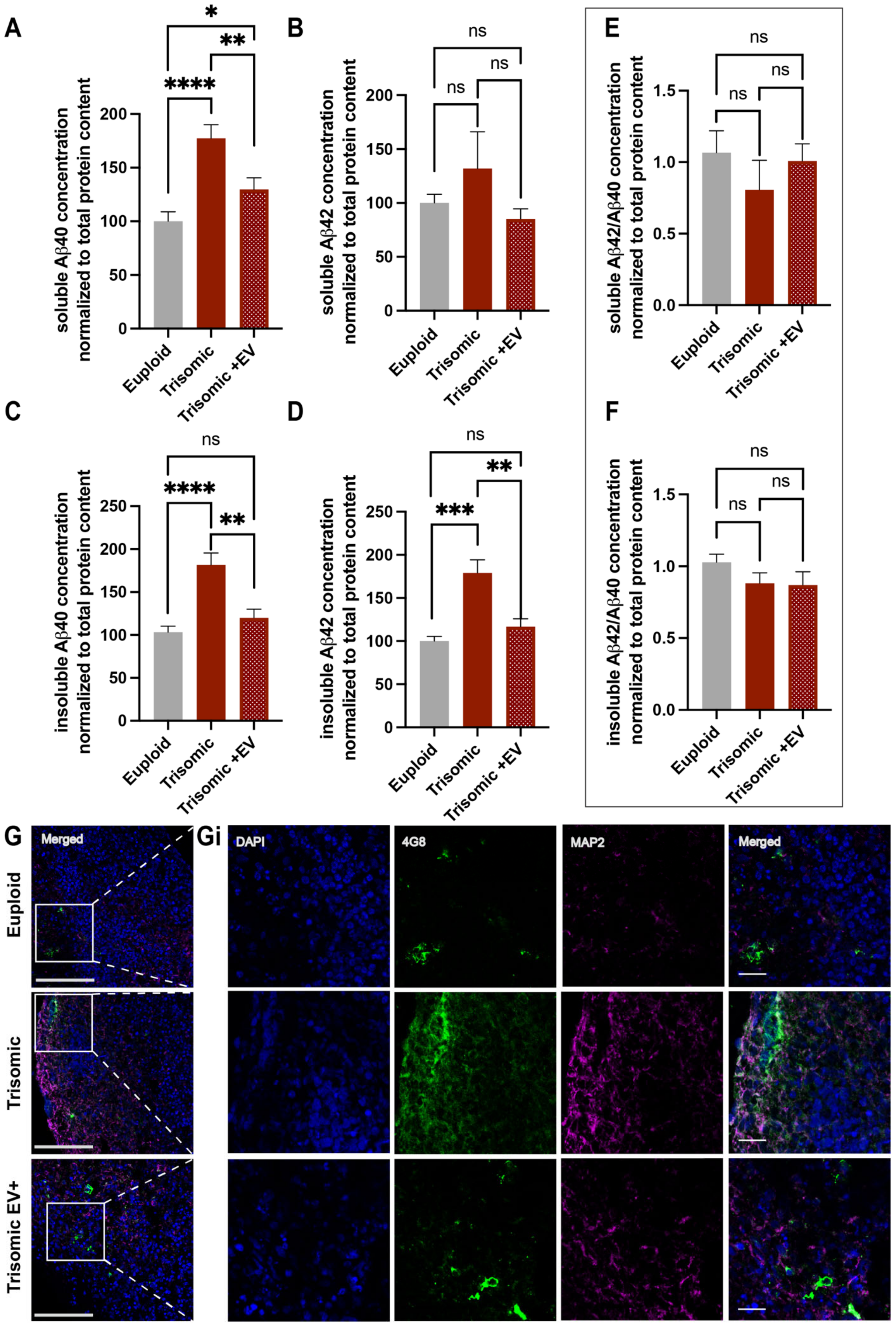
2.6. EVs Alleviated AD Pathology in Trisomic CS
3. Discussion
4. Materials and Methods
4.1. Generation of Cortical Spheroids
4.2. Extracellular Vesicles and Sample Collection
4.3. Cryoprotection and Slice Preparation
4.4. Immunohistochemistry (IHC)
4.5. Imaging and Quantification
4.6. Fractionation of Spheroids and Aβ Enzyme-Linked Immunosorbent Assay (ELISA)
4.7. Western Blotting
4.8. Measuring Spheroid Diameter
4.9. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Irving, C.; Basu, A.; Richmond, S.; Burn, J.; Wren, C. Twenty-year trends in prevalence and survival of Down syndrome. Eur. J. Hum. Genet. 2008, 16, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Kirby, R.S. The prevalence of selected major birth defects in the United States. Semin. Perinatol. 2017, 41, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Mai, C.T.; Isenburg, J.L.; Canfield, M.A.; Meyer, R.E.; Correa, A.; Alverson, C.J.; Lupo, P.J.; Riehle-Colarusso, T.; Cho, S.J.; Aggarwal, D.; et al. National population-based estimates for major birth defects, 2010–2014. Birth Defects Res. 2019, 111, 1420–1435. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, S.E.; Skotko, B.G.; Rafii, M.S.; Strydom, A.; Pape, S.E.; Bianchi, D.W.; Sherman, S.L.; Reeves, R.H. Down syndrome. Nat. Rev. Dis. Primers 2020, 6, 9. [Google Scholar] [CrossRef]
- Head, E.; Lott, I.T.; Wilcock, D.M.; Lemere, C.A. Aging in Down Syndrome and the Development of Alzheimer’s Disease Neuropathology. Curr. Alzheimer Res. 2016, 13, 18–29. [Google Scholar] [CrossRef]
- Levman, J.; MacDonald, A.; Baumer, N.; MacDonald, P.; Stewart, N.; Lim, A.; Cogger, L.; Shiohama, T.; Takahashi, E. Structural magnetic resonance imaging demonstrates abnormal cortical thickness in Down syndrome: Newborns to young adults. Neuroimage Clin. 2019, 23, 101874. [Google Scholar] [CrossRef]
- Pinter, J.D.; Eliez, S.; Schmitt, J.E.; Capone, G.T.; Reiss, A.L. Neuroanatomy of Down’s syndrome: A high-resolution MRI study. Am. J. Psychiatry 2001, 158, 1659–1665. [Google Scholar] [CrossRef]
- Pinter, J.D.; Brown, W.E.; Eliez, S.; Schmitt, J.E.; Capone, G.T.; Reiss, A.L. Amygdala and hippocampal volumes in children with Down syndrome: A high-resolution MRI study. Neurology 2001, 56, 972–974. [Google Scholar] [CrossRef]
- Jernigan, T.L.; Bellugi, U.; Sowell, E.; Doherty, S.; Hesselink, J.R. Cerebral morphologic distinctions between Williams and Down syndromes. Arch. Neurol. 1993, 50, 186–191. [Google Scholar] [CrossRef]
- Bletsch, A.; Mann, C.; Andrews, D.S.; Daly, E.; Tan, G.M.Y.; Murphy, D.G.M.; Ecker, C. Down syndrome is accompanied by significantly reduced cortical grey-white matter tissue contrast. Hum. Brain Mapp. 2018, 39, 4043–4054. [Google Scholar] [CrossRef]
- Lee, N.R.; Adeyemi, E.I.; Lin, A.; Clasen, L.S.; Lalonde, F.M.; Condon, E.; Driver, D.I.; Shaw, P.; Gogtay, N.; Raznahan, A.; et al. Dissociations in Cortical Morphometry in Youth with Down Syndrome: Evidence for Reduced Surface Area but Increased Thickness. Cereb. Cortex 2016, 26, 2982–2990. [Google Scholar] [CrossRef]
- Golden, J.A.; Hyman, B.T. Development of the superior temporal neocortex is anomalous in trisomy 21. J. Neuropathol. Exp. Neurol. 1994, 53, 513–520. [Google Scholar] [CrossRef]
- Mrak, R.E.; Griffin, W.S.T. Trisomy 21 and the brain. J. Neuropathol. Exp. Neurol. 2004, 63, 679–685. [Google Scholar] [CrossRef]
- Engidawork, E.; Gulesserian, T.; Fountoulakis, M.; Lubec, G. Aberrant protein expression in cerebral cortex of fetus with Down syndrome. Neuroscience 2003, 122, 145–154. [Google Scholar] [CrossRef]
- Guidi, S.; Ciani, E.; Bonasoni, P.; Santini, D.; Bartesaghi, R. Widespread proliferation impairment and hypocellularity in the cerebellum of fetuses with down syndrome. Brain Pathol. 2011, 21, 361–373. [Google Scholar] [CrossRef]
- Larsen, K.B.; Laursen, H.; Græm, N.; Samuelsen, G.B.; Bogdanovic, N.; Pakkenberg, B. Reduced cell number in the neocortical part of the human fetal brain in Down syndrome. Ann. Anat. Anat. Anz. 2008, 190, 421–427. [Google Scholar] [CrossRef]
- Rotmensch, S.; Goldstein, I.; Liberati, M.; Shalev, J.; Ben-Rafael, Z.; Copel, J.A. Fetal transcerebellar diameter in Down syndrome. Obstet. Gynecol. 1997, 89, 534–537. [Google Scholar] [CrossRef]
- Sylvester, P.E. The hippocampus in Down’s syndrome. J. Ment. Defic. Res. 1983, 27 Pt 3, 227–236. [Google Scholar] [CrossRef]
- Wisniewski, K.E. Down syndrome children often have brain with maturation delay, retardation of growth, and cortical dysgenesis. Am. J. Med. Genet. 1990, 37, 274–281. [Google Scholar] [CrossRef]
- Schmidt-Sidor, B.; Wisniewski, K.E.; Shepard, T.H.; Sersen, E.A. Brain growth in Down syndrome subjects 15 to 22 weeks of gestational age and birth to 60 months. Clin. Neuropathol. 1990, 9, 181–190. [Google Scholar]
- Stagni, F.; Giacomini, A.; Emili, M.; Guidi, S.; Bartesaghi, R. Neurogenesis impairment: An early developmental defect in Down syndrome. Free Radic. Biol. Med. 2018, 114, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, H.A.; Reeves, R.H. Hippocampal hypocellularity in the Ts65Dn mouse originates early in development. Brain Res. 2006, 1104, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Insausti, A.M.; Megías, M.; Crespo, D.; Cruz-Orive, L.M.; Dierssen, M.; Vallina, I.F.; Insausti, R.; Flórez, J. Hippocampal volume and neuronal number in Ts65Dn mice: A murine model of Down syndrome. Neurosci. Lett. 1998, 253, 175–178. [Google Scholar] [CrossRef]
- Reeves, R.H.; Irving, N.G.; Moran, T.H.; Wohn, A.; Kitt, C.; Sisodia, S.S.; Schmidt, C.; Bronson, R.T.; Davisson, M.T. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat. Genet. 1995, 11, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Ábrahám, H.; Vincze, A.; Veszprémi, B.; Kravják, A.; Gömöri, É.; Kovács, G.G.; Seress, L. Impaired myelination of the human hippocampal formation in Down syndrome. Int. J. Dev. Neurosci. 2012, 30, 147–158. [Google Scholar] [CrossRef]
- Wisniewski, K.E.; Schmidt-Sidor, B. Postnatal delay of myelin formation in brains from Down syndrome infants and children. Clin. Neuropathol. 1989, 8, 55–62. [Google Scholar]
- Olmos-Serrano, J.L.; Kang, H.J.; Tyler, W.A.; Silbereis, J.C.; Cheng, F.; Zhu, Y.; Pletikos, M.; Jankovic-Rapan, L.; Cramer, N.P.; Galdzicki, Z.; et al. Down Syndrome Developmental Brain Transcriptome Reveals Defective Oligodendrocyte Differentiation and Myelination. Neuron 2016, 89, 1208–1222. [Google Scholar] [CrossRef]
- Chan, W.Y.; Lorke, D.E.; Tiu, S.C.; Yew, D.T. Proliferation and apoptosis in the developing human neocortex. Anat. Rec. 2002, 267, 261–276. [Google Scholar] [CrossRef]
- Contestabile, A.; Fila, T.; Ceccarelli, C.; Bonasoni, P.; Bonapace, L.; Santini, D.; Bartesaghi, R.; Ciani, E. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with down syndrome and in Ts65Dn mice. Hippocampus 2007, 17, 665–678. [Google Scholar] [CrossRef]
- Stagni, F.; Bartesaghi, R. Neuroanatomical alterations in higher-order thalamic nuclei of fetuses with Down syndrome. Clin. Neurol. Neurosurg. 2020, 194, 105870. [Google Scholar] [CrossRef]
- Guidi, S.; Bonasoni, P.; Ceccarelli, C.; Santini, D.; Gualtieri, F.; Ciani, E.; Bartesaghi, R. Neurogenesis impairment and increased cell death reduce total neuron number in the hippocampal region of fetuses with Down syndrome. Brain Pathol. 2008, 18, 180–197. [Google Scholar] [CrossRef]
- Hibaoui, Y.; Grad, I.; Letourneau, A.; Sailani, M.R.; Dahoun, S.; Santoni, F.A.; Gimelli, S.; Guipponi, M.; Pelte, M.F.; Béna, F.; et al. Modelling and rescuing neurodevelopmental defect of Down syndrome using induced pluripotent stem cells from monozygotic twins discordant for trisomy 21. EMBO Mol. Med. 2014, 6, 259–277. [Google Scholar] [CrossRef]
- Tang, X.-Y.; Xu, L.; Wang, J.; Hong, Y.; Wang, Y.; Zhu, Q.; Wang, D.; Zhang, X.-Y.; Liu, C.-Y.; Fang, K.-H.; et al. DSCAM/PAK1 pathway suppression reverses neurogenesis deficits in iPSC-derived cerebral organoids from patients with Down syndrome. J. Clin. Investig. 2021, 131, 135763. [Google Scholar] [CrossRef]
- Contestabile, A.; Greco, B.; Ghezzi, D.; Tucci, V.; Benfenati, F.; Gasparini, L. Lithium rescues synaptic plasticity and memory in Down syndrome mice. J. Clin. Investig. 2013, 123, 348–361. [Google Scholar] [CrossRef]
- Li, Z.; Klein, J.A.; Rampam, S.; Kurzion, R.; Campbell, N.B.; Patel, Y.; Haydar, T.F.; Zeldich, E. Asynchronous excitatory neuron development in an isogenic cortical spheroid model of Down syndrome. Front. Neurosci. 2022, 16, 932384. [Google Scholar] [CrossRef]
- Bally, B.P.; Murai, K.K. Astrocytes in Down Syndrome Across the Lifespan. Front. Cell. Neurosci. 2021, 15, 702685. [Google Scholar] [CrossRef]
- Guidi, S.; Giacomini, A.; Stagni, F.; Emili, M.; Uguagliati, B.; Bonasoni, M.P.; Bartesaghi, R. Abnormal development of the inferior temporal region in fetuses with Down syndrome. Brain Pathol. 2018, 28, 986–998. [Google Scholar] [CrossRef]
- Chen, C.; Jiang, P.; Xue, H.; Peterson, S.E.; Tran, H.T.; McCann, A.E.; Parast, M.M.; Li, S.; Pleasure, D.E.; Laurent, L.C.; et al. Role of astroglia in Down’s syndrome revealed by patient-derived human-induced pluripotent stem cells. Nat. Commun. 2014, 5, 4430. [Google Scholar] [CrossRef]
- Zdaniuk, G.; Wierzba-Bobrowicz, T.; Szpak, G.M.; Stępień, T. Astroglia disturbances during development of the central nervous system in fetuses with Down’s syndrome. Folia Neuropathol. 2011, 49, 109–114. [Google Scholar]
- Head, E.; Helman, A.M.; Powell, D.; Schmitt, F.A. Down syndrome, beta-amyloid and neuroimaging. Free Radic. Biol. Med. 2018, 114, 102–109. [Google Scholar] [CrossRef]
- Ovchinnikov, D.A.; Korn, O.; Virshup, I.; Wells, C.A.; Wolvetang, E.J. The Impact of APP on Alzheimer-like Pathogenesis and Gene Expression in Down Syndrome iPSC-Derived Neurons. Stem Cell Rep. 2018, 11, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Sleegers, K.; Brouwers, N.; Gijselinck, I.; Theuns, J.; Goossens, D.; Wauters, J.; Del-Favero, J.; Cruts, M.; van Duijn, C.M.; Van Broeckhoven, C. APP duplication is sufficient to cause early onset Alzheimer’s dementia with cerebral amyloid angiopathy. Brain 2006, 129, 2977–2983. [Google Scholar] [CrossRef] [PubMed]
- Rafii, M.S.; Lukic, A.S.; Andrews, R.D.; Brewer, J.; Rissman, R.A.; Strother, S.C.; Wernick, M.N.; Pennington, C.; Mobley, W.C.; Ness, S.; et al. PET Imaging of Tau Pathology and Relationship to Amyloid, Longitudinal MRI, and Cognitive Change in Down Syndrome: Results from the Down Syndrome Biomarker Initiative (DSBI). J. Alzheimers Dis. 2017, 60, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J. Lessons Learned from Alzheimer Disease: Clinical Trials with Negative Outcomes. Clin. Transl. Sci. 2018, 11, 147–152. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef]
- Coleman, L.G. The emerging world of subcellular biological medicine: Extracellular vesicles as novel biomarkers, targets, and therapeutics. Neural Regen. Res. 2021, 17, 1020–1022. [Google Scholar] [CrossRef]
- Deng, J.; Ji, Y.; Zhu, F.; Liu, L.; Li, L.; Bai, X.; Li, H.; Liu, X.; Luo, Y.; Lin, B.; et al. Mapping secretome-mediated interaction between paired neuron-macrophage single cells. Proc. Natl. Acad. Sci. USA 2022, 119, e2200944119. [Google Scholar] [CrossRef]
- Hamlett, E.D.; Ledreux, A.; Potter, H.; Chial, H.J.; Patterson, D.; Espinosa, J.M.; Bettcher, B.M.; Granholm, A.-C. Exosomal biomarkers in Down syndrome and Alzheimer’s disease. Free Radic. Biol. Med. 2018, 114, 110–121. [Google Scholar] [CrossRef]
- Yuyama, K.; Igarashi, Y. Physiological and pathological roles of exosomes in the nervous system. Biomol. Concepts 2016, 7, 53–68. [Google Scholar] [CrossRef]
- Moore, T.L.; Bowley, B.G.E.; Pessina, M.A.; Calderazzo, S.M.; Medalla, M.; Go, V.; Zhang, Z.G.; Chopp, M.; Finklestein, S.; Harbaugh, A.G.; et al. Mesenchymal derived exosomes enhance recovery of motor function in a monkey model of cortical injury. Restor. Neurol. Neurosci. 2019, 37, 347–362. [Google Scholar] [CrossRef]
- Medalla, M.; Chang, W.; Calderazzo, S.M.; Go, V.; Tsolias, A.; Goodliffe, J.W.; Pathak, D.; De Alba, D.; Pessina, M.; Rosene, D.L.; et al. Treatment with Mesenchymal-Derived Extracellular Vesicles Reduces Injury-Related Pathology in Pyramidal Neurons of Monkey Perilesional Ventral Premotor Cortex. J. Neurosci. 2020, 40, 3385–3407. [Google Scholar] [CrossRef]
- Go, V.; Bowley, B.G.E.; Pessina, M.A.; Zhang, Z.G.; Chopp, M.; Finklestein, S.P.; Rosene, D.L.; Medalla, M.; Buller, B.; Moore, T.L. Extracellular vesicles from mesenchymal stem cells reduce microglial-mediated neuroinflammation after cortical injury in aged Rhesus monkeys. Geroscience 2020, 42, 1–17. [Google Scholar] [CrossRef]
- Go, V.; Sarikaya, D.; Zhou, Y.; Bowley, B.G.E.; Pessina, M.A.; Rosene, D.L.; Zhang, Z.G.; Chopp, M.; Finklestein, S.P.; Medalla, M.; et al. Extracellular vesicles derived from bone marrow mesenchymal stem cells enhance myelin maintenance after cortical injury in aged rhesus monkeys. Exp. Neurol. 2021, 337, 113540. [Google Scholar] [CrossRef]
- Peters, A.; Rosene, D.L.; Moss, M.B.; Kemper, T.L.; Abraham, C.R.; Tigges, J.; Albert, M.S. Neurobiological bases of age-related cognitive decline in the rhesus monkey. J. Neuropathol. Exp. Neurol. 1996, 55, 861–874. [Google Scholar] [CrossRef]
- Stonebarger, G.A.; Urbanski, H.F.; Woltjer, R.L.; Vaughan, K.L.; Ingram, D.K.; Schultz, P.L.; Calderazzo, S.M.; Siedeman, J.A.; Mattison, J.A.; Rosene, D.L.; et al. Amyloidosis increase is not attenuated by long-term calorie restriction or related to neuron density in the prefrontal cortex of extremely aged rhesus macaques. Geroscience 2020, 42, 1733–1749. [Google Scholar] [CrossRef]
- Andrle, M.; Fiedler, W.; Rett, A.; Ambros, P.; Schweizer, D. A case of trisomy 22 in Pongo pygmaeus. Cytogenet. Cell Genet. 1979, 24, 1–6. [Google Scholar] [CrossRef]
- Hirata, S.; Hirai, H.; Nogami, E.; Morimura, N.; Udono, T. Chimpanzee Down syndrome: A case study of trisomy 22 in a captive chimpanzee. Primates 2017, 58, 267–273. [Google Scholar] [CrossRef]
- Madhavan, M.; Nevin, Z.S.; Shick, H.E.; Garrison, E.; Clarkson-Paredes, C.; Karl, M.; Clayton, B.L.L.; Factor, D.C.; Allan, K.C.; Barbar, L.; et al. Induction of myelinating oligodendrocytes in human cortical spheroids. Nat. Methods 2018, 15, 700–706. [Google Scholar] [CrossRef]
- Paşca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.-Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef]
- Gonzalez, C.; Armijo, E.; Bravo-Alegria, J.; Becerra-Calixto, A.; Mays, C.E.; Soto, C. Modeling amyloid beta and tau pathology in human cerebral organoids. Mol. Psychiatry 2018, 23, 2363–2374. [Google Scholar] [CrossRef]
- McCann, B.; Levman, J.; Baumer, N.; Lam, M.Y.; Shiohama, T.; Cogger, L.; MacDonald, A.; Ijner, P.; Takahashi, E. Structural magnetic resonance imaging demonstrates volumetric brain abnormalities in down syndrome: Newborns to young adults. Neuroimage Clin. 2021, 32, 102815. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Dhanasekaran, A.R.; Gardiner, K.J. Mouse models of Down syndrome: Gene content and consequences. Mamm. Genome 2016, 27, 538–555. [Google Scholar] [CrossRef] [PubMed]
- Mouton, P.R.; Martin, L.J.; Calhoun, M.E.; Forno, G.D.; Price, D.L. Cognitive decline strongly correlates with cortical atrophy in Alzheimer’s dementia. Neurobiol. Aging 1998, 19, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Toda, T.; Shinmyo, Y.; Duong, T.A.D.; Masuda, K.; Kawasaki, H. An essential role of SVZ progenitors in cortical folding in gyrencephalic mammals. Sci. Rep. 2016, 6, 29578. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, L.; Galdzicki, Z.; Haydar, T.F. Defects in embryonic neurogenesis and initial synapse formation in the forebrain of the Ts65Dn mouse model of Down syndrome. J. Neurosci. 2007, 27, 11483–11495. [Google Scholar] [CrossRef]
- Lockrow, J.P.; Fortress, A.M.; Granholm, A.-C.E. Age-related neurodegeneration and memory loss in down syndrome. Curr. Gerontol. Geriatr. Res. 2012, 2012, 463909. [Google Scholar] [CrossRef]
- Mito, T.; Becker, L.E. Developmental changes of S-100 protein and glial fibrillary acidic protein in the brain in Down syndrome. Exp. Neurol. 1993, 120, 170–176. [Google Scholar] [CrossRef]
- Reiche, L.; Küry, P.; Göttle, P. Aberrant Oligodendrogenesis in Down Syndrome: Shift in Gliogenesis? Cells 2019, 8, 1591. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, M.; Peng, C.; Zhao, G.; Gu, R. GFAP expression in injured astrocytes in rats. Exp. Ther. Med. 2017, 14, 1905–1908. [Google Scholar] [CrossRef]
- Klein, J.A.; Li, Z.; Rampam, S.; Cardini, J.; Ayoub, A.; Shaw, P.; Rachubinski, A.L.; Espinosa, J.M.; Zeldich, E.; Haydar, T.F. Sonic Hedgehog Pathway Modulation Normalizes Expression of Olig2 in Rostrally Patterned NPCs with Trisomy 21. Front. Cell. Neurosci. 2021, 15, 794675. [Google Scholar] [CrossRef]
- Bin, J.M.; Harris, S.N.; Kennedy, T.E. The oligodendrocyte-specific antibody ‘CC1’ binds Quaking 7. J. Neurochem. 2016, 139, 181–186. [Google Scholar] [CrossRef]
- Contreras, D.; Garcia, G.; Jones, M.K.; Martinez, L.E.; Jayakarunakaran, A.; Gangalapudi, V.; Tang, J.; Wu, Y.; Zhao, J.J.; Chen, Z.; et al. Differential Susceptibility of Fetal Retinal Pigment Epithelial Cells, hiPSC- Retinal Stem Cells, and Retinal Organoids to Zika Virus Infection. Viruses 2023, 15, 142. [Google Scholar] [CrossRef]
- Qian, X.; Su, Y.; Adam, C.D.; Deutschmann, A.U.; Pather, S.R.; Goldberg, E.M.; Su, K.; Li, S.; Lu, L.; Jacob, F.; et al. Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell 2020, 26, 766–781.e9. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef]
- Mesci, P.; Souza, J.S.D.; Martin-Sancho, L.; Macia, A.; Saleh, A.; Yin, X.; Snethlage, C.; Adams, J.W.; Avansini, S.H.; Herai, R.H.; et al. SARS-CoV-2 infects human brain organoids causing cell death and loss of synapses that can be rescued by treatment with Sofosbuvir. PLOS Biol. 2022, 20, e3001845. [Google Scholar] [CrossRef]
- Gomez, W.; Morales, R.; Maracaja-Coutinho, V.; Parra, V.; Nassif, M. Down syndrome and Alzheimer’s disease: Common molecular traits beyond the amyloid precursor protein. Aging 2020, 12, 1011–1033. [Google Scholar] [CrossRef]
- Wang, J.; Dickson, D.W.; Trojanowski, J.Q.; Lee, V.M.-Y. The Levels of Soluble versus Insoluble Brain Aβ Distinguish Alzheimer’s Disease from Normal and Pathologic Aging. Exp. Neurol. 1999, 158, 328–337. [Google Scholar] [CrossRef]
- Dumurgier, J.; Schraen, S.; Gabelle, A.; Vercruysse, O.; Bombois, S.; Laplanche, J.-L.; Peoc’h, K.; Sablonnière, B.; Kastanenka, K.V.; Delaby, C.; et al. Cerebrospinal fluid amyloid-β 42/40 ratio in clinical setting of memory centers: A multicentric study. Alzheimers Res. Ther. 2015, 7, 30. [Google Scholar] [CrossRef]
- Ma, X.; Huang, M.; Zheng, M.; Dai, C.; Song, Q.; Zhang, Q.; Li, Q.; Gu, X.; Chen, H.; Jiang, G.; et al. ADSCs-derived extracellular vesicles alleviate neuronal damage, promote neurogenesis and rescue memory loss in mice with Alzheimer’s disease. J. Control. Release 2020, 327, 688–702. [Google Scholar] [CrossRef]
- Zhang, G.; Zhu, Z.; Wang, H.; Yu, Y.; Chen, W.; Waqas, A.; Wang, Y.; Chen, L. Exosomes derived from human neural stem cells stimulated by interferon gamma improve therapeutic ability in ischemic stroke model. J. Adv. Res. 2020, 24, 435–445. [Google Scholar] [CrossRef]
- Jarmalavičiūtė, A.; Tunaitis, V.; Pivoraitė, U.; Venalis, A.; Pivoriūnas, A. Exosomes from dental pulp stem cells rescue human dopaminergic neurons from 6-hydroxy-dopamine-induced apoptosis. Cytotherapy 2015, 17, 932–939. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, K.E.; Laure-Kamionowska, M.; Wisniewski, H.M. Evidence of arrest of neurogenesis and synaptogenesis in brains of patients with Down’s syndrome. N. Engl. J. Med. 1984, 311, 1187–1188. [Google Scholar] [CrossRef] [PubMed]
- Baburamani, A.A.; Vontell, R.T.; Uus, A.; Pietsch, M.; Patkee, P.A.; Wyatt-Ashmead, J.; Chin-Smith, E.C.; Supramaniam, V.G.; Tournier, J.D.; Deprez, M.; et al. Assessment of radial glia in the frontal lobe of fetuses with Down syndrome. Acta Neuropathol. Commun. 2020, 8, 141. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Nehra, S.; Do, L.H.; Ghosh, A.; Deshpande, A.J.; Singhal, N. Biphasic cell cycle defect causes impaired neurogenesis in down syndrome. Front. Genet. 2022, 13, 1007519. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, L.; Best, T.K.; Cramer, N.P.; Carney, R.S.E.; Isaac, J.T.R.; Galdzicki, Z.; Haydar, T.F. Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat. Neurosci. 2010, 13, 927–934. [Google Scholar] [CrossRef]
- Das, I.; Park, J.-M.; Shin, J.H.; Jeon, S.K.; Lorenzi, H.; Linden, D.J.; Worley, P.F.; Reeves, R.H. Hedgehog Agonist Therapy Corrects Structural and Cognitive Deficits in a Down Syndrome Mouse Model. Sci. Transl. Med. 2013, 5, 201ra120. [Google Scholar] [CrossRef]
- Morris, G.; Walker, A.J.; Berk, M.; Maes, M.; Puri, B.K. Cell Death Pathways: A Novel Therapeutic Approach for Neuroscientists. Mol. Neurobiol. 2018, 55, 5767–5786. [Google Scholar] [CrossRef]
- Joshi, P.; Turola, E.; Ruiz, A.; Bergami, A.; Libera, D.D.; Benussi, L.; Giussani, P.; Magnani, G.; Comi, G.; Legname, G.; et al. Microglia convert aggregated amyloid-β into neurotoxic forms through the shedding of microvesicles. Cell Death Differ. 2014, 21, 582–593. [Google Scholar] [CrossRef]
- Walsh, D.M.; Selkoe, D.J. Aβ Oligomers–a decade of discovery. J. Neurochem. 2007, 101, 1172–1184. [Google Scholar] [CrossRef]
- Asai, H.; Ikezu, S.; Tsunoda, S.; Medalla, M.; Luebke, J.; Haydar, T.; Wolozin, B.; Butovsky, O.; Kügler, S.; Ikezu, T. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 2015, 18, 1584–1593. [Google Scholar] [CrossRef]
- Guo, J.L.; Lee, V.M.Y. Cell-to-cell transmission of pathogenic proteins in neurodegenerative diseases. Nat. Med. 2014, 20, 130–138. [Google Scholar] [CrossRef]
- Saman, S.; Kim, W.; Raya, M.; Visnick, Y.; Miro, S.; Saman, S.; Jackson, B.; McKee, A.C.; Alvarez, V.E.; Lee, N.C.Y.; et al. Exosome-associated Tau Is Secreted in Tauopathy Models and Is Selectively Phosphorylated in Cerebrospinal Fluid in Early Alzheimer Disease. J. Biol. Chem. 2012, 287, 3842–3849. [Google Scholar] [CrossRef]
- Fagerlund, I.; Dougalis, A.; Shakirzyanova, A.; Gómez-Budia, M.; Pelkonen, A.; Konttinen, H.; Ohtonen, S.; Fazaludeen, M.F.; Koskuvi, M.; Kuusisto, J.; et al. Microglia-like Cells Promote Neuronal Functions in Cerebral Organoids. Cells 2021, 11, 124. [Google Scholar] [CrossRef]
- Zourray, C.; Kurian, M.A.; Barral, S.; Lignani, G. Electrophysiological Properties of Human Cortical Organoids: Current State of the Art and Future Directions. Front. Mol. Neurosci. 2022, 15, 839366. [Google Scholar] [CrossRef]
- Giffin-Rao, Y.; Sheng, J.; Strand, B.; Xu, K.; Huang, L.; Medo, M.; Risgaard, K.A.; Dantinne, S.; Mohan, S.; Keshan, A.; et al. Altered patterning of trisomy 21 interneuron progenitors. Stem Cell Rep. 2022, 17, 1366–1379. [Google Scholar] [CrossRef]
- Xin, H.; Li, Y.; Cui, Y.; Yang, J.J.; Zhang, Z.G.; Chopp, M. Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. J. Cereb. Blood Flow Metab. 2013, 33, 1711–1715. [Google Scholar] [CrossRef]
- Ash, P.E.A.; Stanford, E.A.; Al Abdulatif, A.; Ramirez-Cardenas, A.; Ballance, H.I.; Boudeau, S.; Jeh, A.; Murithi, J.M.; Tripodis, Y.; Murphy, G.J.; et al. Dioxins and related environmental contaminants increase TDP-43 levels. Mol. Neurodegener. 2017, 12, 35. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campbell, N.B.; Patel, Y.; Moore, T.L.; Medalla, M.; Zeldich, E. Extracellular Vesicle Treatment Alleviates Neurodevelopmental and Neurodegenerative Pathology in Cortical Spheroid Model of Down Syndrome. Int. J. Mol. Sci. 2023, 24, 3477. https://doi.org/10.3390/ijms24043477
Campbell NB, Patel Y, Moore TL, Medalla M, Zeldich E. Extracellular Vesicle Treatment Alleviates Neurodevelopmental and Neurodegenerative Pathology in Cortical Spheroid Model of Down Syndrome. International Journal of Molecular Sciences. 2023; 24(4):3477. https://doi.org/10.3390/ijms24043477
Chicago/Turabian StyleCampbell, Natalie Baker, Yesha Patel, Tara L. Moore, Maria Medalla, and Ella Zeldich. 2023. "Extracellular Vesicle Treatment Alleviates Neurodevelopmental and Neurodegenerative Pathology in Cortical Spheroid Model of Down Syndrome" International Journal of Molecular Sciences 24, no. 4: 3477. https://doi.org/10.3390/ijms24043477
APA StyleCampbell, N. B., Patel, Y., Moore, T. L., Medalla, M., & Zeldich, E. (2023). Extracellular Vesicle Treatment Alleviates Neurodevelopmental and Neurodegenerative Pathology in Cortical Spheroid Model of Down Syndrome. International Journal of Molecular Sciences, 24(4), 3477. https://doi.org/10.3390/ijms24043477





