MYC2: A Master Switch for Plant Physiological Processes and Specialized Metabolite Synthesis
Abstract
:1. Introduction
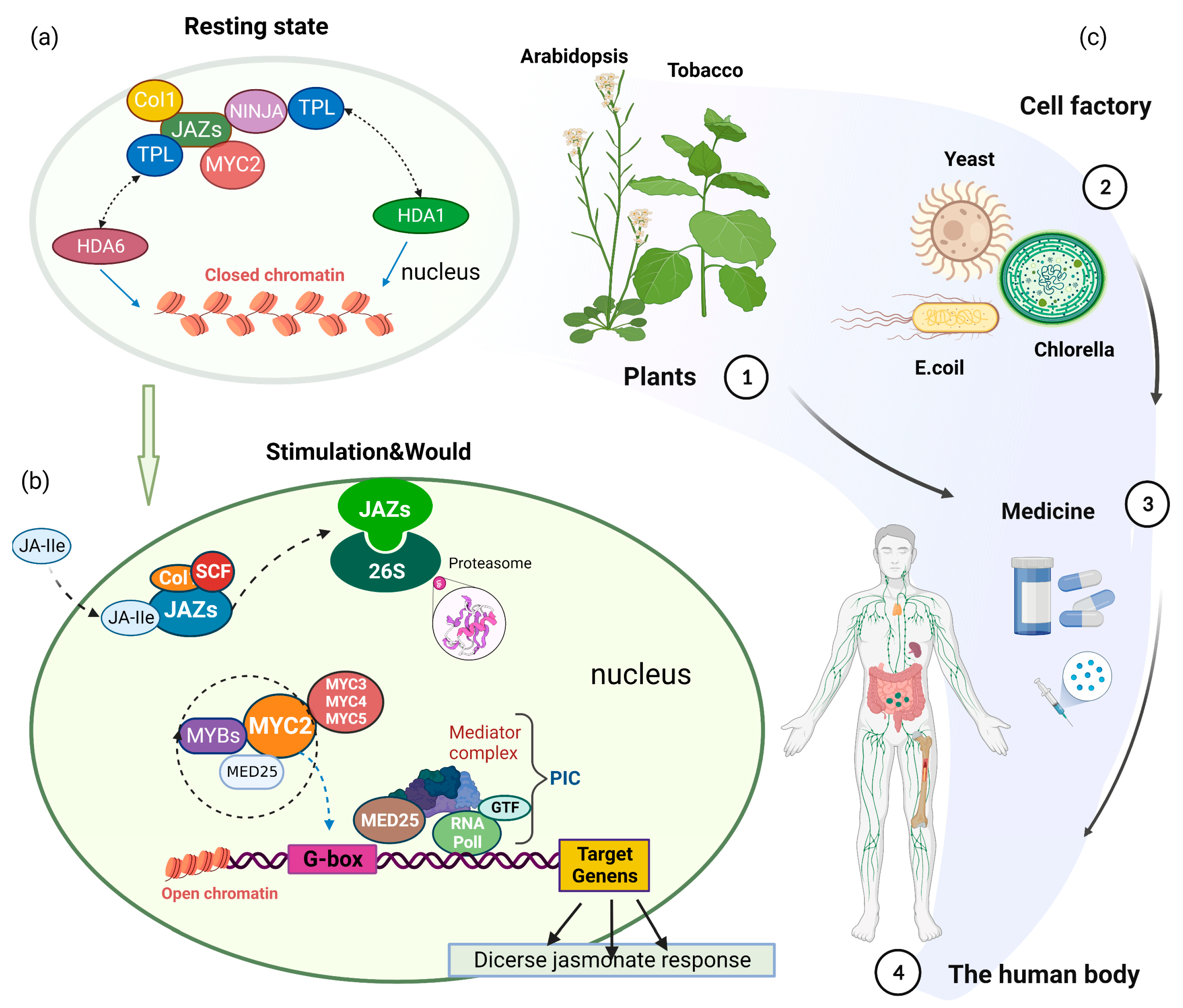
2. MYC2-Mediated JA Signaling
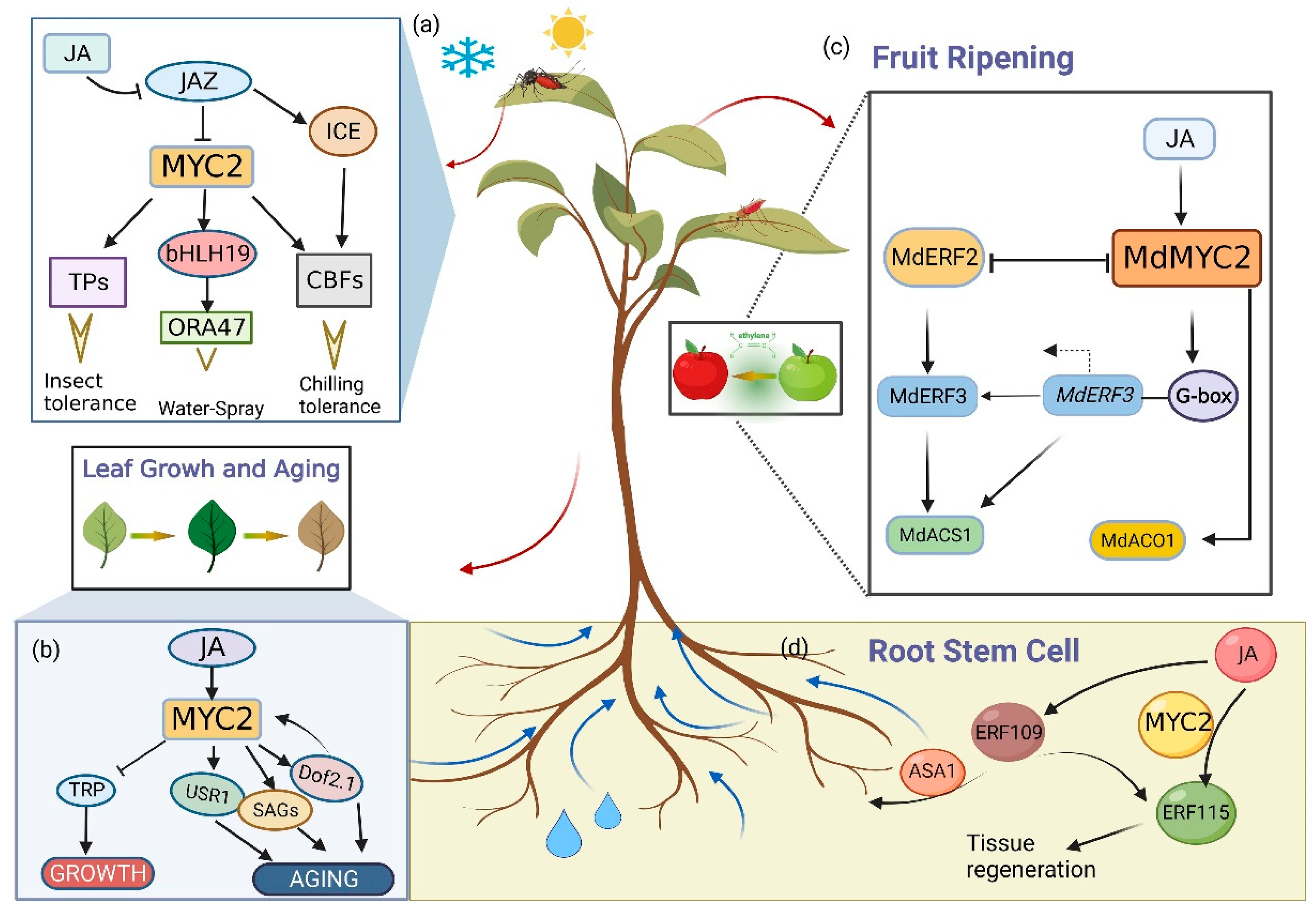
3. MYC Is Involved in the Regulation of Biotic and Abiotic Stress Conditions
4. MYC2 Is Involved in the Regulation of Plant Growth and Development
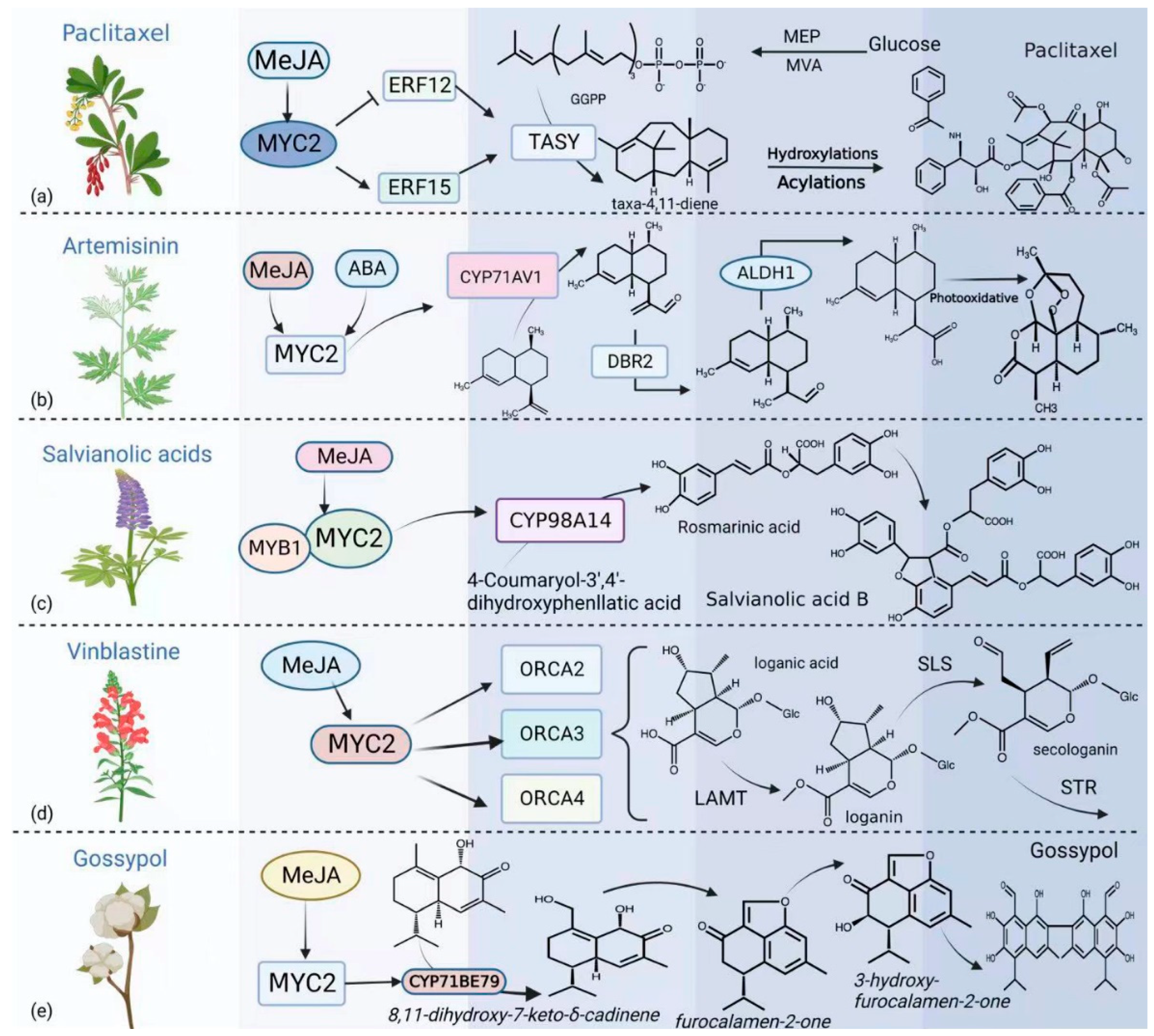
5. MYC2 Is Involved in the Regulation of Specialized Metabolites in Plants
6. Prospects of Application of MYC2 in Chassis-Based Synthesis of Natural Products
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Studham, M.E.; MacIntosh, G.C. Phytohormone Signaling Pathway Analysis Method for Comparing Hormone Responses in Plant-Pest Interactions. BMC Res. Notes 2012, 5, 392. [Google Scholar] [CrossRef]
- Aslam, S.; Gul, N.; Mir, M.A.; Asgher, M.; Al-Sulami, N.; Abulfaraj, A.A.; Qari, S. Role of Jasmonates, Calcium, and Glutathione in Plants to Combat Abiotic Stresses Through Precise Signaling Cascade. Front. Plant Sci. 2021, 12, 668029. [Google Scholar] [CrossRef] [PubMed]
- Hoo, S.C.; Koo, A.J.K.; Gao, X.; Jayanty, S.; Thines, B.; Jones, A.D.; Howe, G.A. Regulation and Function of Arabidopsis JASMONATE ZIM-Domain Genes in Response to Wounding and Herbivory. Plant Physiol. 2008, 146, 952–964. [Google Scholar] [CrossRef]
- Dombrecht, B.; Gang, P.X.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 Differentially Modulates Diverse Jasmonate-Dependent Functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef]
- Yamada, Y.; Koyama, T.; Sato, F. Basic Helix-Loop-Helix Transcription Factors and Regulation of Alkaloid Biosynthesis. Plant Signal. Behav. 2011, 6, 1627–1630. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Dutta, S.; Chattopadhyay, S. MYC2 Regulates ARR16, a Component of Cytokinin Signaling Pathways, in Arabidopsis Seedling Development. Plant Direct 2019, 3, e00177. [Google Scholar] [CrossRef]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef]
- Shin, J.; Heidrich, K.; Sanchez-Villarreal, A.; Parker, J.E.; Davis, S.J. TIME FOR COFFEE Represses Accumulation of the MYC2 Transcription Factor to Provide Time-of-Day Regulation of Jasmonate Signaling in Arabidopsis. Plant Cell 2012, 24, 2470–2482. [Google Scholar] [CrossRef]
- Zhang, Y.; Xing, H.; Wang, H.; Yu, L.; Yang, Z.; Meng, X.; Hu, P.; Fan, H. SlMYC2 Interacted with the SlTOR Promoter and Mediated JA Signaling to Regulate Growth and Fruit Quality in Tomato. Front. Plant Sci. 2022, 13, 1013445. [Google Scholar] [CrossRef]
- Zhang, P.J.; Zhao, C.; Ye, Z.H.; Yu, X.P. Trade-off between Defense Priming by Herbivore-Induced Plant Volatiles and Constitutive Defense in Tomato. Pest Manag. Sci. 2020, 76, 1893–1901. [Google Scholar] [CrossRef]
- Ballhorn, D.J.; Kautz, S.; Schädler, M. Induced Plant Defense via Volatile Production Is Dependent on Rhizobial Symbiosis. Oecologia 2013, 172, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Mao, Y.B.; Liu, Y.Q.; Chen, D.Y.; Chen, F.Y.; Fang, X.; Hong, G.J.; Wang, L.J.; Wang, J.W.; Chen, X.Y. Jasmonate Response Decay and Defense Metabolite Accumulation Contributes to Age-Regulated Dynamics of Plant Insect Resistance. Nat. Commun. 2017, 8, 13925. [Google Scholar] [CrossRef] [PubMed]
- Calabrò, S. Plant Secondary Metabolites. In Rumen Microbiology: From Evolution to Revolution; Springer: Delhi, India, 2015; Volume 10, pp. 153–159. [Google Scholar] [CrossRef]
- Twaij, B.M.; Hasan, M.N. Bioactive Secondary Metabolites from Plant Sources: Types, Synthesis, and Their Therapeutic Uses. Int. J. Plant Biol. 2022, 13, 4–14. [Google Scholar] [CrossRef]
- Greenwell, M.; Rahman, P.K.S.M. Medicinal Plants: Their Use in Anticancer Treatment. Int. J. Pharm. Sci. Res. 2015, 6, 4103–4112. [Google Scholar] [CrossRef]
- Zhang, Q.W.; Lin, L.G.; Ye, W.C. Techniques for Extraction and Isolation of Natural Products: A Comprehensive Review. Chinese Med. 2018, 13, 20. [Google Scholar] [CrossRef]
- Hörner, M.; Weber, W. Molecular Switches in Animal Cells. FEBS Lett. 2012, 586, 2084–2096. [Google Scholar] [CrossRef]
- Eguchi, A.; Lee, G.O.; Wan, F.; Erwin, G.S.; Ansari, A.Z. Controlling Gene Networks and Cell Fate with Precision-Targeted DNA-Binding Proteins and Small-Molecule-Based Genome Readers. Biochem. J. 2014, 462, 397–413. [Google Scholar] [CrossRef]
- Du, M.; Zhao, J.; Tzeng, D.T.W.; Liu, Y.; Deng, L.; Yang, T.; Zhai, Q.; Wu, F.; Huang, Z.; Zhou, M.; et al. MYC2 Orchestrates a Hierarchical Transcriptional Cascade That Regulates Jasmonate-Mediated Plant Immunity in Tomato. Plant Cell 2017, 29, 1883–1906. [Google Scholar] [CrossRef] [Green Version]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ Repressor Proteins Are Targets of the SCFCOI1 Complex during Jasmonate Signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Lian, T.-F.; Xu, Y.-P.; Li, L.-F.; Su, X.-D. Crystal Structure of Tetrameric Arabidopsis MYC2 Reveals the Mechanism of Enhanced Interaction with DNA. Cell Rep. 2017, 19, 1334–1342. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Alonso, M.M.; Sánchez-Parra, B.; Ortiz-García, P.; Santamaría, M.E.; Díaz, I.; Pollmann, S. Jasmonic Acid-Dependent Myc Transcription Factors Bind to a Tandem g-Box Motif in the Yucca8 and Yucca9 Promoters to Regulate Biotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 9768. [Google Scholar] [CrossRef] [PubMed]
- Goossens, J.; Swinnen, G.; Bossche, R.V.; Pauwels, L.; Goossens, A. Change of a Conserved Amino Acid in the MYC2 and MYC3 Transcription Factors Leads to Release of JAZ Repression and Increased Activity. New Phytol. 2015, 206, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Zhang, L.; Zhou, C.; Yu, C.W.; Chaikam, V. HDA6 Is Required for Jasmonate Response, Senescence and Flowering in Arabidopsis. J. Exp. Bot. 2008, 59, 225–234. [Google Scholar] [CrossRef]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate Perception by Inositol-Phosphate-Potentiated COI1-JAZ Co-Receptor. Nature 2010, 468, 400–407. [Google Scholar] [CrossRef]
- Zhang, C.; Lei, Y.; Lu, C.; Wang, L.; Wu, J. MYC2, MYC3, and MYC4 Function Additively in Wounding-Induced Jasmonic Acid Biosynthesis and Catabolism. J. Integr. Plant Biol. 2020, 62, 1159–1175. [Google Scholar] [CrossRef]
- Fu, Z.W.; Li, J.H.; Feng, Y.R.; Yuan, X.; Lu, Y.T. The Metabolite Methylglyoxal-Mediated Gene Expression Is Associated with Histone Methylglyoxalation. Nucleic Acids Res. 2021, 49, 1886–1899. [Google Scholar] [CrossRef]
- Walley, J.W.; Rowe, H.C.; Xiao, Y.; Chehab, E.W.; Kliebenstein, D.J.; Wagner, D.; Dehesh, K. The Chromatin Remodeler SPLAYED Regulates Specific Stress Signaling Pathways. PLoS Pathog. 2008, 4, e1000237. [Google Scholar] [CrossRef]
- You, Y.; Zhai, Q.; An, C.; Li, C. Leunig_homolog Mediates MYC2-Dependent Transcriptional Activation in Cooperation with the Coactivators HAC1 and MED25. Plant Cell 2019, 31, 2187–2205. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Q.; Li, C. The Plant Mediator Complex and Its Role in Jasmonate Signaling. J. Exp. Bot. 2019, 70, 3415–3424. [Google Scholar] [CrossRef]
- Zhai, Q.; Deng, L.; Li, C. Mediator Subunit MED25: At the Nexus of Jasmonate Signaling. Curr. Opin. Plant Biol. 2020, 57, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C. Termination in Jasmonate Signaling by MYC2 and MTBs. Trends Plant Sci. 2019, 24, 667–669. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. Myc2 Regulates the Termination of Jasmonate Signaling via an Autoregulatory Negative Feedback Loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef]
- Aleman, F.; Yazaki, J.; Lee, M.; Takahashi, Y.; Kim, A.Y.; Li, Z.; Kinoshita, T.; Ecker, J.R.; Schroeder, J.I. An ABA-Increased Interaction of the PYL6 ABA Receptor with MYC2 Transcription Factor: A Putative Link of ABA and JA Signaling. Sci. Rep. 2016, 6, 28941. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J. Derepression of Ethylene-Stabilized Transcription Factors (EIN3/EIL1) Mediates Jasmonate and Ethylene Signaling Synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef]
- Hou, X.; Lee, L.Y.C.; Xia, K.; Yan, Y.; Yu, H. DELLAs Modulate Jasmonate Signaling via Competitive Binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of Plant Hormones in Plant Defence Responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Spoel, S.H.; Koornneef, A.; Claessens, S.M.C.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Buchala, A.J.; Métraux, J.P.; Brown, R.; Kazan, K.; et al. NPR1 Modulates Cross-Talk between Salicylate- and Jasmonate-Dependent Defense Pathways through a Novel Function in the Cytosol. Plant Cell 2003, 15, 760–770. [Google Scholar] [CrossRef]
- Nomoto, M.; Skelly, M.J.; Itaya, T.; Mori, T.; Suzuki, T.; Matsushita, T.; Tokizawa, M.; Kuwata, K.; Mori, H.; Yamamoto, Y.Y.; et al. Suppression of MYC Transcription Activators by the Immune Cofactor NPR1 Fine-Tunes Plant Immune Responses. Cell Rep. 2021, 37, 110125. [Google Scholar] [CrossRef]
- Luo, F.; Zhang, Q.; Xin, H.; Liu, H.; Yang, H.; Doblin, M.S.; Bacic, A.; Li, L. A Phytochrome B-PIF4-MYC2/MYC4 Module Inhibits Secondary Cell Wall Thickening in Response to Shaded Light. Plant Commun. 2022, 3, 100416. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Prasad, V.B.R.; Chattopadhyay, S. Functional Interconnection of MYC2 and SPA1 in the Photomorphogenic Seedling Development of Arabidopsis. Plant Physiol. 2010, 154, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Chattopadhyay, S. MYC2, a BHLH Transcription Factor, Modulates the Adult Phenotype of SPA1. Plant Signal. Behav. 2010, 5, 1650–1652. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, V.; Meier, S.; Petersen, L.N.; Ingle, R.A.; Roden, L.C. Defence Responses of Arabidopsis Thaliana to Infection by Pseudomonas Syringae Are Regulated by the Circadian Clock. PLoS ONE 2011, 6, e26968. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Srivastava, A.K.; Roy, D.; Mansi, M.; Gough, C.; Bhagat, P.K.; Zhang, C.; Sadanandom, A. The Conjugation of SUMO to the Transcription Factor MYC2 Functions in Blue Light-Mediated Seedling Development in Arabidopsis. Plant Cell 2022, 34, 2892–2906. [Google Scholar] [CrossRef]
- Chico, J.M.; Lechner, E.; Fernandez-Barbero, G.; Canibano, E.; García-Casado, G.; Franco-Zorrilla, J.M.; Hammann, P.; Zamarreño, A.M.; García-Mina, J.M.; Rubio, V.; et al. CUL3BPM E3 Ubiquitin Ligases Regulate MYC2, MYC3, and MYC4 Stability and JA Responses. Proc. Natl. Acad. Sci. USA 2020, 117, 6205–6215. [Google Scholar] [CrossRef] [PubMed]
- Pesch, M.; Schultheiß, I.; Digiuni, S.; Uhrig, J.F.; Hülskamp, M. Mutual Control of Intracellular Localisation of the Patterning Proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development 2013, 140, 3456–3467. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, X.; Zhu, D.; Cui, S.; Li, X.; Cao, Y.; Ma, L. A Single Amino Acid Substitution in IIIf Subfamily of Basic Helix-Loop-Helix Transcription Factor AtMYC1 Leads to Trichome and Root Hair Patterning Defects by Abolishing Its Interaction with Partner Proteins in Arabidopsis. J. Biol. Chem. 2012, 287, 14109–14121. [Google Scholar] [CrossRef]
- Symonds, V.V.; Hatlestad, G.; Lloyd, A.M. Natural Allelic Variation Defines a Role for ATMYC1: Trichome Cell Fate Determination. PLoS Genet. 2011, 7, e1002069. [Google Scholar] [CrossRef]
- Clay, N.K.; Adio, A.M.; Denoux, C.; Jander, G.; Ausubel, M. Glucosinola Required Fo Innate Immune Response. Science 2016, 323, 95–101. [Google Scholar] [CrossRef] [Green Version]
- Rajniak, J.; Barco, B.; Clay, N.K.; Sattely, E.S. A New Cyanogenic Metabolite in Arabidopsis Required for Inducible Pathogen Defence. Nature 2015, 525, 376–379. [Google Scholar] [CrossRef]
- Andreou, A.; Feussner, I. Lipoxygenases—Structure and Reaction Mechanism. Phytochemistry 2009, 70, 1504–1510. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, Metabolism, and Signaling by Proteins Activating and Repressing Transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef]
- Lee, D.S.; Nioche, P.; Hamberg, M.; Raman, C.S. Structural Insights into the Evolutionary Paths of Oxylipin Biosynthetic Enzymes. Nature 2008, 455, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Stintzi, A.; Browse, J. The Arabidopsis Male-Sterile Mutant, Opr3, Lacks the 12-Oxophytodienoic Acid Reductase Required for Jasmonate Synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef]
- Staswick, P.E.; Tiryaki, I. The Oxylipin Signal Jasmonic Acid Is Activated by an Enzyme That Conjugate It to Isoleucine in Arabidopsis W inside Box Sign. Plant Cell 2004, 16, 2117–2127. [Google Scholar] [CrossRef] [PubMed]
- Zamioudis, C.; Pieterse, C.M.J. Modulation of Host Immunity by Beneficial Microbes. Mol. Plant-Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Li, P.; Chen, Q.Y.; Chen, X.Y.; Yan, Z.W.; Wang, M.Y.; Mao, Y.B. Differential Contributions of MYCs to Insect Defense Reveals Flavonoids Alleviating Growth Inhibition Caused by Wounding in Arabidopsis. Front. Plant Sci. 2021, 12, 700555. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, X.; Gong, Y.; Wang, D.; Xu, D.; Wang, N.; Sun, Y.; Gao, L.; Liu, S.S.; Deng, X.W.; et al. Red-Light Is an Environmental Effector for Mutualism between Begomovirus and Its Vector Whitefly. PLoS Pathog. 2021, 17, e1008770. [Google Scholar] [CrossRef]
- Hichri, I.; Heppel, S.C.; Pillet, J.; Léon, C.; Czemmel, S.; Delrot, S.; Lauvergeat, V.; Bogs, J. The Basic Helix-Loop-Helix Transcription Factor MYC1 Is Involved in the Regulation of the Flavonoid Biosynthesis Pathway in Grapevine. Mol. Plant 2010, 3, 509–523. [Google Scholar] [CrossRef]
- Xu, X.; Fang, P.; Zhang, H.; Chi, C.; Song, L.; Xia, X.; Shi, K.; Zhou, Y.; Zhou, J.; Yu, J. Strigolactones Positively Regulate Defense against Root-Knot Nematodes in Tomato. J. Exp. Bot. 2019, 70, 1325–1337. [Google Scholar] [CrossRef]
- Zhang, X.; Peng, H.; Zhu, S.; Xing, J.; Li, X.; Zhu, Z.; Zheng, J.; Wang, L.; Wang, B.; Chen, J.; et al. Nematode-Encoded RALF Peptide Mimics Facilitate Parasitism of Plants through the FERONIA Receptor Kinase. Mol. Plant 2020, 13, 1434–1454. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, X.; Zu, H.; Zeng, X.; Baldwin, I.T.; Lou, Y.; Li, R. Molecular Dissection of Rice Phytohormone Signaling Involved in Resistance to a Piercing-Sucking Herbivore. New Phytol. 2021, 230, 1639–1652. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, G.; De Meyer, M.; Pollier, J.; Molina-Hidalgo, F.J.; Ceulemans, E.; Venegas-Molina, J.; De Milde, L.; Fernández-Calvo, P.; Ron, M.; Pauwels, L.; et al. The Basic Helix–Loop–Helix Transcription Factors MYC1 and MYC2 Have a Dual Role in the Regulation of Constitutive and Stress-Inducible Specialized Metabolism in Tomato. New Phytol. 2022, 236, 911–928. [Google Scholar] [CrossRef]
- Panda, S.; Jozwiak, A.; Sonawane, P.D.; Szymanski, J.; Kazachkova, Y.; Vainer, A.; Kilambi, H.V.; Almekias-Siegl, E.; Dikaya, V.; Bocobza, S.; et al. Steroidal Alkaloids Defence Metabolism and Plant Growth Are Modulated by the Joint Action of Gibberellin and Jasmonate Signalling. New Phytol. 2022, 233, 1220–1237. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, I.; Takehara, Y.; Ota, M.; Imada, K.; Sasaki, K.; Kajihara, H.; Sakai, S.; Jogaiah, S.; Ito, S.-i. Magnesium Oxide Induces Immunity against Fusarium Wilt by Triggering the Jasmonic Acid Signaling Pathway in Tomato. J. Biotechnol. 2021, 325, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Uji, Y.; Taniguchi, S.; Tamaoki, D.; Shishido, H.; Akimitsu, K.; Gomi, K. Overexpression of OsMYC2 Results in the Up-Regulation of Early JA-Rresponsive Genes and Bacterial Blight Resistance in Rice. Plant Cell Physiol. 2016, 57, 1814–1827. [Google Scholar] [CrossRef]
- Song, S.; Huang, H.; Gao, H.; Wang, J.; Wu, D.; Liu, X.; Yang, S.; Zhai, Q.; Li, C.; Qi, T.; et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 Modulates Antagonism between Jasmonate and Ethylene Signaling in Arabidopsis. Plant Cell 2014, 26, 263–279. [Google Scholar] [CrossRef]
- Woldemariam, M.G.; Baldwin, I.T.; Galis, I. Transcriptional Regulation of Plant Inducible Defenses against Herbivores: A Mini-Review. J. Plant Interact. 2011, 6, 113–119. [Google Scholar] [CrossRef]
- Gautam, J.K.; Giri, M.K.; Singh, D.; Chattopadhyay, S.; Nandi, A.K. MYC2 Influences Salicylic Acid Biosynthesis and Defense against Bacterial Pathogens in Arabidopsis Thaliana. Physiol. Plant. 2021, 173, 2248–2261. [Google Scholar] [CrossRef]
- van Moerkercke, A.; Duncan, O.; Zander, M.; Simura, J.; Broda, M.; Bossche, R.V.; Lewsey, M.G.; Lama, S.; Singh, K.B.; Ljung, K.; et al. A MYC2/MYC3/MYC4-Dependent Transcription Factor Network Regulates Water Spray-Responsive Gene Expression and Jasmonate Levels. Proc. Natl. Acad. Sci. USA 2019, 116, 23345–23356. [Google Scholar] [CrossRef]
- Wei, X.; Mao, L.; Wei, X.; Xia, M.; Xu, C. MYB41, MYB107, and MYC2 Promote ABA-Mediated Primary Fatty Alcohol Accumulation via Activation of AchnFAR in Wound Suberization in Kiwifruit. Hortic. Res. 2020, 7, 86. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (BHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, A.; Zhou, L.; He, P.; Datta, A. Detecting Drought Regulators Using Stochastic Inference in Bayesian Networks. PLoS ONE 2021, 16, e0255486. [Google Scholar] [CrossRef]
- Liu, H.; Cui, P.; Zhang, B.; Zhu, J.; Liu, C.; Li, Q. Binding of the Transcription Factor MYC2-like to the ABRE of the OsCYP2 Promoter Enhances Salt Tolerance in Oryza Sativa. PLoS ONE 2022, 17, e0276075. [Google Scholar] [CrossRef]
- Kashyap, P.; Deswal, R. Two ICE Isoforms Showing Differential Transcriptional Regulation by Cold and Hormones Participate in Brassica Juncea Cold Stress Signaling. Gene 2019, 695, 32–41. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Li, X. Role of Jasmonate Signaling Pathway in Resistance to Dehydration Stress in Arabidopsis. Acta Physiol. Plant. 2019, 41, 100. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, H.; Liu, W.; Wang, N.; Qu, C.; Jiang, S.; Fang, H.; Zhang, Z.; Chen, X. Methyl Jasmonate Enhances Apple’ Cold Tolerance through the JAZ–MYC2 Pathway. Plant Cell Tissue Organ Cult. 2019, 136, 75–84. [Google Scholar] [CrossRef]
- Ming, R.; Zhang, Y.; Wang, Y.; Khan, M.; Dahro, B.; Liu, J.H. The JA-Responsive MYC2-BADH-like Transcriptional Regulatory Module in Poncirus Trifoliata Contributes to Cold Tolerance by Modulation of Glycine Betaine Biosynthesis. New Phytol. 2021, 229, 2730–2750. [Google Scholar] [CrossRef]
- Tiwari, S.; Prasad, V.; Chauhan, P.S.; Lata, C. Bacillus Amyloliquefaciens Confers Tolerance to Various Abiotic Stresses and Modulates Plant Response to Phytohormones through Osmoprotection and Gene Expression Regulation in Rice. Front. Plant Sci. 2017, 8, 1510. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.F.; Yu, C.P.; Wu, Y.H.; Lu, M.Y.J.; Tu, S.L.; Wu, S.H.; Shiu, S.H.; Ku, M.S.B.; Li, W.H. Elevated Auxin Biosynthesis and Transport Underlie High Vein Density in C4 Leaves. Proc. Natl. Acad. Sci. USA 2017, 114, E6884–E6891. [Google Scholar] [CrossRef]
- Zhuo, M.; Sakuraba, Y.; Yanagisawa, S. A Jasmonate-Activated MYC2-Dof2.1-MYC2 Transcriptional Loop Promotes Leaf Senescence in Arabidopsis. Plant Cell 2020, 32, 242–262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ji, T.T.; Li, T.T.; Tian, Y.Y.; Wang, L.F.; Liu, W.C. Jasmonic Acid Promotes Leaf Senescence through MYC2-Mediated Repression of CATALASE2 Expression in Arabidopsis. Plant Sci. 2020, 299, 110604. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Wang, J.; Huang, H.; Liu, B.; Gao, H.; Liu, Y.; Song, S.; Xie, D. Regulation of Jasmonate-Induced Leaf Senescence by Antagonism between BHLH Subgroup IIIe and IIId Factors in Arabidopsis. Plant Cell 2015, 27, 1634–1649. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, M.; Guo, Y. Ring/U-Box Protein AtUSR1 Functions in Promoting Leaf Senescence Through JA Signaling Pathway in Arabidopsis. Front. Plant Sci. 2020, 11, 608589. [Google Scholar] [CrossRef]
- Lee, B.R.; Zaman, R.; La, V.H.; Bae, D.W.; Kim, T.H. Ethephon-Induced Ethylene Enhances Starch Degradation and Sucrose Transport with an Interactive Abscisic Acid-Mediated Manner in Mature Leaves of Oilseed Rape (Brassica Napus L.). Plants 2021, 10, 1670. [Google Scholar] [CrossRef]
- Ding, F.; Wang, C.; Xu, N.; Zhang, S.; Wang, M. SlMYC2 Mediates Jasmonate-Induced Tomato Leaf Senescence by Promoting Chlorophyll Degradation and Repressing Carbon Fixation. Plant Physiol. Biochem. 2022, 180, 27–34. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, J.; Zhai, Q.; Zhou, W.; Qi, L.; Xu, L.; Wang, B.; Chen, R.; Jiang, H.; Qi, J.; et al. The Basic Helix-Loop-Helix Transcription Factor Myc2 Directly Represses Plethora Expression during Jasmonate-Mediated Modulation of the Root Stem Cell Niche in Arabidopsis. Plant Cell 2011, 23, 3335–3352. [Google Scholar] [CrossRef]
- Marhava, P.; Hoermayer, L.; Yoshida, S.; Marhavý, P.; Benková, E.; Friml, J. Re-Activation of Stem Cell Pathways for Pattern Restoration in Plant Wound Healing. Cell 2019, 177, 957–969.e13. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, C.L.; Cui, M.; Zhou, W.J.; Wu, H.L.; Ling, H.Q. Four IVa BHLH Transcription Factors Are Novel Interactors of FIT and Mediate JA Inhibition of Iron Uptake in Arabidopsis. Mol. Plant 2018, 11, 1166–1183. [Google Scholar] [CrossRef] [Green Version]
- Xing, J.; Cao, X.; Zhang, M.; Wei, X.; Zhang, J. Plant Nitrogen Availability and Crosstalk with Phytohormones Signalings and Their Biotechnology Breeding Application in Crops. Plant Biotechnol. 2022, 12, 27. [Google Scholar] [CrossRef]
- Wang, P.; Xu, X.; Tang, Z.; Zhang, W.; Huang, X.Y.; Zhao, F.J. Oswrky28 Regulates Phosphate and Arsenate Accumulation, Root System Architecture and Fertility in Rice. Front. Plant Sci. 2018, 9, 1330. [Google Scholar] [CrossRef] [PubMed]
- Shigeyama, T.; Tominaga, A.; Arima, S.; Sakai, T.; Inada, S.; Jikumaru, Y.; Kamiya, Y.; Uchiumi, T.; Abe, M.; Hashiguchi, M.; et al. Additional Cause for Reduced JA-Ile in the Root of a Lotus Japonicus PhyB Mutant. Plant Signal. Behav. 2012, 7, 746–748. [Google Scholar] [CrossRef]
- Li, T.; Xu, Y.; Zhang, L.; Ji, Y.; Tan, D.; Yuan, H.; Wang, A. The Jasmonate-Activated Transcription Factor MdMYC2 Regulates ETHYLENE RESPONSE FACTOR and Ethylene Biosynthetic Genes to Promote Ethylene Biosynthesis during Apple Fruit Ripening. Plant Cell 2017, 29, 1316–1334. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Sun, H.; Han, Z.; Wang, S.; Wang, T.; Li, Q.; Tian, J.; Wang, Y.; Zhang, X.; Xu, X.; et al. ERF4 Affects Fruit Ripening by Acting as a JAZ Interactor between Ethylene and Jasmonic Acid Hormone Signaling Pathways. Hortic. Plant J. 2022, 8, 689–699. [Google Scholar] [CrossRef]
- Yan, F.; Cai, T.; Wu, Y.; Chen, S.; Chen, J. Physiological and Transcriptomics Analysis of the Effect of Recombinant Serine Protease on the Preservation of Loquat. Genomics 2021, 113, 3750–3761. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, Y.; Wu, X.; Wang, L.; Li, J.; Wan, M.; Jia, B.; Ye, Z.; Liu, L.; Tang, X.; et al. MYB1R1 and MYC2 Regulate ω-3 Fatty Acid Desaturase Involved in ABA-Mediated Suberization in the Russet Skin of a Mutant of ‘Dangshansuli’ (Pyrus Bretschneideri Rehd.). Front. Plant Sci. 2022, 13, 910938. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, S.; Qu, Y.; Gao, R.; Xiao, Y.; Wang, Z.; Zhai, R.; Yang, C.; Xu, L. Jasmonic Acid and Ethylene Participate in the Gibberellin-Induced Ovule Programmed Cell Death Process in Seedless Pear ‘1913’ (Pyrus Hybrid). Int. J. Mol. Sci. 2021, 22, 9844. [Google Scholar] [CrossRef]
- Baud, S.; Lepiniec, L. Regulation of de Novo Fatty Acid Synthesis in Maturing Oilseeds of Arabidopsis. Plant Physiol. Biochem. 2009, 47, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Qi, S.; Liu, K.; Li, D.; Jin, C.; Li, Z.; Huang, G.; Hai, J.; Zhang, M.; Chen, M. MYC2, MYC3, and MYC4 Function Redundantly in Seed Storage Protein Accumulation in Arabidopsis. Plant Physiol. Biochem. 2016, 108, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Du, X.; Zhu, Y.; Wang, Z.; Hua, S.; Li, Z.; Guo, W.; Zhang, G.; Peng, J.; Jiang, L. Seed Fatty Acid Reducer Acts Downstream of Gibberellin Signalling Pathway to Lower Seed Fatty Acid Storage in Arabidopsis. Plant Cell Environ. 2012, 35, 2155–2169. [Google Scholar] [CrossRef]
- Cai, Q.; Yuan, Z.; Chen, M.; Yin, C.; Luo, Z.; Zhao, X.; Liang, W.; Hu, J.; Zhang, D. Jasmonic Acid Regulates Spikelet Development in Rice. Nat. Commun. 2014, 5, 3476. [Google Scholar] [CrossRef] [PubMed]
- Bian, S.; Tian, T.; Ding, Y.; Yan, N.; Zhang, Z.; Zhang, H.; Wang, C.; Fang, N.; Liu, Y. BHLH Transcription Factor NtMYC2a Regulates Carbohydrate Metabolism during the Pollen Development of Tobacco (Nicotiana Tabacum L. Cv. TN90). Plants 2022, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Zha, S.; Luo, Y.; Li, L.; Wang, S.; Wu, S.; Cheng, S.; Li, L. JAZ1-3 and MYC2-1 Synergistically Regulate the Transformation from Completely Mixed Flower Buds to Female Flower Buds in Castanea Mollisima. Int. J. Mol. Sci. 2022, 23, 6452. [Google Scholar] [CrossRef]
- Huang, H.; Gao, H.; Liu, B.; Qi, T.; Tong, J.; Xiao, L.; Xie, D.; Song, S. Arabidopsis MYB24 Regulates Jasmonate-Mediated Stamen Development. Front. Plant Sci. 2017, 8, 1525. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Gong, Y.; Liu, B.; Wu, D.; Zhang, M.; Xie, D.; Song, S. The Della Proteins Interact with MYB21 and MYB24 to Regulate Filament Elongation in Arabidopsis. BMC Plant Biol. 2020, 20, 64. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of Jasmonate-Mediated Stamen Development and Seed Production by a BHLH-MYB Complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef]
- Chen, X.; Wang, D.D.; Fang, X.; Chen, X.Y.; Mao, Y.B. Plant Specialized Metabolism Regulated by Jasmonate Signaling. Plant Cell Physiol. 2019, 60, 2638–2647. [Google Scholar] [CrossRef]
- Afrin, S.; Huang, J.J.; Luo, Z.Y. JA-Mediated Transcriptional Regulation of Secondary Metabolism in Medicinal Plants. Sci. Bull. 2015, 60, 1062–1072. [Google Scholar] [CrossRef]
- Zhou, M.; Memelink, J. Jasmonate-Responsive Transcription Factors Regulating Plant Secondary Metabolism. Biotechnol. Adv. 2016, 34, 441–449. [Google Scholar] [CrossRef]
- Zhang, M.; Jin, X.; Chen, Y.; Wei, M.; Liao, W.; Zhao, S.; Fu, C.; Yu, L. TcMYC2a, a Basic Helix–Loop–Helix Transcription Factor, Transduces JA-Signals and Regulates Taxol Biosynthesis in Taxus Chinensis. Front. Plant Sci. 2018, 9, 863. [Google Scholar] [CrossRef]
- Yu, Z.X.; Li, J.X.; Yang, C.Q.; Hu, W.L.; Wang, L.J.; Chen, X.Y. The Jasmonate-Responsive AP2/ERF Transcription Factors AaERF1 and AaERF2 Positively Regulate Artemisinin Biosynthesis in Artemisia Annua L. Mol. Plant 2012, 5, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Huang, H.; Xie, L.; Hao, X.; Kayani, S.I.; Liu, H.; Qin, W.; Chen, T.; Pan, Q.; Liu, P.; et al. Basic Helix-Loop-Helix Transcription Factors AabHLH2 and AabHLH3 Function Antagonistically With AaMYC2 and Are Negative Regulators in Artemisinin Biosynthesis. Front. Plant Sci. 2022, 13, 885622. [Google Scholar] [CrossRef]
- Kayani, S.I.; Shen, Q.; Ma, Y.; Fu, X.; Xie, L.; Zhong, Y.; Tiantian, C.; Pan, Q.; Li, L.; Rahman, S.U.; et al. The YABBY Family Transcription Factor AaYABBY5 Directly Targets Cytochrome P450 Monooxygenase (CYP71AV1) and Double-Bond Reductase 2 (DBR2) Involved in Artemisinin Biosynthesis in Artemisia Annua. Front. Plant Sci. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Du, T.; Niu, J.; Su, J.; Li, S.; Guo, X.; Li, L.; Cao, X.; Kang, J. SmbHLH37 Functions Antagonistically with SmMYC2 in Regulating Jasmonate-Mediated Biosynthesis of Phenolic Acids in Salvia Miltiorrhiza. Front. Plant Sci. 2018, 871, 1720. [Google Scholar] [CrossRef]
- Zhang, H.; Hedhili, S.; Montiel, G.; Zhang, Y.; Chatel, G.; Pré, M.; Gantet, P.; Memelink, J. The Basic Helix-Loop-Helix Transcription Factor CrMYC2 Controls the Jasmonate-Responsive Expression of the ORCA Genes That Regulate Alkaloid Biosynthesis in Catharanthus Roseus. Plant J. 2011, 67, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Sunilkumar, G.; Campbell, L.A.M.; Puckhaber, L.; Stipanovic, R.D.; Rathore, K.S. Engineering Cottonseed for Use in Human Nutrition by Tissue-Specific Reduction of Toxic Gossypol. Proc. Natl. Acad. Sci. USA 2006, 103, 18054–18059. [Google Scholar] [CrossRef]
- Han, X.; Xing, Y.; Zhu, Y.; Luo, L.; Liu, L.; Zhai, Y.; Wang, W.; Shao, R.; Ren, M.; Li, F.; et al. GhMYC2 Activates Cytochrome P450 Gene CYP71BE79 to Regulate Gossypol Biosynthesis in Cotton. Planta 2022, 256, 63. [Google Scholar] [CrossRef]
- Huo, Y.; Zhang, J.; Zhang, B.; Chen, L.; Zhang, X.; Zhu, C. Myc2 Transcription Factors Twmyc2a and Twmyc2b Negatively Regulate Triptolide Biosynthesis in Tripterygium Wilfordii Hairy Roots. Plants 2021, 10, 679. [Google Scholar] [CrossRef]
- Ribeiro, B.; Lacchini, E.; Bicalho, K.U.; Mertens, J.; Arendt, P.; Bossche, R.V.; Calegario, G.; Gryffroy, L.; Ceulemans, E.; Buitink, J.; et al. A Seed-Specific Regulator of Triterpene Saponin Biosynthesis in Medicago Truncatula. Plant Cell 2020, 32, 2020–2042. [Google Scholar] [CrossRef]
- Zhai, J.; Hao, H.; Xiao, H.; Cao, Y.; Lin, X.; Huang, X. Identification of JAZ-Interacting MYC Transcription Factors Involved in Latex Drainage in Hevea Brasiliensis. Sci. Rep. 2018, 8, 909. [Google Scholar] [CrossRef]
- Guo, D.; Li, H.L.; Wang, Y.; Zhu, J.H.; Peng, S.Q. A Myelocytomatosis Transcription Factor from Hevea Brasiliensis Positively Regulates the Expression of the Small Rubber Particle Protein Gene. Ind. Crops Prod. 2019, 133, 90–97. [Google Scholar] [CrossRef]
- Ke, J.; Wang, B.; Yoshikuni, Y. Microbiome Engineering: Synthetic Biology of Plant-Associated Microbiomes in Sustainable Agriculture. Trends Biotechnol. 2021, 39, 244–261. [Google Scholar] [CrossRef] [PubMed]
- Petzold, C.J.; Chan, L.J.G.; Nhan, M.; Adams, P.D. Analytics for Metabolic Engineering. Front. Bioeng. Biotechnol. 2015, 3, 135. [Google Scholar] [CrossRef] [PubMed]
- Garner, K.L. Principles of Synthetic Biology. Essays Biochem. 2021, 65, 791–811. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, P.; Zhao, Q.; Huang, A.C. Making Small Molecules in Plants: A Chassis for Synthetic Biology-based Production of Plant Natural Products. J. Integr. Plant Biol. 2022, 4, 29. [Google Scholar] [CrossRef]
- Yang, Y.; Alexander, T.; Ahkami, A.H.; Neal, C.; Stewart, C.N., Jr.; Blumwald, E. Plant Synthetic Biology Innovations for Biofuels and Bioproducts. Trends in Biotech. 2022, 40, 1454–1468. [Google Scholar] [CrossRef]
- Belcher, M.S.; Vuu, K.M.; Zhou, A.; Mansoori, N.; Ramos, A.A.; Thompson, M.G.; Scheller, H.V.; Loqué, D.; Shih, P.M. Design of Orthogonal Regulatory Systems for Modulating Gene Expression in Plants. Nat. Chem. Biol. 2020, 16, 857–865. [Google Scholar] [CrossRef]
- Boehm, C.R.; Bock, R. Recent Advances and Current Challenges in Synthetic Biology of the Plastid Genetic System and Metabolism. Plant Physiol. 2019, 179, 794–802. [Google Scholar] [CrossRef]
- Chae, L.; Kim, T.; Nilo-poyanco, R.; Rhee, S.Y.; Science, S.; Series, N.; May, N.; Chae, L.; Kim, T.; Nilo-poyanco, R.; et al. Genomic Signatures of of Specialized Metabolism in Plants. Science 2014, 344, 510–513. [Google Scholar] [CrossRef]
- Drábková, L.Z.; Dobrev, P.I.; Motyka, V. Phytohormone Profiling across the Bryophytes. PLoS ONE 2015, 10, e0125411. [Google Scholar] [CrossRef]
- Abdala, G.; Miersch, O.; Correa, N.; Rosas, S. Detection of Jasmonic Acid in Cultures of Escherichia Coli and Saccharomyces Cerevisiae. Nat. Prod. Lett. 1999, 14, 55–63. [Google Scholar] [CrossRef]
- Hu, Y.J.; Gu, C.C.; Wang, X.F.; Min, L.; Li, C.C. Asymmetric Total Synthesis of Taxol. J. Am. Chem. Soc. 2021, 143, 17862–17870. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.A.K.; Der, B.S.; Shin, J.; Vaidyanathan, P.; Paralanov, V.; Strychalski, E.A.; Ross, D.; Densmore, D.; Voigt, C.A. Genetic Circuit Design Automation. Science 2016, 352, aac7341. [Google Scholar] [CrossRef]
- Zhang, J.; Hansen, L.G.; Gudich, O.; Viehrig, K.; Lassen, L.M.M.; Schrübbers, L.; Adhikari, K.B.; Rubaszka, P.; Carrasquer-Alvarez, E.; Chen, L.; et al. A Microbial Supply Chain for Production of the Anti-Cancer Drug Vinblastine. Nature 2022, 609, 341–347. [Google Scholar] [CrossRef]
- Ro, D.K.; Paradise, E.M.; Quellet, M.; Fisher, K.J.; Newman, K.L.; Ndungu, J.M.; Ho, K.A.; Eachus, R.A.; Ham, T.S.; Kirby, J.; et al. Production of the Antimalarial Drug Precursor Artemisinic Acid in Engineered Yeast. Nature 2006, 440, 940–943. [Google Scholar] [CrossRef]
- Xiong, X.; Gou, J.; Liao, Q.; Li, Y.; Zhou, Q.; Bi, G.; Li, C.; Du, R.; Wang, X.; Sun, T.; et al. The Taxus Genome Provides Insights into Paclitaxel Biosynthesis. Nat. Plants 2021, 7, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Mutanda, I.; Wang, K.; Yang, L.; Wang, J.; Wang, Y. Chloroplastic Metabolic Engineering Coupled with Isoprenoid Pool Enhancement for Committed Taxanes Biosynthesis in Nicotiana Benthamiana. Nat. Commun. 2019, 10, 4850. [Google Scholar] [CrossRef]
- Zhou, K.; Qiao, K.; Edgar, S.; Stephanopoulos, G. Distributing a Metabolic Pathway among a Microbial Consortium Enhances Production of Natural Products. Nat. Biotechnol. 2015, 33, 377–383. [Google Scholar] [CrossRef]
- Zhu, Q.; Yu, S.; Zeng, D.; Liu, H.; Wang, H.; Yang, Z.; Xie, X.; Shen, R.; Tan, J.; Li, H.; et al. Development of “Purple Endosperm Rice” by Engineering Anthocyanin Biosynthesis in the Endosperm with a High-Efficiency Transgene Stacking System. Mol. Plant 2017, 10, 918–929. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.J.; Kim, Y.M.; Lee, J.Y.; Kim, S.J.; Cho, K.C.; Chandrasekhar, T.; Song, P.S.; Woo, Y.M.; Kim, J.-I. Production of Purple-Colored Creeping Bentgrass Using Maize Transcription Factor Genes Pl and Lc through Agrobacterium-Mediated Transformation. Plant Cell Rep. 2009, 28, 397–406. [Google Scholar] [CrossRef]
- Bovy, A.; De Vos, R.; Kemper, M.; Schijlen, E.; Pertejo, M.A.; Muir, S.; Collins, G.; Robinson, S.; Verhoeyen, M.; Hughes, S.; et al. High-Flavonol Tomatoes Resulting from the Heterologous Expression of the Maize Transcription Factor Genes LC and C1. Plant Cell 2002, 14, 2509–2526. [Google Scholar] [CrossRef] [PubMed]
- Moses, T.; Mehrshahi, P.; Smith, A.G.; Goossens, A. Synthetic Biology Approaches for the Production of Plant Metabolites in Unicellular Organisms. J. Exp. Bot. 2017, 68, 4057–4074. [Google Scholar] [CrossRef]
- Yao, L.; Zhang, H.; Liu, Y.; Ji, Q.; Xie, J.; Zhang, R.; Huang, L.; Mei, K.; Wang, J.; Gao, W. Engineering of Triterpene Metabolism and Overexpression of the Lignin Biosynthesis Gene PAL Promotes Ginsenoside Rg3 Accumulation in Ginseng Plant Chassis. J. Integr. Plant Biol. 2022, 64, 1739–1754. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.; Clastre, M.; Courdavault, V.; O’Connor, S.E. De Novo Production of the Plant-Derived Alkaloid Strictosidine in Yeast. Proc. Natl. Acad. Sci. USA 2015, 112, 3205–3210. [Google Scholar] [CrossRef] [PubMed]
- van Herpen, T.W.J.M.; Cankar, K.; Nogueira, M.; Bosch, D.; Bouwmeester, H.J.; Beekwilder, J. Nicotiana Benthamiana as a Production Platform for Artemisinin Precursors. PLoS ONE 2010, 5, e14222. [Google Scholar] [CrossRef]
- Hasan, M.M.; Kim, H.S.; Jeon, J.H.; Kim, S.H.; Moon, B.K.; Song, J.Y.; Shim, S.H.; Baek, K.H. Metabolic Engineering of Nicotiana Benthamiana for the Increased Production of Taxadiene. Plant Cell Rep. 2014, 33, 895–904. [Google Scholar] [CrossRef]
- Miettinen, K.; Dong, L.; Navrot, N.; Schneider, T.; Burlat, V.; Pollier, J.; Woittiez, L.; Van Der Krol, S.; Lugan, R.; Ilc, T.; et al. The Seco-Iridoid Pathway from Catharanthus Roseus. Nat. Commun. 2014, 5, 3606. [Google Scholar] [CrossRef]
- Mukherjee, A.; Sarkar, D.; Sasmal, S. A Review of Green Synthesis of Metal Nanoparticles Using Algae. Front. Microbiol. 2021, 12, 693899. [Google Scholar] [CrossRef]
- Ramanan, R.; Kim, B.H.; Cho, D.H.; Oh, H.M.; Kim, H.S. Algae-Bacteria Interactions: Evolution, Ecology and Emerging Applications. Biotechnol. Adv. 2016, 34, 14–29. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Gong, Y.; He, Y.; Xin, Y.; Lv, N.; Du, X.; Li, Y.; Jeong, B.-R.; Xu, J. Genome Engineering of Nannochloropsis with Hundred-Kilobase Fragment Deletions by Cas9 Cleavages. Plant J. 2021, 106, 1148–1162. [Google Scholar] [CrossRef]
- Heiss, S.; Hörmann, A.; Tauer, C.; Sonnleitner, M.; Egger, E.; Grabherr, R.; Heinl, S. Evaluation of Novel Inducible Promoter/Repressor Systems for Recombinant Protein Expression in Lactobacillus Plantarum. Microb. Cell Factories 2016, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.B.; Zeng, A.P. Engineering a Lysine-ON Riboswitch for Metabolic Control of Lysine Production in Corynebacterium Glutamicum. ACS Synth. Biol. 2015, 4, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Gossen, M.; Bujardt, H. Tight Control of Gene Expression in Mammalian Cells by Tetracycline-Responsive Promoters Author (s): Manfred Gossen and Hermann Bujard Source: Proceedings of the National Academy of Sciences of the United States of America, Published by: National Aca. Proc. Natl. Acad. Sci. USA 1992, 89, 5547–5551. [Google Scholar]
- Urlinger, S.; Udo, B.; Marion, T.; Mazahir, H. Exploring the Sequence Space for Tetracyline-Dependent Transcriptional Activators: Novel Mutations Yield Expanded range and Sensitivity. Proc. Natl. Acad. Sci. USA 2000, 14, 7963–7968. [Google Scholar] [CrossRef] [Green Version]
| Type of Function | List of Genes | Reference | |
|---|---|---|---|
| Biotic and Abiotic Stress Conditions | Insect tolerance | TPs MGAIs SGA PPC PR PDF1.2 | [59] [58] [64,65] [64,65] [67] [68] |
| Water spray | bHLH19 ERF109 ORA47 | [71] [71] [71] | |
| Drought stress | rd22 ADH | [73] [73] | |
| Chilling tolerance | MdCBF1 | [78] | |
| Plant Growth and Development | Leaf growth and aging | Dof2.1 SAG12/13/29/113 CAB1 RBCS AtUSR1 AMY3 BAM1 SUTI/4 SWEET11 SIPAO | [82] [82,83] [82,83] [82,83] [85] [86] [86] [86] [86] [87] |
| Root stem cell | PLT1/2 ERF115/109 | [88] [89] | |
| Fruit ripening and pollen maturation | MdERF2/3 MdACO1 AGPs SS2 BAM1 | [95] [94] [103] [103] [103] | |
| Specialized Metabolites | Paclitaxel biosynthesis | TcERF12/15 | [111] |
| Artemisinin synthesis | CYP71AV1 DBR2 | [112,113] [112,113] | |
| Salvia miltiorrhiza | CYP98A14 SmGGPP | [115] [115] | |
| Vinblastine biosynthesis | ORCA2/3/4 | [116] | |
| Gossypol synthesis | CYP71BE79 | [118] | |
| Psammosilene tunicoides synthesis | TwTPS27a/b | [119] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, L.; Wang, Y.; Qiu, L.; Han, X.; Zhu, Y.; Liu, L.; Man, M.; Li, F.; Ren, M.; Xing, Y. MYC2: A Master Switch for Plant Physiological Processes and Specialized Metabolite Synthesis. Int. J. Mol. Sci. 2023, 24, 3511. https://doi.org/10.3390/ijms24043511
Luo L, Wang Y, Qiu L, Han X, Zhu Y, Liu L, Man M, Li F, Ren M, Xing Y. MYC2: A Master Switch for Plant Physiological Processes and Specialized Metabolite Synthesis. International Journal of Molecular Sciences. 2023; 24(4):3511. https://doi.org/10.3390/ijms24043511
Chicago/Turabian StyleLuo, Lei, Ying Wang, Lu Qiu, Xingpei Han, Yaqian Zhu, Lulu Liu, Mingwu Man, Fuguang Li, Maozhi Ren, and Yadi Xing. 2023. "MYC2: A Master Switch for Plant Physiological Processes and Specialized Metabolite Synthesis" International Journal of Molecular Sciences 24, no. 4: 3511. https://doi.org/10.3390/ijms24043511
APA StyleLuo, L., Wang, Y., Qiu, L., Han, X., Zhu, Y., Liu, L., Man, M., Li, F., Ren, M., & Xing, Y. (2023). MYC2: A Master Switch for Plant Physiological Processes and Specialized Metabolite Synthesis. International Journal of Molecular Sciences, 24(4), 3511. https://doi.org/10.3390/ijms24043511






