Cumulus Cells Accelerate Postovulatory Oocyte Aging through IL1–IL1R1 Interaction in Mice
Abstract
1. Introduction
2. Results
2.1. POA Induces Preimplantation Embryo Development Failure
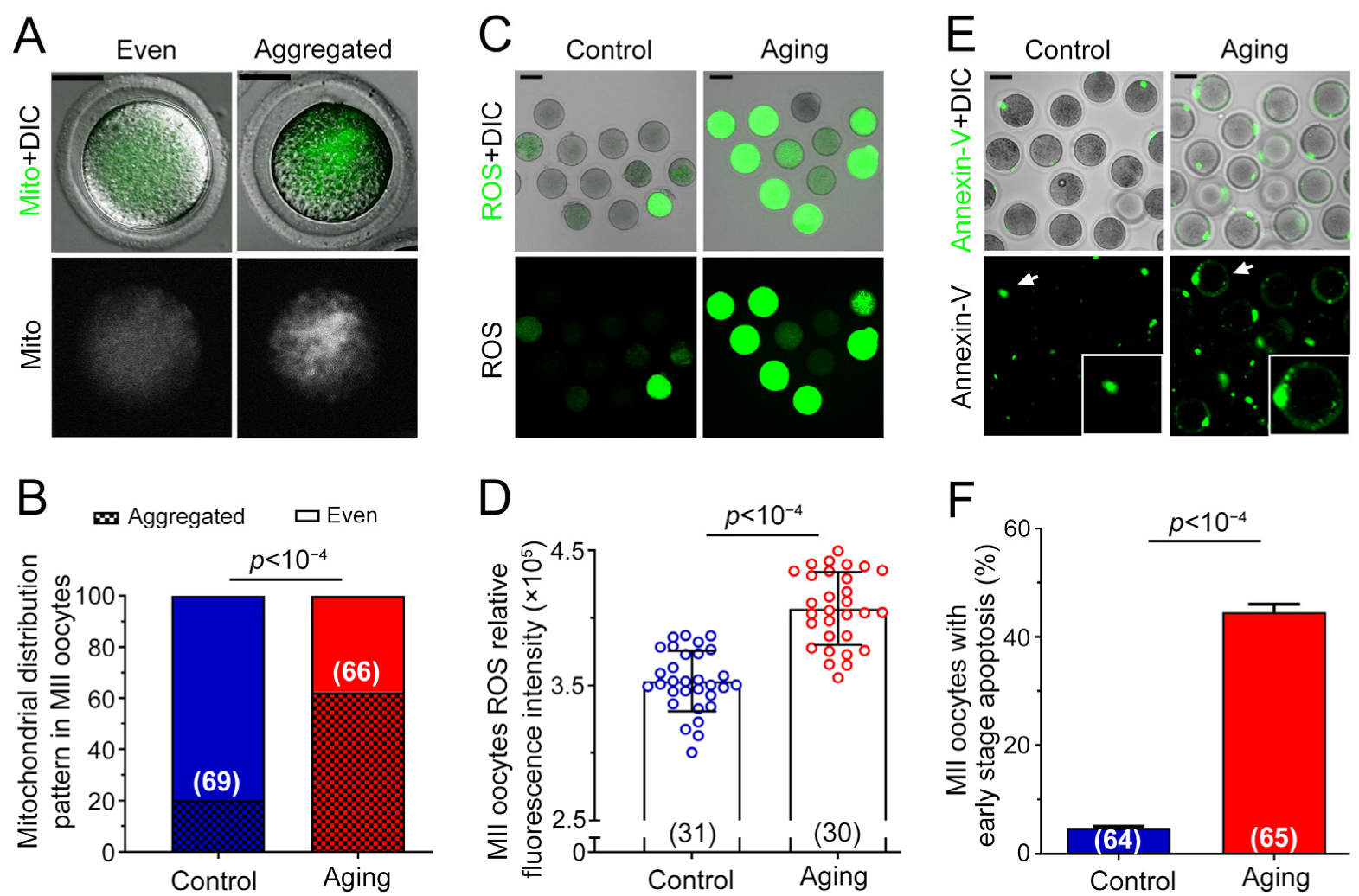
2.2. Various Abnormalities of Oocytes Were Observed in POA
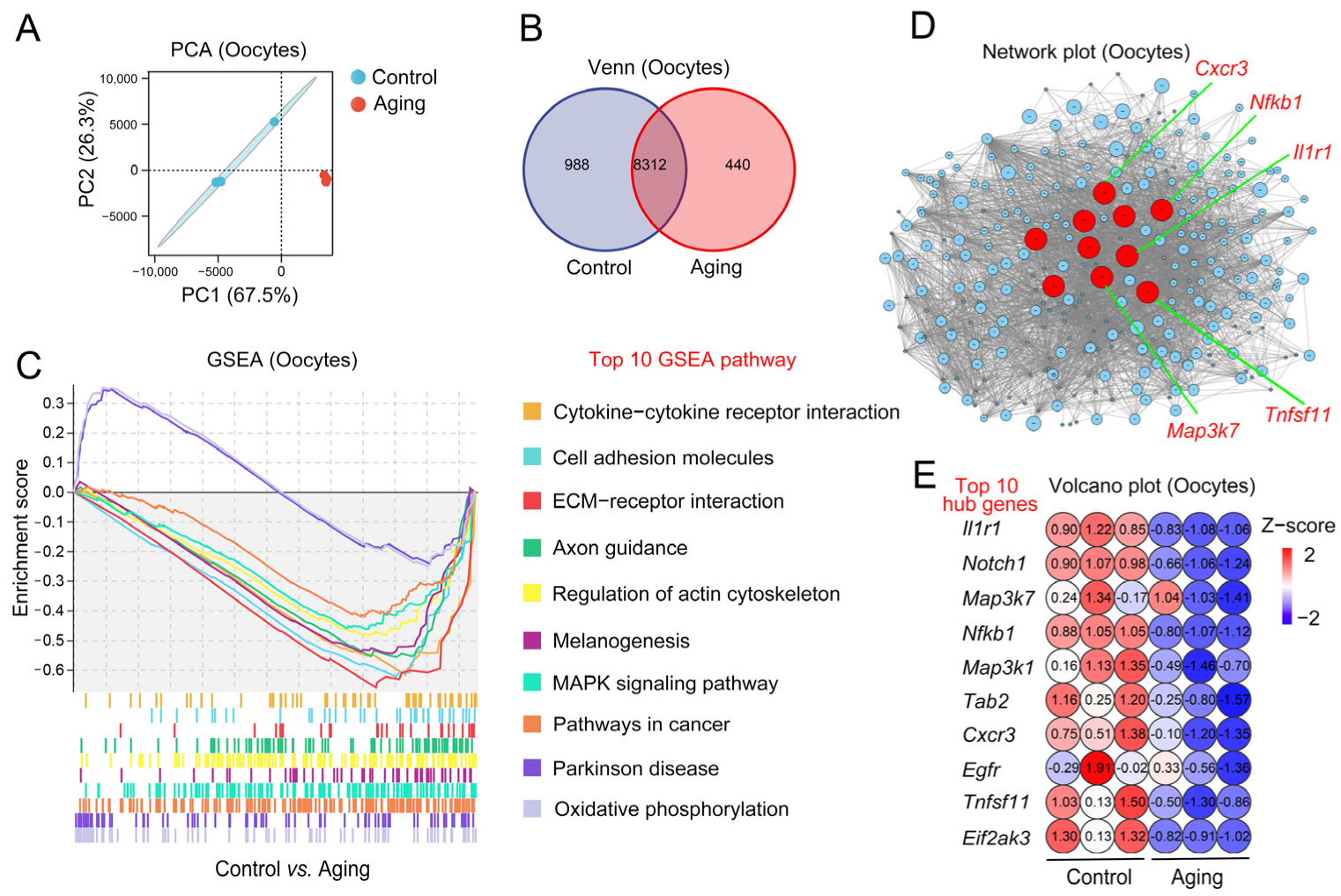
2.3. Changes of Aging-Related Transcription Level in Oocytes
2.4. Changes to Aging-Related Transcription Levels in Cumulus Cells
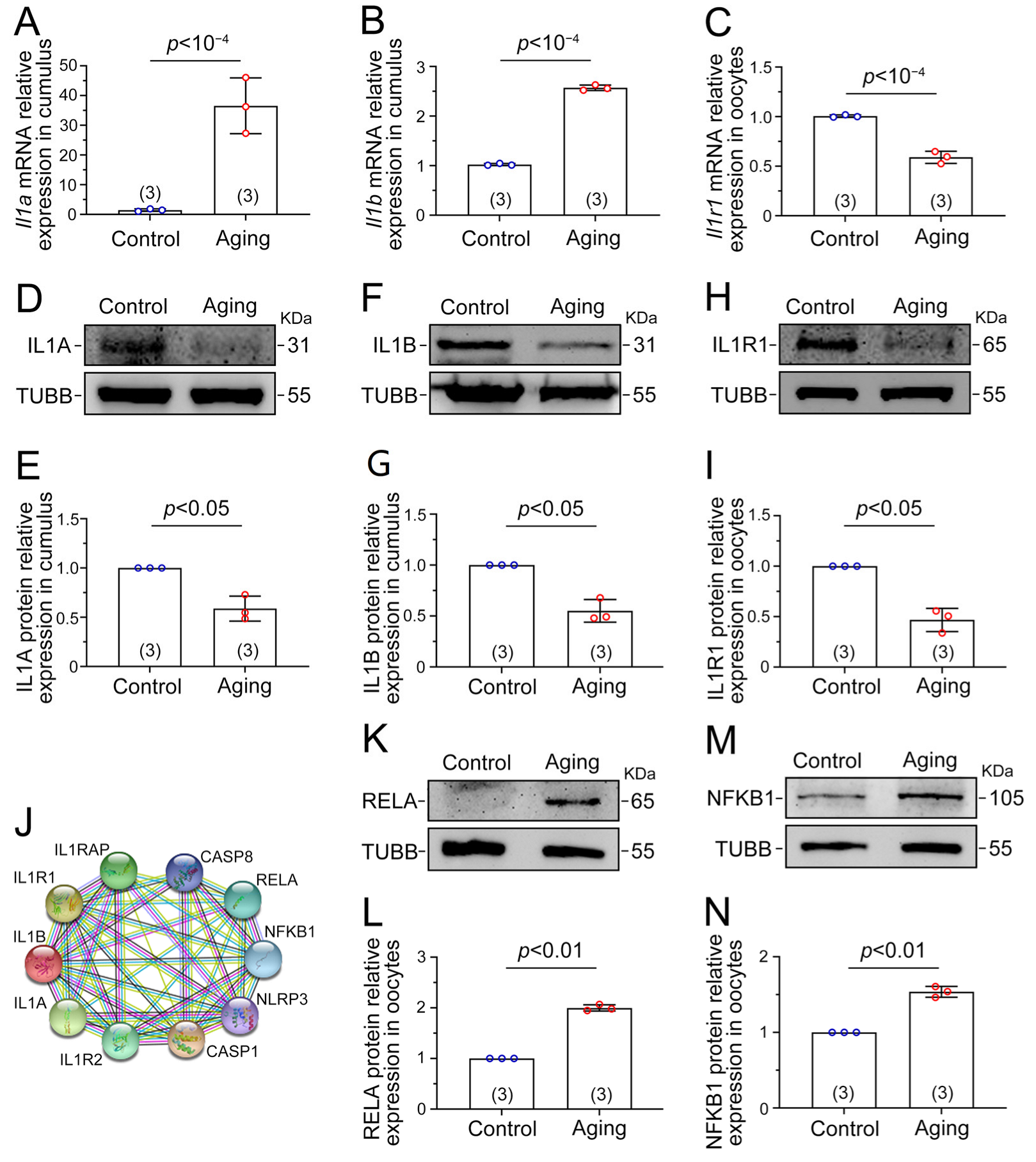
2.5. Il1–Il1r1 Participate in Interactions between the Cumulus Cells and Oocytes
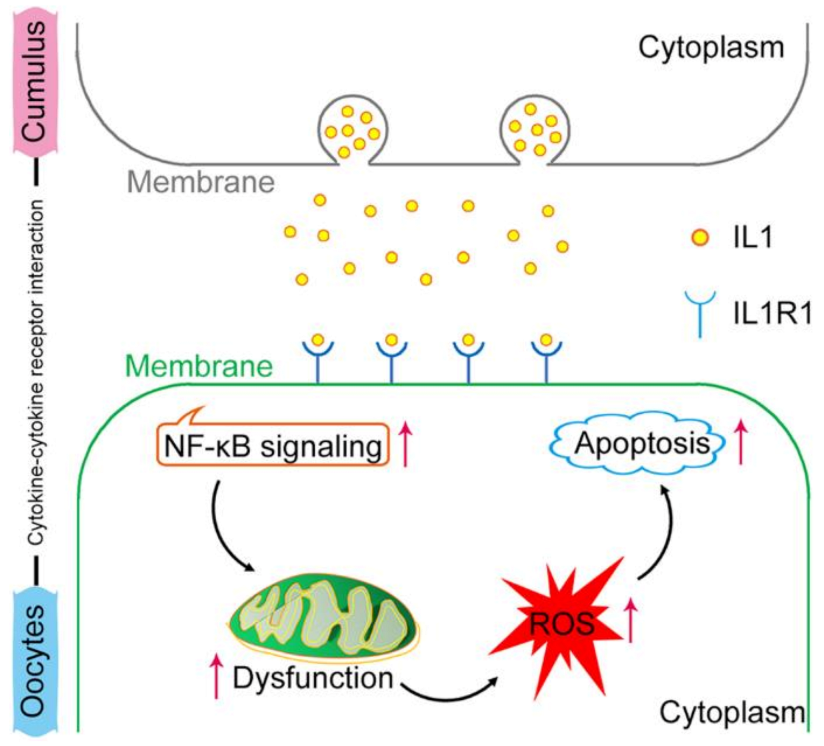
2.6. NF-κB Signaling Is Activated by the IL1–IL1R1 Interaction
3. Discussion
4. Materials and Methods
4.1. Animals and Experimental Design
4.2. Cumulus Cells and Oocytes Collection
4.3. IVF Experiment and Embryo Culture
4.4. Mitochondria Distribution Detection
4.5. ROS Assay
4.6. Early Apoptosis Detection
4.7. Transcriptome Sequencing of Cumulus Cells and Oocytes
4.8. Cell Communication Inference
4.9. Real-Time Quantitative PCR
4.10. Western Blotting
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, T.; Zhou, Y.; Li, L.; Wang, H.H.; Ma, X.S.; Qian, W.P.; Shen, W.; Schatten, H.; Sun, Q.Y. SIRT1, 2, 3 protect mouse oocytes from postovulatory aging. Aging 2016, 8, 685–696. [Google Scholar] [CrossRef]
- Jeon, H.J.; Cui, X.S.; Guo, J.; Lee, J.M.; Kim, J.S.; Oh, J.S. TCTP regulates spindle assembly during postovulatory aging and prevents deterioration in mouse oocyte quality. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1328–1334. [Google Scholar] [CrossRef] [PubMed]
- Di Nisio, V.; Antonouli, S.; Damdimopoulou, P.; Salumets, A.; Cecconi, S. In vivo and in vitro postovulatory aging: When time works against oocyte quality? J. Assist. Reprod. Genet. 2022, 39, 905–918. [Google Scholar] [CrossRef]
- Ono, T.; Mizutani, E.; Li, C.; Yamagata, K.; Wakayama, T. Offspring from intracytoplasmic sperm injection of aged mouse oocytes treated with caffeine or MG132. Genesis 2011, 49, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Moghadam, A.R.E.; Moghadam, M.T.; Hemadi, M.; Saki, G. Oocyte quality and aging. JBRA Assist. Reprod. 2022, 26, 105–122. [Google Scholar] [CrossRef]
- Takahashi, T.; Igarashi, H.; Amita, M.; Hara, S.; Matsuo, K.; Kurachi, H. Molecular mechanism of poor embryo development in postovulatory aged oocytes: Mini review. J. Obstet. Gynaecol. Res. 2013, 39, 1431–1439. [Google Scholar] [CrossRef]
- Prasad, S.; Tiwari, M.; Koch, B.; Chaube, S.K. Morphological, cellular and molecular changes during postovulatory egg aging in mammals. J. Biomed. Sci. 2015, 22, 36. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Igarashi, H.; Amita, M.; Hara, S.; Kurachi, H. Cellular and molecular mechanisms of various types of oocyte aging. Reprod. Med. Biol. 2011, 10, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Lord, T.; Aitken, R.J. Oxidative stress and ageing of the post-ovulatory oocyte. Reproduction 2013, 146, R217–R227. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.J.; Schatten, H.; Zhang, C.L.; Sun, Q.Y. Oocyte ageing and epigenetics. Reproduction 2015, 149, R103–R114. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Xu, J.; Zeng, X.; Sun, Y.P.; Yang, Q. Nicotinamide Riboside supplementation ameliorated postovulatory oocyte quality decline. Reproduction 2023, 165, 103–111. [Google Scholar] [CrossRef]
- Barrett, S.L.; Albertini, D.F. Cumulus cell contact during oocyte maturation in mice regulates meiotic spindle positioning and enhances developmental competence. J. Assist. Reprod. Genet. 2010, 27, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Moulavi, F.; Hosseini, S.M. Diverse patterns of cumulus cell expansion during in vitro maturation reveal heterogeneous cellular and molecular features of oocyte competence in dromedary camel. Theriogenology 2018, 119, 259–267. [Google Scholar] [CrossRef]
- Melo, E.O.; Cordeiro, D.M.; Pellegrino, R.; Wei, Z.; Daye, Z.J.; Nishimura, R.C.; Dode, M.A. Identification of molecular markers for oocyte competence in bovine cumulus cells. Anim. Genet. 2017, 48, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.S. Ovulation: New factors that prepare the oocyte for fertilization. Mol. Cell. Endocrinol. 2005, 234, 75–79. [Google Scholar] [CrossRef]
- Tiwari, M.; Tripathi, A.; Chaube, S.K. Presence of encircling granulosa cells protects against oxidative stress-induced apoptosis in rat eggs cultured in vitro. Apoptosis 2017, 22, 98–107. [Google Scholar] [CrossRef]
- Miao, Y.L.; Liu, X.Y.; Qiao, T.W.; Miao, D.Q.; Luo, M.J.; Tan, J.H. Cumulus cells accelerate aging of mouse oocytes. Biol. Reprod. 2005, 73, 1025–1031. [Google Scholar] [CrossRef][Green Version]
- Kong, Q.Q.; Wang, J.; Xiao, B.; Lin, F.H.; Zhu, J.; Sun, G.Y.; Luo, M.J.; Tan, J.H. Cumulus cell-released tumor necrosis factor (TNF)-α promotes post-ovulatory aging of mouse oocytes. Aging 2018, 10, 1745–1757. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Lin, F.H.; Zhang, J.; Lin, J.; Li, H.; Li, Y.W.; Tan, X.W.; Tan, J.H. The signaling pathways by which the Fas/FasL system accelerates oocyte aging. Aging 2016, 8, 291–303. [Google Scholar] [CrossRef]
- Lim, R.; Barker, G.; Menon, R.; Lappas, M. A Novel Role for SIRT3 in Regulating Mediators Involved in the Terminal Pathways of Human Labor and Delivery. Biol. Reprod. 2016, 95, 95. [Google Scholar] [CrossRef] [PubMed]
- Nishikimi, A.; Mukai, J.; Yamada, M. Nuclear translocation of nuclear factor kappa B in early 1-cell mouse embryos. Biol. Reprod. 1999, 60, 1536–1541. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rangan, G.; Wang, Y.; Harris, D. NF-kappaB signalling in chronic kidney disease. Front. Biosci. 2009, 14, 3496–3522. [Google Scholar] [CrossRef] [PubMed]
- Apte, R.N.; Dotan, S.; Elkabets, M.; White, M.R.; Reich, E.; Carmi, Y.; Song, X.; Dvozkin, T.; Krelin, Y.; Voronov, E. The involvement of IL-1 in tumorigenesis, tumor invasiveness, metastasis and tumor-host interactions. Cancer Metastasis Rev. 2006, 25, 387–408. [Google Scholar] [CrossRef]
- Shafique, S.; Winn, L.M. Characterizing the effects of in utero valproic acid exposure on NF-κB signaling in CD-1 mouse embryos during neural tube closure. Neurotoxicol. Teratol. 2021, 83, 106941. [Google Scholar] [CrossRef] [PubMed]
- Arend, W.P.; Palmer, G.; Gabay, C. IL-1, IL-18, and IL-33 families of cytokines. Immunol. Rev. 2008, 223, 20–38. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, H.Y.; Cho, S.; Yoo, S.J.; Kim, W.J.; Yeon, H.R.; Choi, K.; Choi, J.M.; Kang, S.W.; Lee, W.W. Induction of the IL-1RII decoy receptor by NFAT/FOXP3 blocks IL-1β-dependent response of Th17 cells. eLife 2021, 10, e61841. [Google Scholar] [CrossRef]
- de los Santos, M.J.; Anderson, D.J.; Racowsky, C.; Simón, C.; Hill, J.A. Expression of interleukin-1 system genes in human gametes. Biol. Reprod. 1998, 59, 1419–1424. [Google Scholar] [CrossRef]
- Xu, F.; Liu, R.; Cao, X. Hyperandrogenism stimulates inflammation and promote apoptosis of cumulus cells. Cell. Mol. Biol. 2017, 63, 64–68. [Google Scholar] [CrossRef]
- Deyerle, K.L.; Sims, J.E.; Dower, S.K.; Bothwell, M.A. Pattern of IL-1 receptor gene expression suggests role in noninflammatory processes. J. Immunol. 1992, 149, 1657–1665. [Google Scholar] [CrossRef]
- Martoriati, A.; Lalmanach, A.C.; Goudet, G.; Gérard, N. Expression of interleukin-1 (IL-1) system genes in equine cumulus-oocyte complexes and influence of IL-1beta during in vitro maturation. Biol. Reprod. 2002, 67, 630–636. [Google Scholar] [CrossRef]
- Simón, C.; Frances, A.; Piquette, G.; Polan, M.L. Immunohistochemical localization of the interleukin-1 system in the mouse ovary during follicular growth, ovulation, and luteinization. Biol. Reprod. 1994, 50, 449–457. [Google Scholar] [CrossRef]
- Bellehumeur, C.; Blanchet, J.; Fontaine, J.Y.; Bourcier, N.; Akoum, A. Interleukin 1 regulates its own receptors in human endometrial cells via distinct mechanisms. Hum. Reprod. 2009, 24, 2193–2204. [Google Scholar] [CrossRef] [PubMed]
- Jantra, S.; Bigliardi, E.; Brizzi, R.; Ietta, F.; Bechi, N.; Paulesu, L. Interleukin 1 in oviductal tissues of viviparous, oviparous, and ovuliparous species of amphibians. Biol. Reprod. 2007, 76, 1009–1015. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lord, T.; Martin, J.H.; Aitken, R.J. Accumulation of electrophilic aldehydes during postovulatory aging of mouse oocytes causes reduced fertility, oxidative stress, and apoptosis. Biol. Reprod. 2015, 92, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; ShiYang, X.; Zhang, Y.; Miao, Y.; Chen, Y.; Cui, Z.; Xiong, B. Coenzyme Q10 ameliorates the quality of postovulatory aged oocytes by suppressing DNA damage and apoptosis. Free Radic. Biol. Med. 2019, 143, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.L.; Kikuchi, K.; Sun, Q.Y.; Schatten, H. Oocyte aging: Cellular and molecular changes, developmental potential and reversal possibility. Hum. Reprod. Update 2009, 15, 573–585. [Google Scholar] [CrossRef]
- Prasad, S.; Chaube, S.K. Increased Telomerase Reverse Transcriptase Expression Associates with Spontaneous Exit from M-II Arrest in Rat Eggs. Cell Reprogram. 2017, 19, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Lord, T.; Nixon, B.; Jones, K.T.; Aitken, R.J. Melatonin prevents postovulatory oocyte aging in the mouse and extends the window for optimal fertilization in vitro. Biol. Reprod. 2013, 88, 67. [Google Scholar] [CrossRef]
- Luciano, A.M.; Lodde, V.; Beretta, M.S.; Colleoni, S.; Lauria, A.; Modina, S. Developmental capability of denuded bovine oocyte in a co-culture system with intact cumulus-oocyte complexes: Role of cumulus cells, cyclic adenosine 3′,5′-monophosphate, and glutathione. Mol. Reprod. Dev. 2005, 71, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Tanghe, S.; Van Soom, A.; Nauwynck, H.; Coryn, M.; de Kruif, A. Minireview: Functions of the cumulus oophorus during oocyte maturation, ovulation, and fertilization. Mol. Reprod. Dev. 2002, 61, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.J.; Wu, S.N.; Shen, J.P.; Wang, D.H.; Kong, X.W.; Lu, A.; Li, Y.J.; Zhou, H.X.; Zhao, Y.F.; Liang, C.G. The beneficial effects of cumulus cells and oocyte-cumulus cell gap junctions depends on oocyte maturation and fertilization methods in mice. PeerJ 2016, 4, e1761. [Google Scholar] [CrossRef]
- Russell, D.L.; Gilchrist, R.B.; Brown, H.M.; Thompson, J.G. Bidirectional communication between cumulus cells and the oocyte: Old hands and new players? Theriogenology 2016, 86, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Qiao, T.W.; Liu, N.; Miao, D.Q.; Zhang, X.; Han, D.; Ge, L.; Tan, J.H. Cumulus cells accelerate aging of mouse oocytes by secreting a soluble factor(s). Mol. Reprod. Dev. 2008, 75, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Labrecque, R.; Sirard, M.A. The study of mammalian oocyte competence by transcriptome analysis: Progress and challenges. Mol. Hum. Reprod. 2014, 20, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Molinari, E.; Bar, H.; Pyle, A.M.; Patrizio, P. Transcriptome analysis of human cumulus cells reveals hypoxia as the main determinant of follicular senescence. Mol. Hum. Reprod. 2016, 22, 866–876. [Google Scholar] [CrossRef]
- Mishina, T.; Tabata, N.; Hayashi, T.; Yoshimura, M.; Umeda, M.; Mori, M.; Ikawa, Y.; Hamada, H.; Nikaido, I.; Kitajima, T.S. Single-oocyte transcriptome analysis reveals aging-associated effects influenced by life stage and calorie restriction. Aging Cell 2021, 20, e13428. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Zhang, Z.; Wang, Y. T cell receptor signaling pathway and cytokine-cytokine receptor interaction affect the rehabilitation process after respiratory syncytial virus infection. PeerJ 2019, 7, e7089. [Google Scholar] [CrossRef] [PubMed]
- Da Luz, C.M.; Da Broi, M.G.; Koopman, L.O.; Plaça, J.R.; da Silva-Jr, W.A.; Ferriani, R.A.; Meola, J.; Navarro, P.A. Transcriptomic analysis of cumulus cells shows altered pathways in patients with minimal and mild endometriosis. Sci. Rep. 2022, 12, 5775. [Google Scholar] [CrossRef]
- Fang, T.; Wu, Z.W.; Wang, Y.; Wang, F.; Du, Z.Q.; Yang, C.X. Comparative transcriptome analysis identifies important maternal molecules and associated biological pathways for pig and human mature oocytes. Reprod. Domest. Anim. 2022, 57, 643–652. [Google Scholar] [CrossRef]
- Biase, F.H.; Kimble, K.M. Functional signaling and gene regulatory networks between the oocyte and the surrounding cumulus cells. BMC Genom. 2018, 19, 351. [Google Scholar] [CrossRef]
- Vorländer, M.K.; Pacheco-Fiallos, B.; Plaschka, C. Structural basis of mRNA maturation: Time to put it together. Curr. Opin. Struct. Biol. 2022, 75, 102431. [Google Scholar] [CrossRef] [PubMed]
- Frisch, S.M. Interleukin-1α: Novel functions in cell senescence and antiviral response. Cytokine 2022, 154, 155875. [Google Scholar] [CrossRef]
- Piccioli, P.; Rubartelli, A. The secretion of IL-1β and options for release. Semin. Immunol. 2013, 25, 425–429. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, C.; Li, W.; Kuang, J.; Qiu, X.Y.; Min, L.; Zhu, L. Targeted protein degradation in mammalian cells: A promising avenue toward future. Comput. Struct. Biotechnol. J. 2022, 20, 5477–5489. [Google Scholar] [CrossRef] [PubMed]
- Ainscough, J.S.; Frank Gerberick, G.; Zahedi-Nejad, M.; Lopez-Castejon, G.; Brough, D.; Kimber, I.; Dearman, R.J. Dendritic cell IL-1α and IL-1β are polyubiquitinated and degraded by the proteasome. J. Biol. Chem. 2014, 289, 35582–35592. [Google Scholar] [CrossRef] [PubMed]
- Brachova, P.; Alvarez, N.S.; Christenson, L.K. Loss of Cnot6l Impairs Inosine RNA Modifications in Mouse Oocytes. Int. J. Mol. Sci. 2021, 22, 1191. [Google Scholar] [CrossRef] [PubMed]
- Teruel, M.; Smith, R.; Catalano, R. Growth factors and embryo development. Biocell 2000, 24, 107–122. [Google Scholar] [PubMed]
- McNiven, M.A. Big gulps: Specialized membrane domains for rapid receptor-mediated endocytosis. Trends Cell Biol. 2006, 16, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Kornilova, E.S. Receptor-mediated endocytosis and cytoskeleton. Biochemistry 2014, 79, 865–878. [Google Scholar] [CrossRef]
- Cartwright, T.; Perkins, N.D.; Wilson, C.L. NFKB1: A suppressor of inflammation, ageing and cancer. FEBS J. 2016, 283, 1812–1822. [Google Scholar] [CrossRef] [PubMed]
- Gehrke, N.; Hövelmeyer, N.; Waisman, A.; Straub, B.K.; Weinmann-Menke, J.; Wörns, M.A.; Galle, P.R.; Schattenberg, J.M. Hepatocyte-specific deletion of IL1-RI attenuates liver injury by blocking IL-1 driven autoinflammation. J. Hepatol. 2018, 68, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Catheline, S.E.; Bell, R.D.; Oluoch, L.S.; James, M.N.; Escalera-Rivera, K.; Maynard, R.D.; Chang, M.E.; Dean, C.; Botto, E.; Ketz, J.P.; et al. IKKβ-NF-κB signaling in adult chondrocytes promotes the onset of age-related osteoarthritis in mice. Sci. Signal. 2021, 14, eabf3535. [Google Scholar] [CrossRef] [PubMed]
- Rolova, T.; Dhungana, H.; Korhonen, P.; Valonen, P.; Kolosowska, N.; Konttinen, H.; Kanninen, K.; Tanila, H.; Malm, T.; Koistinaho, J. Deletion of Nuclear Factor kappa B p50 Subunit Decreases Inflammatory Response and Mildly Protects Neurons from Transient Forebrain Ischemia-induced Damage. Aging Dis. 2016, 7, 450–465. [Google Scholar] [CrossRef]
- Li, J.; Deng, Y.; Peng, D.; Zhao, L.; Fang, Y.; Zhu, X.; Li, S.; Aschner, M.; Ou, S.; Jiang, Y. Sodium P-aminosalicylic Acid Attenuates Manganese-Induced Neuroinflammation in BV2 Microglia by Modulating NF-κB Pathway. Biol. Trace Elem. Res. 2021, 199, 4688–4699. [Google Scholar] [CrossRef]
- Fielder, E.; Tweedy, C.; Wilson, C.; Oakley, F.; LeBeau, F.E.N.; Passos, J.F.; Mann, D.A.; von Zglinicki, T.; Jurk, D. Anti-inflammatory treatment rescues memory deficits during aging in nfkb1(-/-) mice. Aging Cell 2020, 19, e13188. [Google Scholar] [CrossRef]
- Gabellini, C.; Castellini, L.; Trisciuoglio, D.; Kracht, M.; Zupi, G.; Del Bufalo, D. Involvement of nuclear factor-kappa B in bcl-xL-induced interleukin 8 expression in glioblastoma. J. Neurochem. 2008, 107, 871–882. [Google Scholar] [CrossRef]
- Nisr, R.B.; Shah, D.S.; Ganley, I.G.; Hundal, H.S. Proinflammatory NFkB signalling promotes mitochondrial dysfunction in skeletal muscle in response to cellular fuel overloading. Cell. Mol. Life Sci. 2019, 76, 4887–4904. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Lee, H.; Lim, J.W.; Kim, H. Inhibitory effect of alpha-lipoic acid on mitochondrial dysfunction and interleukin-8 expression in interleukin-1beta-stimulated ataxia teleangiectasia fibroblasts. J. Physiol. Pharmacol. 2020, 71, 155–165. [Google Scholar]
- Ge, L.; Gao, Y.Q.; Han, Z.; Liu, S.J.; Wang, X.Y.; Zhang, X.J.; Tang, R.H.; Zhang, R.F.; Sun, D.; Feng, B.; et al. Administration of olaquindox impairs spermatogenesis and sperm quality by increasing oxidative stress and early apoptosis in mice. Ecotoxicol. Environ. Saf. 2022, 234, 113396. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, X.; Yang, Q.; Sun, D.; Jiang, Z.-Y.; Wang, T.; Liu, H.-R.; Han, Z.; Wang, L.; Liang, C.-G. Cumulus Cells Accelerate Postovulatory Oocyte Aging through IL1–IL1R1 Interaction in Mice. Int. J. Mol. Sci. 2023, 24, 3530. https://doi.org/10.3390/ijms24043530
Wen X, Yang Q, Sun D, Jiang Z-Y, Wang T, Liu H-R, Han Z, Wang L, Liang C-G. Cumulus Cells Accelerate Postovulatory Oocyte Aging through IL1–IL1R1 Interaction in Mice. International Journal of Molecular Sciences. 2023; 24(4):3530. https://doi.org/10.3390/ijms24043530
Chicago/Turabian StyleWen, Xin, Qi Yang, Dui Sun, Zhao-Yu Jiang, Teng Wang, Hao-Ran Liu, Zhe Han, Lu Wang, and Cheng-Guang Liang. 2023. "Cumulus Cells Accelerate Postovulatory Oocyte Aging through IL1–IL1R1 Interaction in Mice" International Journal of Molecular Sciences 24, no. 4: 3530. https://doi.org/10.3390/ijms24043530
APA StyleWen, X., Yang, Q., Sun, D., Jiang, Z.-Y., Wang, T., Liu, H.-R., Han, Z., Wang, L., & Liang, C.-G. (2023). Cumulus Cells Accelerate Postovulatory Oocyte Aging through IL1–IL1R1 Interaction in Mice. International Journal of Molecular Sciences, 24(4), 3530. https://doi.org/10.3390/ijms24043530




