Structural Characteristics of High-Mobility Group Proteins HMGB1 and HMGB2 and Their Interaction with DNA
Abstract
:1. Introduction
2. Results and Discussion
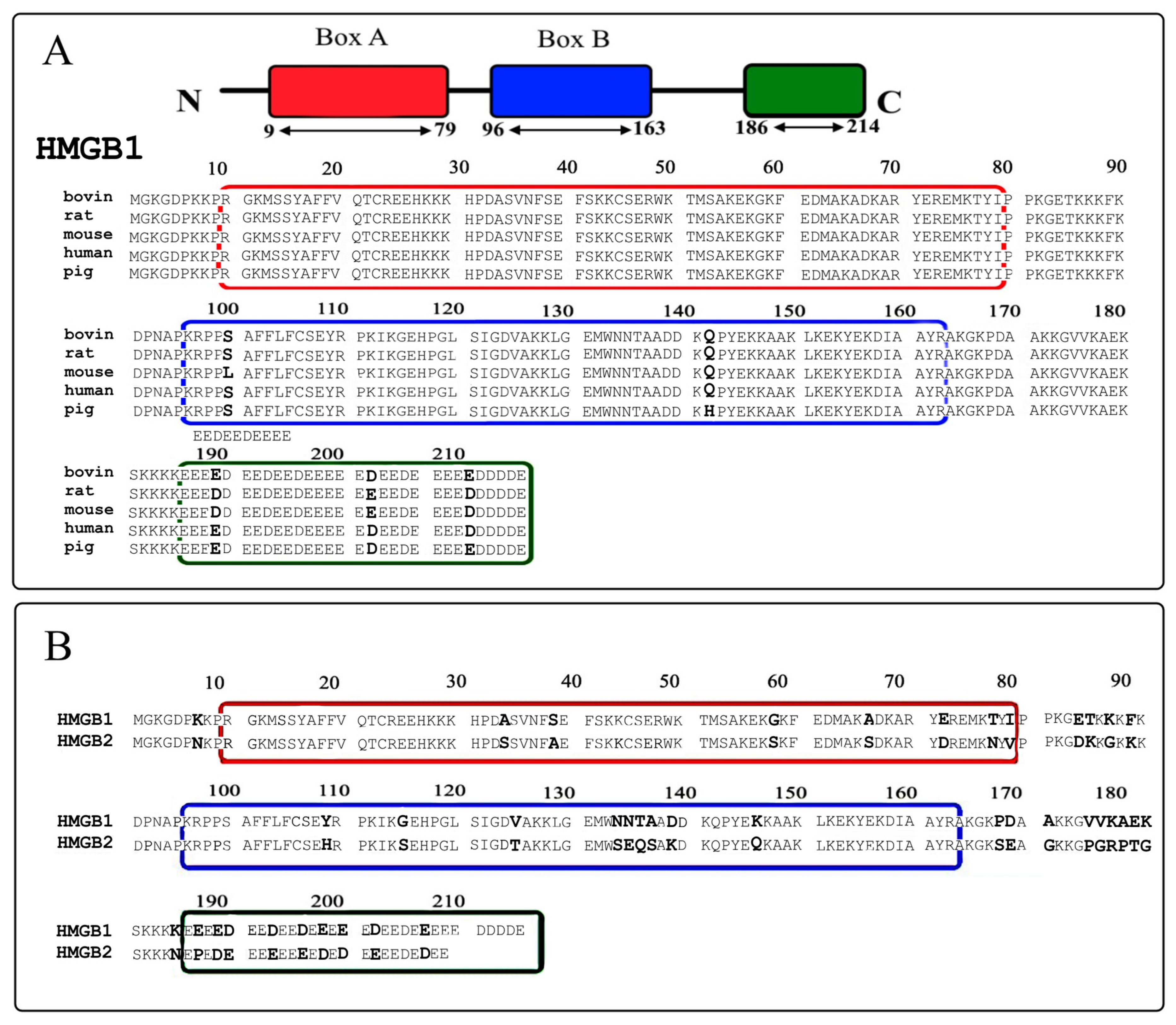
2.1. Secondary Structure of HMGB1 and HMGB2 Proteins
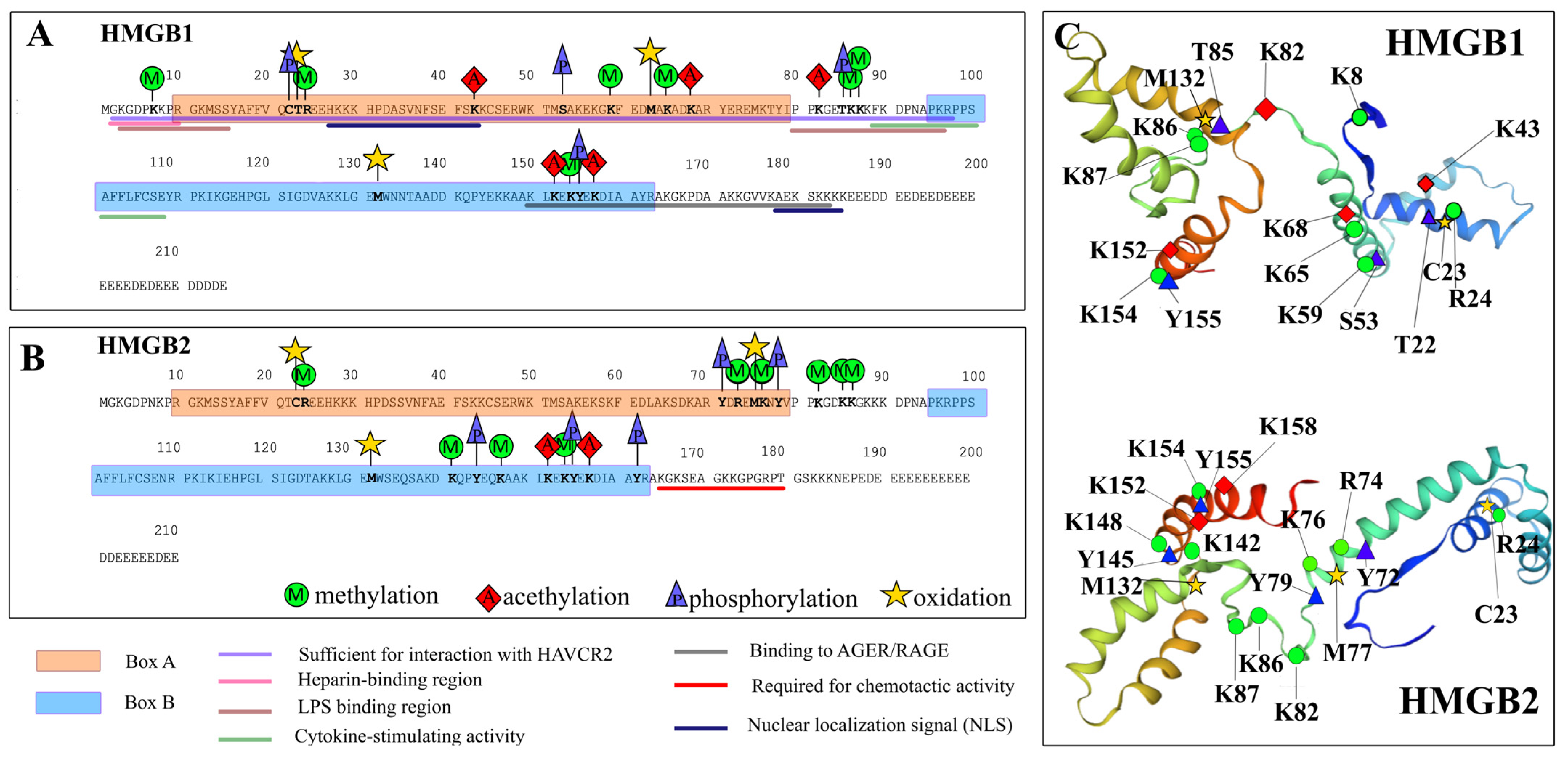
2.2. Post-Translational Modifications of HMGB1 and HMGB2 Proteins in the Calf Thymus
2.2.1. Acetylation
2.2.2. Methylation
2.2.3. Phosphorylation
2.2.4. Glycosylation
2.2.5. ADP-Ribosylation
2.2.6. Oxidation
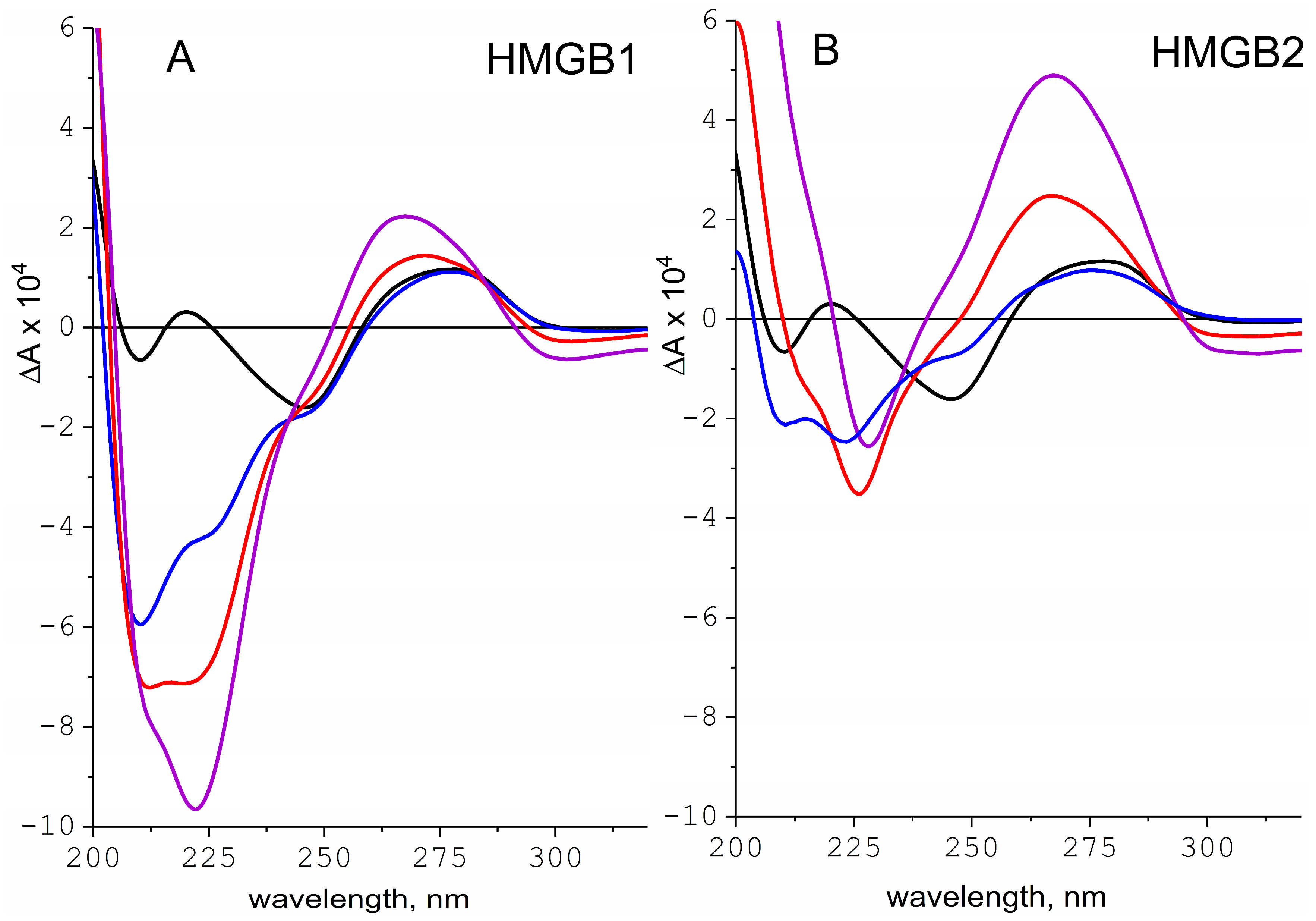
2.3. Interaction of HMGB1 and HMGB2 Proteins with DNA
3. Materials and Methods
3.1. Proteins and DNA
3.2. Enzymatic Hydrolysis and MALDI-FT-ICR-MS Analysis
3.3. Mass Spectrometry
3.4. Western Blotting
3.5. Circular Dichroism Spectroscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bustin, M.; Reeves, R. High-mobility-group chromosomal proteins: Architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 1996, 54, 35–100. [Google Scholar] [CrossRef] [PubMed]
- Bustin, M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999, 19, 5237–5246. [Google Scholar] [CrossRef] [PubMed]
- Stros, M. HMGB proteins: Interactions with DNA and chromatin. Biochim. Biophys. Acta 2010, 1799, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Chikhirzhina, E.; Starkova, T.; Beljajev, A.; Polyanichko, A.M.; Tomilin, A.N. Functional Diversity of Non-Histone Chromosomal Protein HmgB1. Inter. J. Mol. Sci. 2020, 21, 7948. [Google Scholar] [CrossRef]
- Ozturk, N.; Singh, I.; Mehta, A.; Braun, T.; Barreto, G. HMGA proteins as modulators of chromatin structure during transcriptional activation. Front. Cell Dev. Biol. 2014, 2, 5. [Google Scholar] [CrossRef]
- Chiefari, E.; Foti, D.P.; Sgarra, R.; Pegoraro, S.; Arcidiacono, B.; Brunetti, F.S.; Greco, M.; Manfioletti, G.; Brunetti, A. Transcriptional Regulation of Glucose Metabolism: The Emerging Role of the HMGA1 Chromatin Factor. Front. Endocrin. (Lausanne) 2018, 9, 357. [Google Scholar] [CrossRef]
- Postnikov, Y.V.; Trieschmann, L.; Rickers, A.; Bustin, M. Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. J. Mol. Biol. 1995, 252, 423–432. [Google Scholar] [CrossRef]
- Murphy, K.J.; Cutter, A.R.; Fang, H.; Postnikov, Y.V.; Bustin, M.; Hayes, J.J. HMGN1 and 2 remodel core and linker histone tail domains within chromatin. Nucl. Acids Res. 2017, 45, 9917–9930. [Google Scholar] [CrossRef]
- Reeves, R. High mobility group (HMG) proteins, modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair 2015, 36, 122–136. [Google Scholar] [CrossRef]
- Read, C.M.; Cary, P.D.; Crane-Robinson, C.; Driscoll, P.C.; Norman, D.G. Solution structure of a DNA-binding domain from HMG1. Nucl. Acids Res. 1993, 21, 3427–3436. [Google Scholar] [CrossRef] [Green Version]
- Weir, H.M.; Kraulis, P.J.; Hill, C.S.; Raine, A.R.; Laue, E.D.; Thomas, J.O. Structure of the HMG box motif in the B-domain of HMG. EMBO J. 1993, 12, 1311–1319. [Google Scholar] [CrossRef] [PubMed]
- Chikhirzhina, E.; Starkova, T.; Polyanichko, A. The structural organization of the nuclear protein HMGB1 and its effect on the formation of the ordered supramolecular complexes. Biophysics 2021, 66, 373–378. [Google Scholar] [CrossRef]
- Johnstone, T.C.; Wilson, J.J.; Lippard, S.J. Monofunctional and higher-valent platinum anticancer agents. Inorg. Chem. 2013, 52, 12234–12249. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, R.H.; Goodwin, G.H. Isolation and Analysis. In The HMG Chromosomal Proteins; Johns, E.W., Ed.; Academic Press Inc.: London, UK, 1982; pp. 43–45, 59–60. ISBN 9780323158749. [Google Scholar]
- Kohlstaedt, L.A.; Sung, E.C.; Fujishige, A.; Cole, R.D. Non-histone chromosomal protein HMG1 modulates the histone H1-induced condensation of DNA. J. Biol. Chem. 1987, 262, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Kohlstaedt, L.A.; Cole, R.D. Specific interaction between H1 histone and high mobility protein HMG1. Biochemistry 1994, 3, 570–575. [Google Scholar] [CrossRef]
- Cato, L.; Stott, K.; Watson, M.; Thomas, J.O. The Interaction of HMGB1 and Linker Histones Occurs Through their Acidic and Basic Tails. J. Mol. Biol. 2008, 384, 1262–1272. [Google Scholar] [CrossRef]
- Polanska, E.; Pospisilova, S.; Stros, M. Binding of histone H1 to DNA is differentially modulated by redox state of HMGB1. PLoS ONE 2014, 9, e89070. [Google Scholar] [CrossRef]
- Stros, M.; Polanska, E.; Kucirek, M.; Pospisilova, S. Histone H1 differentially inhibits DNA bending by reduced and oxidized HMGB1 protein. PLoS ONE 2015, 10, e0138774. [Google Scholar] [CrossRef]
- Postnikov, Y.V.; Bustin, M. Functional interplay between histone H1 and HMG proteins in chromatin. Biochim. Biophys. Acta 2016, 1859, 462–467. [Google Scholar] [CrossRef]
- Chikhirzhina, E.V.; Polyanichko, A.M.; Skvortsov, A.N.; Kostyleva, E.I.; Houssier, C.; Vorobyev, V.I. HMG1 Domains: The Victims of the Circumstances. Mol. Biol. (Mosk.) 2002, 36, 412–418. [Google Scholar] [CrossRef]
- Chikhirzhina, E.; Polyanichko, A.; Leonenko, Z.; Wieser, H.; Vorob’ev, V. C-terminal domain of nonhistone protein HMGB1 as a modulator of HMGB1-DNA structural interactions. Spectroscopy 2010, 24, 361–366. [Google Scholar] [CrossRef]
- Chikhirzhina, E.V.; Polyanichko, A.M.; Starkova, T.Y. Extranuclear functions of nonhistone protein HMGB1. Tsitologiya 2020, 62, 716–725. [Google Scholar] [CrossRef]
- Knapp, S.; Muller, S.; Digilio, G.; Bonaldi, T.; Bianchi, M.E.; Musco, G. The Long Acidic Tail of High Mobility Group Box 1 (HMGB1) Protein Forms an Extended and Flexible Structure That Interacts with Specific Residues within and between the HMG Boxes. Biochemistry 2004, 43, 11992–11997. [Google Scholar] [CrossRef]
- Stott, K.; Watson, M.; Howe, F.S.; Grossmann, J.G.; Thomas, J.O. Tail-mediated collapse of HMGB1 is dynamic and occurs via differential binding of the acidic tail to the A and B domains. J. Mol. Biol. 2010, 403, 706–722. [Google Scholar] [CrossRef] [PubMed]
- Kuznik, B.I.; Khavinson, V.K.; Linkova, N.S.; Sall, T.S. Alarmin1 (HMGB1) and Age-Related Pathologies. Epygenetic Regulatory Mechanisms. Usp. Fiziol. Nauk 2017, 48, 40–55. [Google Scholar]
- Raucci, A.; Di Maggio, S.; Scavello, F.; D’Ambrosio, A.; Bianchi, M.E.; Capogrossi, M.C. The Janus face of HMGB1 in heart disease: A necessary update. Cell Mol. Life Sci. 2019, 76, 211–229. [Google Scholar] [CrossRef]
- Brien, M.-E.; Baker, B.; Duval, C.; Gaudreault, V.; Jones, R.L.; Girard, S. Alarmins at the maternal-fetal interface: Involvement of inflammation in placental dysfunction and pregnancy complications. Can. Pfysiol. Pharmacor. 2019, 96, 206–212. [Google Scholar] [CrossRef]
- Venereau, E.; Casalgrandi, M.; Schiraldi, M.; Antoine, D.J.; Cattaneo, A.; De Marchis, F.; Liu, J.; Antonelli, A.; Preti, A.; Raeli, L.; et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J. Exp. Med. 2012, 209, 1519–1528. [Google Scholar] [CrossRef] [PubMed]
- Richard, S.A.; Jiang, Y.; Xiang, L.H.; Zhou, S.; Wang, J.; Su, Z.; Xu, H. Post-translational modifications of high mobility group box 1 and cancer. Am. J. Transl. Res. 2017, 9, 5181–5196. [Google Scholar]
- Ito, I.; Fukazawa, J.; Yoshida, M. Post-translational methylation of high mobility group box 1 (HMGB1) causes its cytoplasmic localization in neutrophils. J. Biol. Chem. 2007, 282, 16336–16344. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Livesey, K.M.; Zeh, H.J., III; Lotze, M.T. High Mobility Group Box 1 (HMGB1) Activates an Autophagic Response to Oxidative Stress. Antioxid. Redox Signal. 2011, 15, 2185–2195. [Google Scholar] [CrossRef]
- Stark, K.; Philippi, V.; Stockhausen, S.; Busse, J.; Antonelli, A.; Miller, M.; Schubert, I.; Hoseinpour, P.; Chandraratne, S.; von Brühl, M.L.; et al. Disulfide HMGB1 derived from platelets coordinates venous thrombosis in mice. Blood 2016, 128, 2435–2449. [Google Scholar] [CrossRef]
- Huang, Z.; Zhong, Z.; Zhang, L.; Wang, X.; Xu, R.; Zhu, L.; Wang, Z.; Hu, S.; Zhao, X. Down-regulation of HMGB1 expression by shRNA constructs inhibits the bioactivity of urothelial carcinoma cell lines via the NF-κB pathway. Sci. Rep. 2015, 5, 12807. [Google Scholar] [CrossRef]
- Chen, G.Y.; Tang, J.; Zheng, P.; Liu, Y. CD24 and Siglec-10 selectively repress tissue damageinduced immune responses. Science 2009, 323, 1722–1725. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.K.; Wang, B.; Lu, Q.H.; Zhang, W.; Qin, W.-D.; Liu, X.-J.; Liu, X.-Q.; An, F.-S.; Zhang, Y.; Zhang, M.-X. Inhibition of highmobility group box 1 improves myocardial fibrosis and dysfunction in diabetic cardiomyopathy. Int. J. Cardiol. 2014, 172, 202–212. [Google Scholar] [CrossRef]
- Song, J.; Liu, Q.; Tang, H.; Tao, A.; Wang, H.; Kao, R.; Rui, T. Activation of PI3Kgamma/Akt pathway increases cardiomyocyte HMGB1 expression in diabetic environment. Oncotarget 2016, 7, 80803–80810. [Google Scholar] [CrossRef]
- Lee, G.; Santo, A.I.E.; Zwingenberger, S.; Cai, L.; Vogl, T.; Feldmann, M.; Horwood, N.J.; Chan, J.K.; Nanchahal, J. Fully reduced HMGB1 accelerates the regeneration of multiple tissues by transitioning stem cells to GAlert. Proc. Natl. Acad. Sci. USA 2018, 115, E4463–E4472. [Google Scholar] [CrossRef] [PubMed]
- Tirone, M.; Tran, N.L.; Ceriotti, C.; Gorzanelli, A.; Canepari, M.; Bottinelli, R.; Raucci, A.; Di Maggio, S.; Santiago, C.; Mellado, M.; et al. High mobility group box 1 orchestrates tissue regeneration via CXCR4. J. Exp. Med. 2018, 215, 303–318. [Google Scholar] [CrossRef]
- Volz, H.C.; Seidel, C.; Laohachewin, D.; Kaya, Z.; Müller, O.J.; Pleger, S.T.; Lasitschka, F.; Bianchi, M.E.; Remppis, A.; Bierhaus, A.; et al. HMGB1: The missing link between diabetes mellitus and heart failure. Basic Res. Cardiol. 2010, 105, 805–820. [Google Scholar] [CrossRef]
- Venereau, E.; Ceriotti, C.; Bianchi, M.E. DAMPs from cell death to new life. Front. Immunol. 2015, 6, 442. [Google Scholar] [CrossRef]
- Zhu, L.; Ren, L.; Chen, Y.; Fang, J.; Ge, Z.; Li, X. Redox status of highmobility group box 1 performs a dual role in angiogenesis of colorectal carcinoma. J. Cell Mol. Med. 2015, 19, 2128–2135. [Google Scholar] [CrossRef]
- Yang, H.; Lundback, P.; Ottosson, L.; Erlandsson-Harris, H.; Venereau, E.; Bianchi, M.E.; Al-Abed, Y.; Andersson, U.; Tracey, K.J.; Antoine, D.J. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol. Med. 2012, 18, 250. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. HMGB1 in cancer: Good, bad, or both? Clin. Cancer Res. 2013, 19, 4046. [Google Scholar]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, Y.; Zhang, J.; Wu, J.; Shi, Y. Structural and Dynamic Characterization of the Acid-Unfolded State of hUBF HMG Box 1 Provides Clues for the Early Events in Protein Folding. Biochemistry 2005, 44, 8117–8125. [Google Scholar] [CrossRef]
- Uversky, V.N. A decade and a half of protein intrinsic disorder, biology still waits for physics. Protein Sci. 2013, 22, 693–724. [Google Scholar] [CrossRef] [PubMed]
- Breydo, L.; Redington, J.M.; Uversky, V.N. Effects of Intrinsic and Extrinsic Factors on Aggregation of Physiologically Important Intrinsically Disordered Proteins. Int. Rev. Cell Mol. Biol. 2017, 329, 145–185. [Google Scholar] [CrossRef] [PubMed]
- Buck, M. Trifluoroethanol and colleagues: Cosolvents come of age. Recent studies with peptides and proteins. Q Rev. Biophys. 1998, 31, 297–355. [Google Scholar] [CrossRef]
- Roccatano, D.; Colombo, G.; Fioroni, M.; Mark, A.E. Mechanism by which 2,2,2-trifluoroethanol/water mixtures stabilize secondary-structure formation in peptides: A molecular dynamics study. Proc. Natl. Acad. Sci. USA 2002, 99, 12179–12184. [Google Scholar] [CrossRef]
- Arakawa, T.; Timasheff, S.N. The Interactions of Proteins with Salts, Amino Acids, and Sugars at High Concentration. In Advances in Comparative and Environmental Physiology; Gilles, R., Hoffmann, E.K., Bolis, L., Eds.; Springer: Berlin/Heidelberg, Germany, 1991; Volume 9, pp. 226–245. [Google Scholar] [CrossRef]
- Polyanichko, A.M.; Leonenko, Z.V.; Cramb, D.; Wieser, H.; Vorob’ev, V.I.; Chikhirzhina, E.V. Visualization of DNA complexes with HMGB1 and its C-truncated form HMGB1(A+B). Biophysics 2008, 53, 202–206. [Google Scholar] [CrossRef]
- Available online: http://cbdm-01.zdv.uni-mainz.de/~andrade/k2d3/ (accessed on 27 January 2023).
- Louis-Jeune, C.; Andrade-Navarro, M.A.; Perez-Iratxeta, C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins. Struct. Funct. Bioinform. 2012, 80, 374–381. [Google Scholar] [CrossRef]
- Morrow, J.A.; Segall, M.L.; Lund-Katz, S.; Phillips, M.C.; Knapp, M.; Rupp, B.; Weisgraber, K.H. Differences in Stability among the Human Apolipoprotein E Isoforms Determined by the Amino-Terminal Domain. Biochemistry 2000, 39, 11657–11666. [Google Scholar] [CrossRef]
- Bonaldi, T.; Talamo, F.; Scaffidi, P.; Ferrera, D.; Porto, A.; Bachi, A.; Rubartelli, A.; Agresti, A.; Bianchi, M.E. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003, 22, 5551–5560. [Google Scholar] [CrossRef]
- Gardella, S.; Andrei, C.; Ferrera, D.; Lotti, L.V.; Torrisi, M.R.; Bianchi, M.E.; Rubartelli, A. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep. 2002, 3, 995–1001. [Google Scholar] [CrossRef]
- Blott, E.J.; Grifths, G.M. Secretory lysosomes. Nat. Rev. Mol. Cell Biol. 2002, 3, 122–131. [Google Scholar] [CrossRef]
- Pasheva, E.; Sarov, M.; Bidjekov, K.; Ugrinova, I.; Sarg, B.; Lindner, H.; Pashev, I.G. In Vitro Acetylation of HMGB-1 and -2 Proteins by CBP: The Role of the Acidic Tail. Biochemistry 2004, 43, 2935–2940. [Google Scholar] [CrossRef] [PubMed]
- Watson, M.; Stott, K.; Thomas, J.O. Mapping Intramolecular Interaction between Domains in HMGB1 using Tail-truncation Approach. J. Mol. Biol. 2007, 374, 1286–1297. [Google Scholar] [CrossRef] [PubMed]
- Rodionova, T.Y.; Chikhirzhina, E.V.; Vorob’yov, V.I.; Polyanichko, A.M. Changes in the secondary structure of HMGB1 protein bonded to DNA. J. Struct. Chem. 2010, 50, 976–981. [Google Scholar] [CrossRef]
- Chikhirzhina, E.V.; Tatiana, S.J.; Polyanichko, A.M. Interaction between Chromosomal Protein HMGB1 and DNA Studied by DNA-Melting Analysis. J. Spectrosc. 2014, 2014, 387138. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, Z.-H.; Ding, S.-T.; Wu, H.-H.; Lu, J.-J. High Mobility Group Box 1 Protein Is Methylated and Transported to Cytoplasm in Clear Cell Renal Cell Carcinoma. Asian Pac. J. Cancer Prev. 2013, 14, 5789–5795. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.J.; Youn, J.H.; Ji, Y.; Lee, S.E.; Lim, K.J.; Choi, J.E.; Shin, J.-S. HMGB1 is phosphorylated by classical protein kinase C and is secreted by a calcium dependent mechanism. J. Immunol. 2009, 182, 5800–5809. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Peng, X.; Li, H.; Chen, F.; Chen, Y.; Zhang, Y.; Le, K. The performance of the alarmin HMGB1 in pediatric diseases: From lab to clinic. Immun Inflamm Dis. 2021, 9, 8–30. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kwak, M.S.; Park, J.B.; Lee, S.A.; Choi, J.E.; Cho, H.S.; Shin, J.S. N-linked glycosylation plays a crucial role in the secretion of HMGB1. J. Cell Sci. 2016, 129, 29–38. [Google Scholar] [CrossRef]
- Zong, W.X.; Ditsworth, D.; Bauer, D.E.; Wang, Z.Q.; Thompson, C.B. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004, 18, 1272–1282. [Google Scholar] [CrossRef]
- Ditsworth, D.; Zong, W.X.; Thompson, C.B. Activation of poly (ADP)-ribose polymerase (PARP-1) induces release of the pro-inflammatory mediator HMGB1 from the nucleus. J. Biol. Chem. 2007, 282, 17845–17854. [Google Scholar] [CrossRef]
- Kang, R.; Chen, R.; Zhang, Q.; Hou, W.; Wu, S.; Cao, L.; Huang, J.; Yu, Y.; Fan, X.G.; Yan, Z.; et al. HMGB1 in health and disease. Mol. Aspects Med. 2014, 4, 1–116. [Google Scholar] [CrossRef]
- Sinenko, S.A.; Starkova, T.Y.; Kuzmin, A.K.; Tomilin, A.N. Physiological signaling functions of reactive oxygen species in stem cells: From flies to man. Front. Cell Dev. Biol. 2021, 9, 714370. [Google Scholar] [CrossRef]
- Kwak, M.S.; Kim, H.S.; Lkhamsuren, K.; Kim, Y.H.; Han, M.G.; Shin, J.M.; Park, I.H.; Rhee, W.J.; Lee, S.K.; Rhee, S.G.; et al. Peroxiredoxin-mediated disulfide bond formation is required for nucleocytoplasmic translocation and secretion of HMGB1 in response to inflammatory stimuli. Redox Biol. 2019, 24, 101203. [Google Scholar] [CrossRef]
- Kwak, M.S.; Rhee, W.J.; Lee, Y.J.; Kim, H.S.; Kim, Y.H.; Kwon, M.K.; Shin, J.-S. Reactive oxygen species induce Cys106-mediated anti-386 parallel HMGB1 dimerization that protects against DNA damage. Redox Biol. 2021, 40, 101858. [Google Scholar] [CrossRef] [PubMed]
- Lerman, L.S. A transition to a compact form of DNA in polymer solutions. Proc. Natl. Acad. Sci. USA 1971, 68, 1886–1890. [Google Scholar] [CrossRef]
- Jordan, C.F.; Lerman, L.S.; Venable, J.H. Structure and circular dichroism of DNA in concentrated polymer solutions. Nat. New Biol. 1972, 236, 67–70. [Google Scholar] [CrossRef]
- Zlatanova, J.; Yaneva, J. Histone H1-DNA interactions and their relation to chromatin structure and function. DNA Cell Biol. 1991, 10, 239. [Google Scholar] [CrossRef]
- Polyanichko, A.; Wieser, H. Fourier transform infrared/vibrational circular dichroism spectroscopy as an informative tool for the investigation of large supramolecular complexes of biological macromolecules. Biopolymers 2005, 78, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Polyanichko, A.M.; Chikhirzhina, E.V.; Andrushchenko, V.V.; Vorob’ev, V.; Wieser, H. The Effect of Manganese(II) on the Structure of DNA/HMGB1/H1 Complexes: Electronic and Vibrational Circular Dichroism Studies. Biopolymers 2006, 83, 182. [Google Scholar] [CrossRef]
- Chikhirzhina, E.V.; Polyanichko, A.M.; Kostyleva, E.I.; Vorob’ev, V.I. The structure of the complexes of DNA with chromosomal protein HMGB1 and histone H1 in the presence of manganese ions. I. Circular dicroism spectroscopy. Molecular Biology (Mosk.) 2011, 45, 318–326. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, Y. High mobility group proteins and their post-translational modifications. Biochim. Biophys. Acta 2008, 1784, 1159–1166. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins During the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Spirin, A.S. Spectrophotometric determination of total nucleic acids. Biokhimiya 1958, 23, 656–662. [Google Scholar]
- Starkova, T.Y.; Polyanichko, A.M.; Artamonova, T.O.; Khodorkovskii, M.A.; Kostyleva, E.I.; Chikhirzhina, E.V.; Tomilin, A.N. Post-translational modifications of linker histone H1 variants in mammals. Phys. Biol. 2017, 14, 016005. [Google Scholar] [CrossRef]
- Starkova, T.Y.; Artamonova, T.O.; Ermakova, V.V.; Chikhirzhina, E.V.; Khodorkovskii, M.A.; Tomilin, A.N. The Profile of Post-translational Modifications of Histone H1 in Chromatin of Mouse Embryonic Stem Cells. Acta Naturae 2019, 11, 82–91. [Google Scholar] [CrossRef]
- Available online: http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msdigest (accessed on 20 February 2019).
- Available online: http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=SQ (accessed on 20 February 2019).
- McCauley, M.J.; Zimmerman, J.; Maher, L.J., III; Williams, M.C. HMGB binding to DNA: Single and double box motifs. J. Mol. Biol. 2007, 374, 993–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
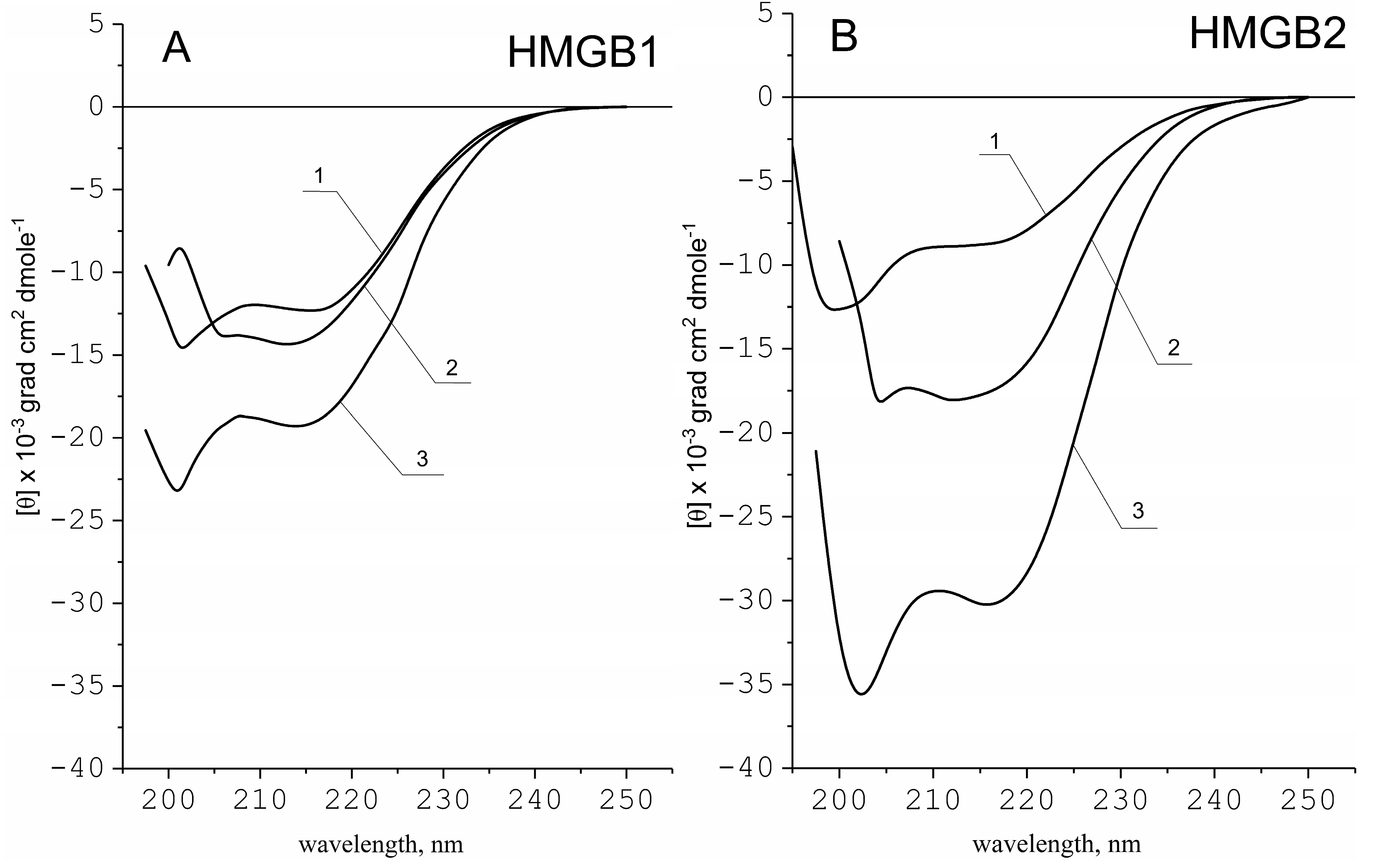
| Protein | α-Helicity (%), [θ222]/K2D3 * | ||
|---|---|---|---|
| Water | 80% Alcohol | 1.5 M NaCl | |
| HMGB1 | 25/30 | 30/30 | 42/46 |
| HMGB2 | 15/14 | 40/42 | 70/70 |
| Protein | Modification | Peptide | Sequence |
|---|---|---|---|
| HMGB1 | 1Oxidation | 58–65 | (K)GKFEDMAK(A) |
| 1Methyl 1Oxidation | 58–65 | (K)GKFEDMAK(A) | |
| 2Acetyl 1Methyl 1Phospho | 151–157 | (K)LKEKYEK(D) | |
| 1Acetyl 1Methyl 1Oxidation | 60–70 | (K)FEDMAKADKAR(Y) | |
| 1Acetyl 1Phospho | 77–87 | (K)TYIPPKGETKK(K) | |
| 2Phospho | 77–87 | (K)TYIPPKGETKK(K) | |
| 1Oxidation | 13–24 | (K)MSSYAFFVQTCR(E) | |
| 1Acetyl | 31–48 | (K)HPDASVNFSEFSKKCSER(W) | |
| 1Oxidation | 129–146 | (K)LGEMWNNTAADDKQPYEK(K) | |
| 2Methyl 1Oxidation 1Phospho | 8–24 | (K)KPRGKMSSYAFFVQTCR(E) | |
| 1Methyl 1Phospho | 13–29 | (K)MSSYAFFVQTCREEHKK(K) | |
| 1Oxidation | 58–65 | (K)GKFEDMAK(A) | |
| 1Oxidation | 71–76 | (R)YDREMK(N) | |
| HMGB2 | 1Phospho | 164–172 | (R)AKGKSEAGK(K) |
| 2Acetyl 1Methyl 1Phospho | 151–157 | (K)LKEKYEK(D) | |
| 1Dimethyl 2Methyl | 77–86 | (K)NYVPPKGDKK(G) | |
| 1Oxidation | 129–139 | (K)LGEMWSEQSAK(D) | |
| 1Acetyl 2Dimethyl 1Phospho | 77–86 | (K)NYVPPKGDKK(G) | |
| 1Oxidation | 128–139 | (K)KLGEMWSEQSAK(D) | |
| 1Oxidation | 13–24 | (K)MSSYAFFVQTCR(E) | |
| 1Acetyl 2Methyl 1Phospho | 140–150 | (K)DKQPYEQKAAK(L) | |
| 1Dimethyl 1Methyl 2Phospho | 71–82 | (R)YDREMKNYVPPK(G) | |
| 1Methyl 1Phospho | 13–29 | (K)MSSYAFFVQTCREEHKK(K) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Starkova, T.Y.; Polyanichko, A.M.; Artamonova, T.O.; Tsimokha, A.S.; Tomilin, A.N.; Chikhirzhina, E.V. Structural Characteristics of High-Mobility Group Proteins HMGB1 and HMGB2 and Their Interaction with DNA. Int. J. Mol. Sci. 2023, 24, 3577. https://doi.org/10.3390/ijms24043577
Starkova TY, Polyanichko AM, Artamonova TO, Tsimokha AS, Tomilin AN, Chikhirzhina EV. Structural Characteristics of High-Mobility Group Proteins HMGB1 and HMGB2 and Their Interaction with DNA. International Journal of Molecular Sciences. 2023; 24(4):3577. https://doi.org/10.3390/ijms24043577
Chicago/Turabian StyleStarkova, Tatiana Y., Alexander M. Polyanichko, Tatiana O. Artamonova, Anna S. Tsimokha, Alexey N. Tomilin, and Elena V. Chikhirzhina. 2023. "Structural Characteristics of High-Mobility Group Proteins HMGB1 and HMGB2 and Their Interaction with DNA" International Journal of Molecular Sciences 24, no. 4: 3577. https://doi.org/10.3390/ijms24043577







