Isolation and Characterization of the First Zobellviridae Family Bacteriophage Infecting Klebsiella pneumoniae
Abstract
:1. Introduction
2. Results
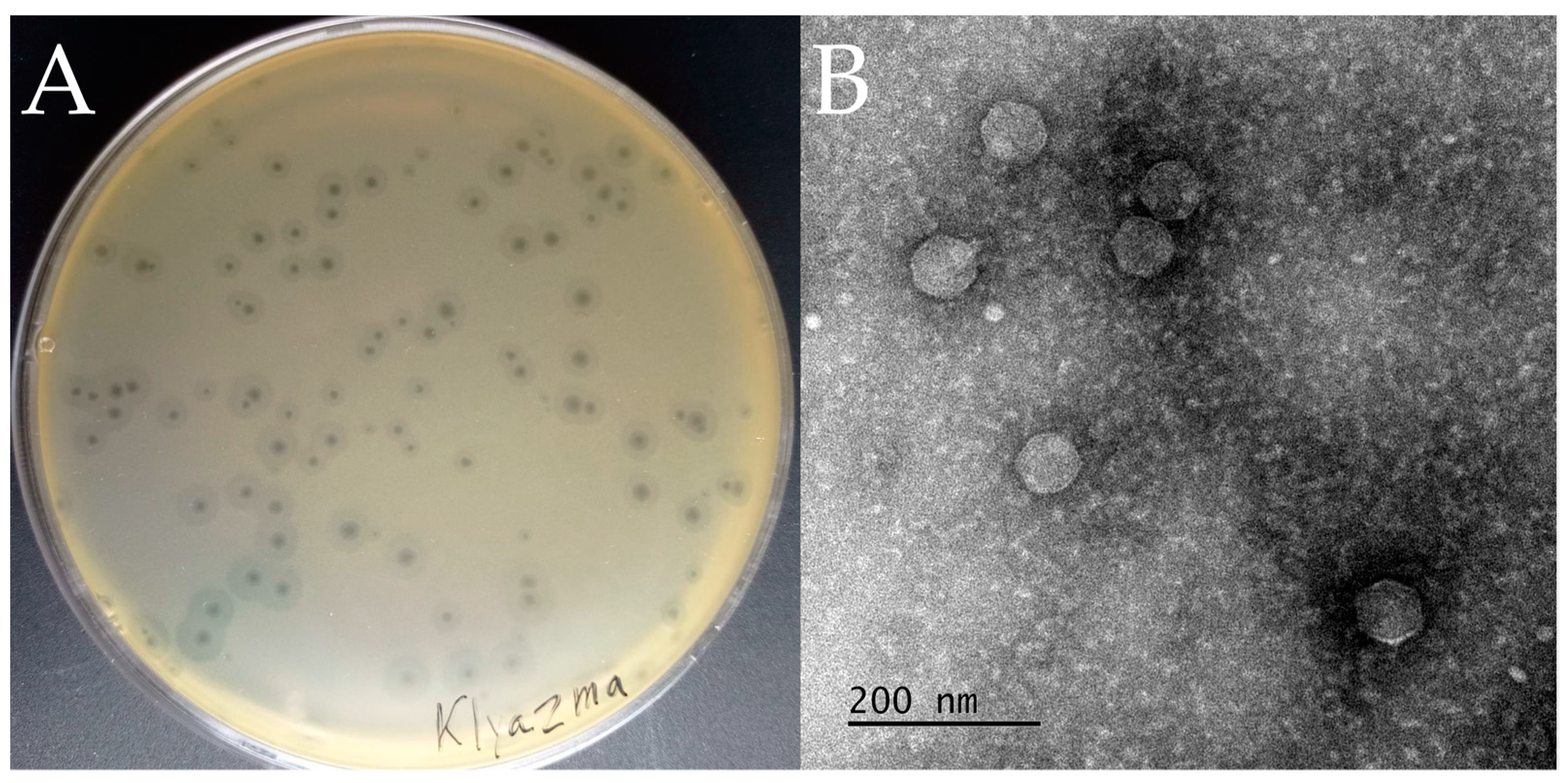
2.1. Isolation and Phenotypic Characterization of vB_KpnP_Klyazma
2.2. Genetic Organization and Phylogenetic Affiliation of vB_KpnP_Klyazma
2.3. Host Range of vB_KpnP_Klyazma and Its Depolymerase Activity
3. Discussion
4. Materials and Methods
4.1. Collection of K. pneumoniae Clinical Strains
4.2. Phage Isolation and Purification
4.3. Electron Microscopy of Phage Particles
4.4. Host Range Determination
4.5. Biological Characterization of vB_KpnP_Klyazma
4.6. DNA Sequencing and Genome Analysis
4.7. Phylogenetic Analysis
4.8. Cloning of Coding Sequence of Depolymerase
4.9. Protein Expression and Purification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Touati, A.; Mairi, A.; Baloul, Y.; Lalaoui, R.; Bakour, S.; Thighilt, L.; Gharout, A.; Rolain, J.-M. First detection of Klebsiella pneumoniae producing OXA-48 in fresh vegetables from Béjaïa city, Algeria. J. Glob. Antimicrob. Resist. 2017, 9, 17–18. [Google Scholar] [CrossRef] [PubMed]
- Asri, N.A.M.; Ahmad, S.; Mohamud, R.; Hanafi, N.M.; Zaidi, N.F.M.; Irekeola, A.A.; Shueb, R.H.; Yee, L.C.; Noor, N.M.; Mustafa, F.H.; et al. Global Prevalence of Nosocomial Multidrug-Resistant Klebsiella Pneumoniae: A Systematic Review and Meta-Analysis. Antibiotics 2021, 10, 1508. [Google Scholar] [CrossRef]
- Reardon, S. WHO warns against “post-antibiotic” era. Nature 2014, 15, 135–138. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Antimicrobial resistance in the EU/EEA (EARS-Net) AER for 2021; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2022; Available online: https://atlas.ecdc.europa.eu/ (accessed on 5 December 2022).
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Liu, Y.C.; Cheng, D.L.; Lin, C.L. Klebsiella pneumoniae Liver, Abscess Associated with Septic Endophthalmitis. Arch. Intern. Med. 1986, 146, 1913–1916. [Google Scholar] [CrossRef]
- Nassif, X.; Fournier, J.M.; Arondel, J.; Sansonetti, P.J. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect. Immun. 1989, 57, 546–552. [Google Scholar] [CrossRef] [Green Version]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. Clin. Microbiol. Rev. 2019, 32, e00001-19. [Google Scholar] [CrossRef] [Green Version]
- Gu, D.; Dong, N.; Zheng, Z.; Lin, D.; Huang, M.; Wang, L.; Chan, E.W.-C.; Shu, L.; Yu, J.; Zhang, R.; et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: A molecular epidemiological study. Lancet Infect. Dis. 2018, 18, 37–46. [Google Scholar] [CrossRef]
- Wyres, K.L.; Wick, R.R.; Judd, L.M.; Froumine, R.; Tokolyi, A.; Gorrie, C.L.; Lam, M.M.C.; Duchêne, S.; Jenney, A.; Holt, K.E. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019, 15, e1008114. [Google Scholar] [CrossRef] [Green Version]
- Górski, A.; Międzybrodzki, R.; Węgrzyn, G.; Jończyk-Matysiak, E.; Borysowski, J.; Weber-Dąbrowska, B. Phage therapy: Current status and perspectives. Med. Res. Rev. 2020, 40, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.R.J.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Herelle, M.F. On an invisible microbe antagonistic to dysentery bacilli. C. R. Acad. Sci. 1917, 165, 373–375. [Google Scholar]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Fabijan, A.P.; Lin, R.C.Y.; Ho, J.; Maddocks, S.; Zakour, N.L.B.; Iredell, J.R.; Westmead Bacteriophage Therapy Team. Safety of bacteriophage therapy in severe Staphylococcus aureus infection. Nat. Microbiol. 2020, 5, 465–472. [Google Scholar] [CrossRef]
- Aslam, S.; Lampley, E.; Wooten, D.; Karris, M.; Benson, C.; Strathdee, S.; Schooley, R.T. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. Open Forum Infect. Dis. 2020, 7, ofaa389. [Google Scholar] [CrossRef]
- Verbeken, G.; Pirnay, J.P. European regulatory aspects of phage therapy: Magistral phage preparations. Curr. Opin. Virol. 2022, 52, 24–29. [Google Scholar] [CrossRef]
- Kuptsov, N.S.; Kornienko, M.A.; Gorodnichev, R.B.; Danilov, D.I.; Malakhova, M.V.; Parfenova, T.V.; Makarenko, G.; Shitikov, E.; Ilina, E. Efficacy of commercial bacteriophage products against eskape pathogens. Bull. Russ. State Med. Univ. 2020, 18–24. [Google Scholar] [CrossRef]
- Danis-Wlodarczyk, K.M.; Wozniak, D.J.; Abedon, S.T. Treating bacterial infections with bacteriophage-based enzybiotics: In vitro, in vivo and clinical application. Antibiotics 2021, 10, 1497. [Google Scholar] [CrossRef]
- Komisarova, E.V.; Krasilnikova, V.M.; Volozhantsev, N.V. Bacteriophages, phage polysaccharide depolymerases and the possibility of their use for the treatment of bacterial infections. Bacteriology 2019, 4, 7–14. [Google Scholar] [CrossRef]
- Dams, D.; Briers, Y. Enzybiotics: Enzyme-Based Antibacterials as Therapeutics. Ther. Enzym. Funct. Clin. Implic. 2019, 1148, 233–253. [Google Scholar]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansal, S.; Harjai, K.; Chhibber, S. Depolymerase improves gentamicin efficacy during Klebsiella pneumoniae induced murine infection. BMC Infect. Dis. 2014, 14, 456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baykara, B.; Cimentepe, M.; Kandemir, T.; Koksal, F. Investigation of the Relationship between Colistin Resistance and Capsule Serotypes in Carbapenem Resistant Klebsiella pneumoniae Strains—PubMed. New Microbiol. 2022, 45, 124–129. Available online: https://pubmed.ncbi.nlm.nih.gov/35699561/ (accessed on 13 February 2023). [PubMed]
- Majkowska-Skrobek, G.; Łątka, A.; Berisio, R.; Maciejewska, B.; Squeglia, F.; Romano, M.; Lavigne, R.; Struve, C.; Drulis-Kawa, Z. Capsule-targeting depolymerase, derived from klebsiella KP36 phage, as a tool for the development of anti-virulent strategy. Viruses 2016, 8, 324. [Google Scholar] [CrossRef] [Green Version]
- Pires, D.P.; Oliveira, H.; Melo, L.D.R.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [Green Version]
- Russo, T.A.; Olson, R.; Fang, C.T.; Stoesser, N.; Miller, M.; MacDonald, U.; Hutson, A.; Barker, J.H.; La Hoz, R.M.; Johnson, J.R.; et al. Identification of biomarkers for differentiation of hypervirulent klebsiella pneumoniae from classical K. pneumoniae. J. Clin. Microbiol. 2018, 56, 776–794. [Google Scholar] [CrossRef] [Green Version]
- Abedon, S.T. Detection of bacteriophages: Phage plaques. In Bacteriophages: Biology, Technology, Therapy; Springer: Cham, Switzerland, 2021; pp. 507–538. [Google Scholar] [CrossRef]
- Ackermann, H.W. Basic phage electron microscopy. Methods Mol. Biol. 2009, 501, 113–126. [Google Scholar] [CrossRef]
- Solovieva, E.V.; Myakinina, V.P.; Kislichkina, A.A.; Krasilnikova, V.M.; Verevkin, V.V.; Mochalov, V.V.; Lev, A.I.; Fursova, N.K.; Volozhantsev, N.V. Comparative genome analysis of novel Podoviruses lytic for hypermucoviscous Klebsiella pneumoniae of K1, K2, and K57 capsular types. Virus Res. 2018, 243, 10–18. [Google Scholar] [CrossRef]
- Volozhantsev, N.V.; Myakinina, V.P.; Popova, A.V.; Kislichkina, A.A.; Komisarova, E.V.; Knyazeva, A.I.; Krasilnikova, V.M.; Fursova, N.K.; Svetoch, E.A. Complete genome sequence of novel T7-like virus vB_KpnP_KpV289 with lytic activity against Klebsiella pneumoniae. Arch. Virol. 2016, 161, 499–501. [Google Scholar] [CrossRef]
- Gorodnichev, R.; Kornienko, M.; Kuptsov, N.; Malakhova, M.; Bespiatykh, D.; Veselovsky, V.; Shitikov, E.; Ilina, E. Molecular genetic characterization of three new Klebsiella pneumoniae bacteriophages suitable for phage therapy. Med. Extrem Situat. 2021, 3, 113–119. [Google Scholar] [CrossRef]
- Alanin, K.W.S.; Olsen, N.S.; Djurhuus, A.M.; Carstens, A.B.; Nielsen, T.K.; Wagner, N.; Bak, F.; Hennessy, R.C.; Nicolaisen, M.H.; Kot, W.; et al. Three Novel Erwinia Billingiae Phages Represent Three New Genera Isolated from Organic Waste; Research Square: Durham, NC, USA, 2022. [Google Scholar] [CrossRef]
- Tikhe, C.V.; Martin, T.M.; Gissendanner, C.R.; Husseneder, C. Complete genome sequence of Citrobacter phage CVT22 isolated from the gut of the Formosan subterranean termite, Coptotermes formosanus Shiraki. Genome Announc. 2015, 3, e00408-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bischoff, V.; Adriaenssens, E.M.; Kropinski, A.M.; Duhaime, M.; Moraru, C. Create One New Family (Zobellviridae) Including One New Subfamily (Cobavirinae), Seven New Genera and 12 New Species (Caudovirales). 2020. Available online: https://ictv.global/ictv/proposals/2020.187B.R.Zobellviridae.zip (accessed on 13 February 2023).
- Hardies, S.C.; Hwang, Y.J.; Hwang, C.Y.; Jang, G.I.; Cho, B.C. Morphology, Physiological Characteristics, and Complete Sequence of Marine Bacteriophage ϕRIO-1 Infecting Pseudoalteromonas marina. J Virol. 2013, 87, 9189–9198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, P.S.; Domek, M.J.; Sanz-García, E.; Makaju, A.; Taylor, R.M.; Hoggan, R.; Culumber, M.D.; Oberg, C.J.; Breakwell, D.P.; Prince, J.T.; et al. Sequence and Structural Characterization of Great Salt Lake Bacteriophage CW02, a Member of the T7-Like Supergroup. J. Virol. 2012, 86, 7907–7917. [Google Scholar] [CrossRef] [Green Version]
- Tang, C.; Deng, C.; Zhang, Y.; Xiao, C.; Wang, J.; Rao, X.; Hu, F.; Lu, S. Characterization and genomic analyses of Pseudomonas aeruginosa podovirus TC6: Establishment of genus Pa11virus. Front. Microbiol. 2018, 9, 2561. [Google Scholar] [CrossRef] [Green Version]
- Hardies, S.C.; Thomas, J.A.; Black, L.; Weintraub, S.T.; Hwang, C.Y.; Cho, B.C. Identification of structural and morphogenesis genes of Pseudoalteromonas phage φRIO-1 and placement within the evolutionary history of Podoviridae. Virology. 2016, 489, 116–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savalia, D.; Westblade, L.F.; Goel, M.; Florens, L.; Kemp, P.; Akulenko, N.; Pavlova, O.; Padovan, J.C.; Chait, B.T.; Washburn, M.P.; et al. Genomic and Proteomic Analysis of phiEco32, a Novel Escherichia coli Bacteriophage. J. Mol. Biol. 2008, 377, 774–789. [Google Scholar] [CrossRef] [Green Version]
- Hardies, S.C.; Comeau, A.M.; Serwer, P.; Suttle, C.A. The complete sequence of marine bacteriophage VpV262 infecting vibrio parahaemolyticus indicates that an ancestral component of a T7 viral supergroup is widespread in the marine environment. Virology 2003, 310, 359–371. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Huang, K.; Yang, X.; Sun, L.; You, J.; Pan, X.; Cui, X.; Yang, H. Characterization of a novel lytic podovirus O4 of Pseudomonas aeruginosa. Arch. Virol. 2018, 163, 2377–2383. [Google Scholar] [CrossRef]
- Cheng, Y.H.; Huang, T.W.; Juan, C.H.; Chou, S.H.; Tseng, Y.Y.; Chen, T.W.; Yang, T.-C.; Lin, Y.-T. Tigecycline-non-susceptible hypervirulent Klebsiella pneumoniae strains in Taiwan. J. Antimicrob. Chemother. 2020, 75, 309–317. [Google Scholar] [CrossRef]
- Kakuta, N.; Nakano, R.; Nakano, A.; Suzuki, Y.; Masui, T.; Horiuchi, S.; Kakuta, R.; Tsubaki, K.; Ogawa, M.; Yano, H. Molecular characteristics of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in Japan: Predominance of CTX-M-15 and emergence of hypervirulent clones. Int. J. Infect. Dis. 2020, 98, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Berglund, B.; Chen, Y.; Zhou, Y.; Xiao, T.; Xiao, Y.; Zhou, K. Hypervirulence Markers Among Non-ST11 Strains of Carbapenem- and Multidrug-Resistant Klebsiella pneumoniae Isolated from Patients with Bloodstream Infections. Front. Microbiol. 2020, 11, 1199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, C.; Wang, Q.; Wang, X.; Chen, H.; Li, H.; Zhang, F.; Li, S.; Wang, R.; Wang, H. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: Geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob. Agents Chemother. 2016, 60, 6115–6120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorodnichev, R.B.; Volozhantsev, N.V.; Krasilnikova, V.M.; Bodoev, I.N.; Kornienko, M.A.; Kuptsov, N.S.; Popova, A.V.; Makarenko, G.I.; Manolov, A.I.; Slukin, P.V.; et al. Novel Klebsiella pneumoniae K23-Specific Bacteriophages from Different Families: Similarity of Depolymerases and Their Therapeutic Potential. Front. Microbiol. 2021, 12, 669618. [Google Scholar] [CrossRef] [PubMed]
- Volozhantsev, N.V.; Borzilov, A.I.; Shpirt, A.M.; Krasilnikova, V.M.; Verevkin, V.V.; Denisenko, E.A.; Kombarova, T.I.; Shashkov, A.S.; Knirel, Y.A.; Dyatlov, I.A. Comparison of the therapeutic potential of bacteriophage KpV74 and phage-derived depolymerase (β-glucosidase) against Klebsiella pneumoniae capsular type K2. Virus Res. 2022, 322, 198951. [Google Scholar] [CrossRef]
- Liu, Y.; Leung, S.S.Y.; Huang, Y.; Guo, Y.; Jiang, N.; Li, P.; Chen, J.; Wang, R.; Bai, C.; Mi, Z.; et al. Identification of Two Depolymerases from Phage IME205 and Their Antivirulent Functions on K47 Capsule of Klebsiella pneumoniae. Front. Microbiol. 2020, 11, 218. [Google Scholar] [CrossRef]
- Li, P.; Ma, W.; Shen, J.; Zhou, X. Characterization of Novel Bacteriophage vB_KpnP_ZX1 and Its Depolymerases with Therapeutic Potential for K57 Klebsiella pneumoniae Infection. Pharmaceutics 2022, 14, 1916. [Google Scholar] [CrossRef]
- Lin, T.L.; Yang, F.L.; Ren, C.T.; Pan, Y.J.; Liao, K.S.; Tu, I.F.; Chang, Y.-P.; Cheng, Y.-Y.; Wu, C.-Y.; Wu, S.-H.; et al. Development of Klebsiella pneumoniae Capsule Polysaccharide-Conjugated Vaccine Candidates Using Phage Depolymerases. Front. Immunol. 2022, 13, 843183. [Google Scholar] [CrossRef]
- Latka, A.; Drulis-Kawa, Z. Advantages and limitations of microtiter biofilm assays in the model of antibiofilm activity of Klebsiella phage KP34 and its depolymerase. Sci. Rep. 2020, 10, 20338. [Google Scholar] [CrossRef]
- Abedon, S.T.; Danis-Wlodarczyk, K.M.; Wozniak, D.J. Phage cocktail development for bacteriophage therapy: Toward improving spectrum of activity breadth and depth. Pharmaceuticals 2021, 14, 1019. [Google Scholar] [CrossRef]
- Pelfrene, E.; Sebris, Z.; Cavaleri, M. Regulatory aspects of the therapeutic use of bacteriophages: Europe. Bacteriophages Biol. Technol. Ther. 2021, 1165–1177. [Google Scholar] [CrossRef]
- Kornienko, M.; Ilina, E.; Lubasovskaya, L.; Priputnevich, T.; Falova, O.; Sukhikh, G.; Govorun, V. Analysis of nosocomial Staphylococcus haemolyticus by MLST and MALDI-TOF mass spectrometry. Infect. Genet. Evol. 2016, 39, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Fang, C.T.; Chuang, Y.P.; Shun, C.T.; Chang, S.C.; Wang, J.T. A Novel Virulence Gene in Klebsiella pneumoniae Strains Causing Primary Liver Abscess and Septic Metastatic Complications. J. Exp. Med. 2004, 199, 697–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Twest, R.; Kropinski, A.M. Bacteriophage enrichment from water and soil. Methods Mol. Biol. 2009, 501, 15–21. [Google Scholar] [CrossRef]
- Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages Using the Small Drop Plaque Assay System. Bacteriophages 2009, 81–85. [Google Scholar] [CrossRef]
- Kutter, E. Phage host range and efficiency of plating. Methods Mol. Biol. 2009, 501, 141–149. [Google Scholar] [CrossRef]
- Mirzaei, K.M.; Nilsson, A.S. Isolation of Phages for Phage Therapy: A Comparison of Spot Tests and Efficiency of Plating Analyses for Determination of Host Range and Efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef] [Green Version]
- Kropinski, A.M. Measurement of the Rate of Attachment of Bacteriophage to Cells. In Bacteriophages Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: New York, NY, USA, 2009; pp. 151–157. [Google Scholar] [CrossRef]
- Diancourt, L.; Passet, V.; Verhoef, J.; Grimont, P.A.D.; Brisse, S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005, 43, 4178–4182. [Google Scholar] [CrossRef] [Green Version]
- Brisse, S.; Passet, V.; Haugaard, A.B.; Babosan, A.; Kassis-Chikhani, N.; Struve, C.; Decré, D. Wzi gene sequencing, a rapid method for determination of capsulartype for Klebsiella strains. J. Clin. Microbiol. 2013, 51, 4073–4078. [Google Scholar] [CrossRef] [Green Version]
- Candan, E.D.; Aksöz, N. Klebsiella pneumoniae: Characteristics of carbapenem resistance and virulence factors. Acta Biochim. Pol. 2015, 62, 867–874. [Google Scholar] [CrossRef]
- Green, M.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor: New York, NY, USA, 2012. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garneau, J.R.; Depardieu, F.; Fortier, L.C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA genes in genomic sequences. In Methods in Molecular Biology; Kollmar, M., Ed.; Humana Press Inc.: New York, NY, USA, 2019; pp. 1–14. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.L.M.; Chooi, Y.H. Clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Liu, B.; Pop, M. ARDB—Antibiotic resistance genes database. Nucleic Acids Res. 2009, 37, 443–447. [Google Scholar] [CrossRef] [Green Version]
- GitHub—Kblin/ncbi-acc-download: Download Files from NCBI Entrez by Accession. Available online: https://github.com/kblin/ncbi-acc-download (accessed on 5 December 2022).
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Dai, Z.; Guo, P.; Fu, X.; Liu, S.; Zhou, L.; Tang, W.; Feng, T.; Chen, M.; Zhan, L.; et al. ggtreeExtra: Compact Visualization of Richly Annotated Phylogenetic Data. Mol. Biol. Evol. 2021, 38, 4039–4042. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Campitelli, E. ggnewscale: Multiple Fill and Color Scales in ggplot2; GitHub: San Francisco, CA, USA, 2022. [Google Scholar] [CrossRef]
- Xu, S. Multiple Geometric Shape Point Layer for “ggplot2” [R Package ggstar Version 1.0.4]. Comprehensive R Archive Network (CRAN). 2022. Available online: https://cran.r-project.org/package=ggstar (accessed on 15 December 2022).
- Aphalo, P.J. Explore the Innards of “ggplot2” Objects [R Package gginnards Version 0.1.1]. Comprehensive R Archive Network (CRAN). 2022. Available online: https://cran.r-project.org/package=gginnards (accessed on 15 December 2022).
- Larsson, J. Automatic Generation of Qualitative Color Palettes [R Package qualpalr Version 0.4.3]. Comprehensive R Archive Network (CRAN). 2018. Available online: https://cran.r-project.org/package=qualpalr (accessed on 15 December 2022).
- Müller, K. A Simpler Way to Find Your Files [R Package here Version 1.0.1]. Comprehensive R Archive Network (CRAN). 2020. Available online: https://cran.r-project.org/package=here (accessed on 15 December 2022).



| Strain | MLST | CPS Type | Meropenem Resistance | String Test | hvKp Genes | EOP | Kl-Dep Activity |
|---|---|---|---|---|---|---|---|
| L2-1B | 268 | KL20 | S | + | + | highly productive | + |
| Kl1886 | 268 | KL20 | S | + | + | inefficient | + |
| Kp-25-1 | 268 | KL20 | R | + | + | lysis from without | + |
| KL1877 | 268 | KL20 | R | − | + | lysis from without | + |
| Kp-G2-6 | 147 | KL20 | S | − | − | lysis from without | + |
| Kp1977 | 147 | KL20 | R | − | + | lysis from without | + |
| KBKp7 | 147 | KL20 | R | − | − | lysis from without | + |
| KL2071 | 147 | KL20 | R | − | − | lysis from without | + |
| KL1909 | 147 | KL20 | R | − | − | lysis from without | + |
| Kp2307 | 147 | KL20 | R | − | + | lysis from without | + |
| Kp9 | 1544 | KL20 | S | + | + | lysis from without | + |
| Kp-40 | no data | KL2 | S | − | no data | no activity | − |
| Kp-28p | no data | KL19 | R | − | no data | no activity | − |
| Kp2432 | no data | KL62 | R | − | no data | no activity | − |
| Kp2066 | no data | KL107 | R | − | no data | no activity | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorodnichev, R.B.; Kornienko, M.A.; Malakhova, M.V.; Bespiatykh, D.A.; Manuvera, V.A.; Selezneva, O.V.; Veselovsky, V.A.; Bagrov, D.V.; Zaychikova, M.V.; Osnach, V.A.; et al. Isolation and Characterization of the First Zobellviridae Family Bacteriophage Infecting Klebsiella pneumoniae. Int. J. Mol. Sci. 2023, 24, 4038. https://doi.org/10.3390/ijms24044038
Gorodnichev RB, Kornienko MA, Malakhova MV, Bespiatykh DA, Manuvera VA, Selezneva OV, Veselovsky VA, Bagrov DV, Zaychikova MV, Osnach VA, et al. Isolation and Characterization of the First Zobellviridae Family Bacteriophage Infecting Klebsiella pneumoniae. International Journal of Molecular Sciences. 2023; 24(4):4038. https://doi.org/10.3390/ijms24044038
Chicago/Turabian StyleGorodnichev, Roman B., Maria A. Kornienko, Maja V. Malakhova, Dmitry A. Bespiatykh, Valentin A. Manuvera, Oksana V. Selezneva, Vladimir A. Veselovsky, Dmitry V. Bagrov, Marina V. Zaychikova, Veronika A. Osnach, and et al. 2023. "Isolation and Characterization of the First Zobellviridae Family Bacteriophage Infecting Klebsiella pneumoniae" International Journal of Molecular Sciences 24, no. 4: 4038. https://doi.org/10.3390/ijms24044038
APA StyleGorodnichev, R. B., Kornienko, M. A., Malakhova, M. V., Bespiatykh, D. A., Manuvera, V. A., Selezneva, O. V., Veselovsky, V. A., Bagrov, D. V., Zaychikova, M. V., Osnach, V. A., Shabalina, A. V., Goloshchapov, O. V., Bespyatykh, J. A., Dolgova, A. S., & Shitikov, E. A. (2023). Isolation and Characterization of the First Zobellviridae Family Bacteriophage Infecting Klebsiella pneumoniae. International Journal of Molecular Sciences, 24(4), 4038. https://doi.org/10.3390/ijms24044038








