Insights into the Transport Cycle of LAT1 and Interaction with the Inhibitor JPH203
Abstract
:1. Introduction
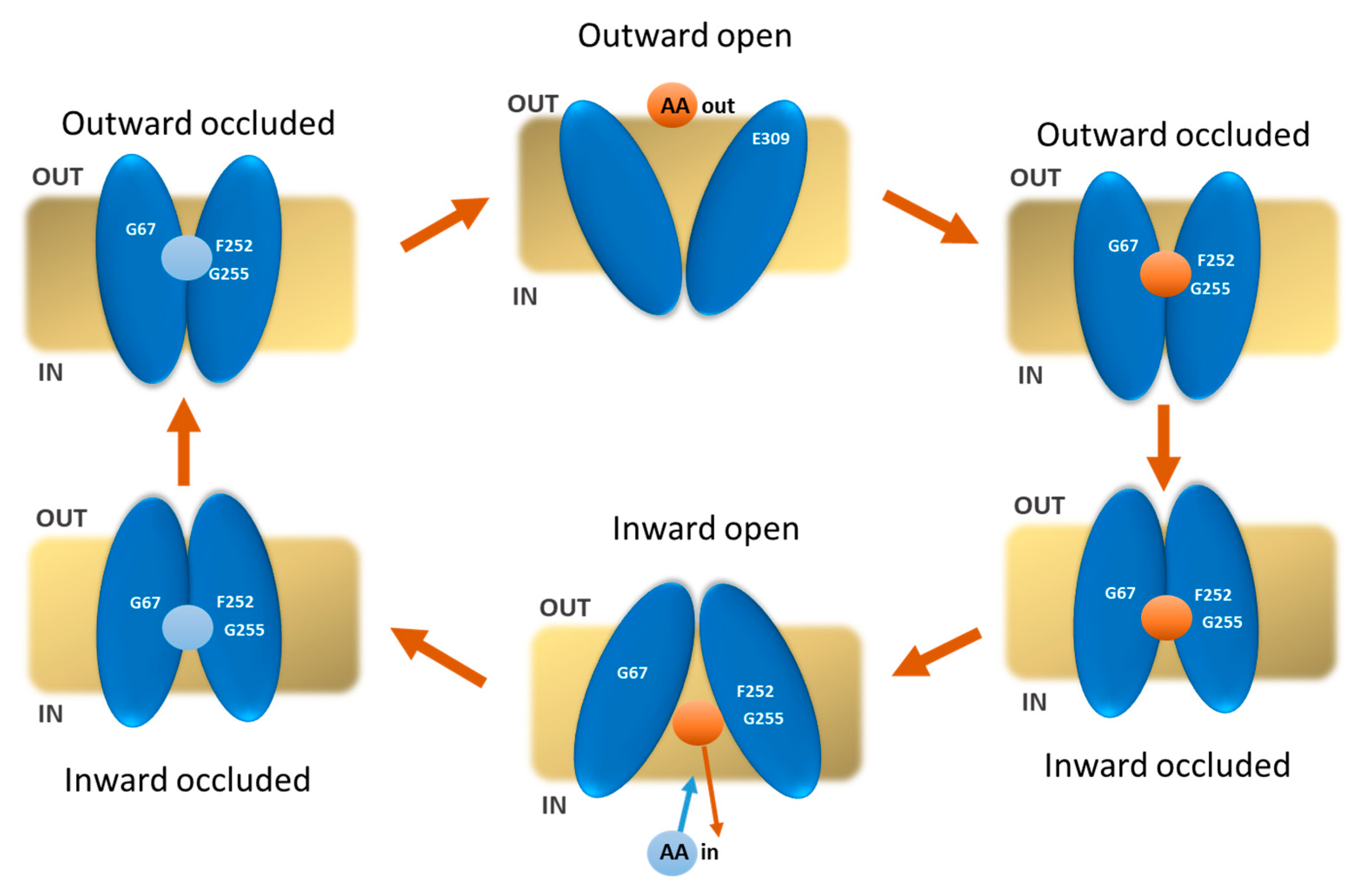
2. Results
2.1. Binding Site Characterization
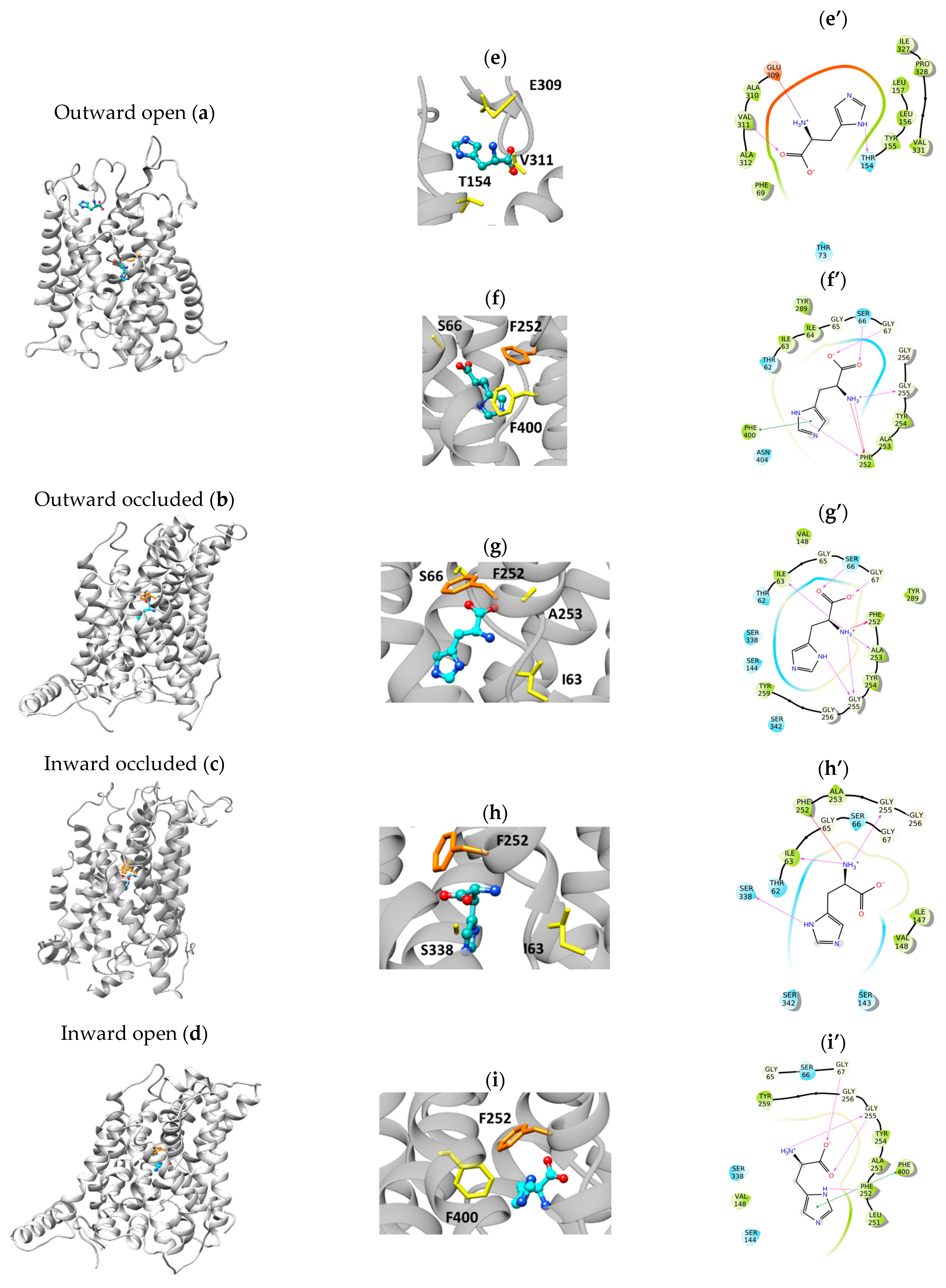
2.2. Docking Analysis
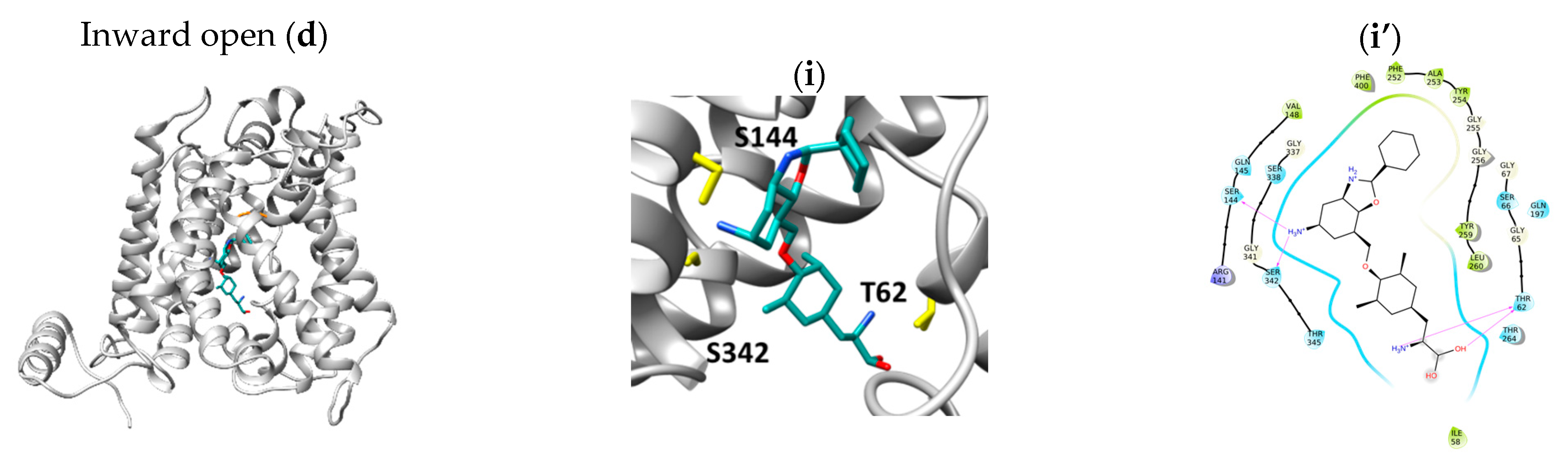
2.3. Docking of the Inhibitor JPH203
3. Discussion
4. Materials and Methods
4.1. Homology Modelling
4.2. Blind Docking
4.3. LAT1 Binding Sites Determination and Standard Precision Docking Were Carried Out Using the Schrodinger Maestro Platform
4.4. Visualization of Docking Results
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fotiadis, D.; Kanai, Y.; Palacin, M. The SLC3 and SLC7 families of amino acid transporters. Mol. Asp. Med. 2013, 34, 139–158. [Google Scholar] [CrossRef]
- Mastroberardino, L.; Spindler, B.; Pfeiffer, R.; Skelly, P.J.; Loffing, J.; Shoemaker, C.B.; Verrey, F. Amino-acid transport by heterodimers of 4F2hc/CD98 and members of a permease family. Nature 1998, 395, 288–291. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Segawa, H.; Miyamoto, K.; Uchino, H.; Takeda, E.; Endou, H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J. Biol. Chem. 1998, 273, 23629–23632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verrey, F.; Closs, E.I.; Wagner, C.A.; Palacin, M.; Endou, H.; Kanai, Y. CATs and HATs: The SLC7 family of amino acid transporters. Pflug. Arch. 2004, 447, 532–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palacin, M.; Nunes, V.; Font-Llitjos, M.; Jimenez-Vidal, M.; Fort, J.; Gasol, E.; Pineda, M.; Feliubadalo, L.; Chillaron, J.; Zorzano, A. The genetics of heteromeric amino acid transporters. Physiology 2005, 20, 112–124. [Google Scholar] [CrossRef]
- Console, L.; Scalise, M.; Salerno, S.; Scanga, R.; Giudice, D.; De Bartolo, L.; Tonazzi, A.; Indiveri, C. N-glycosylation is crucial for trafficking and stability of SLC3A2 (CD98). Sci. Rep. 2022, 12, 14570. [Google Scholar] [CrossRef]
- Palacin, M.; Kanai, Y. The ancillary proteins of HATs: SLC3 family of amino acid transporters. Pflug. Arch. 2004, 447, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, L.; Scalise, M.; Galluccio, M.; Pochini, L.; Albanese, L.M.; Indiveri, C. LAT1 is the transport competent unit of the LAT1/CD98 heterodimeric amino acid transporter. Int. J. Biochem. Cell Biol. 2015, 67, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Scalise, M.; Galluccio, M.; Console, L.; Pochini, L.; Indiveri, C. The Human SLC7A5 (LAT1): The Intriguing Histidine/Large Neutral Amino Acid Transporter and Its Relevance to Human Health. Front. Chem. 2018, 6, 243. [Google Scholar] [CrossRef]
- Kantipudi, S.; Jeckelmann, J.M.; Ucurum, Z.; Bosshart, P.D.; Fotiadis, D. The Heavy Chain 4F2hc Modulates the Substrate Affinity and Specificity of the Light Chains LAT1 and LAT2. Int. J. Mol. Sci. 2020, 21, 7573. [Google Scholar] [CrossRef]
- Tarlungeanu, D.C.; Deliu, E.; Dotter, C.P.; Kara, M.; Janiesch, P.C.; Scalise, M.; Galluccio, M.; Tesulov, M.; Morelli, E.; Sonmez, F.M.; et al. Impaired Amino Acid Transport at the Blood Brain Barrier Is a Cause of Autism Spectrum Disorder. Cell 2016, 167, 1481–1494.e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milkereit, R.; Persaud, A.; Vanoaica, L.; Guetg, A.; Verrey, F.; Rotin, D. LAPTM4b recruits the LAT1-4F2hc Leu transporter to lysosomes and promotes mTORC1 activation. Nat. Commun. 2015, 6, 7250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scalise, M.; Pochini, L.; Galluccio, M.; Console, L.; Indiveri, C. Glutamine Transport and Mitochondrial Metabolism in Cancer Cell Growth. Front. Oncol. 2017, 7, 306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosco, J.; Scalise, M.; Colas, C.; Galluccio, M.; Martini, R.; Rovella, F.; Mazza, T.; Ecker, G.F.; Indiveri, C. ATP modulates SLC7A5 (LAT1) synergistically with cholesterol. Sci. Rep. 2020, 10, 16738. [Google Scholar] [CrossRef] [PubMed]
- Bhutia, Y.D.; Ganapathy, V. Glutamine transporters in mammalian cells and their functions in physiology and cancer. Biochim. Biophys. Acta 2016, 1863, 2531–2539. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, B.C.; Bode, B.P. Amino acid transporters ASCT2 and LAT1 in cancer: Partners in crime? Semin. Cancer Biol. 2005, 15, 254–266. [Google Scholar] [CrossRef]
- Scalise, M.; Scanga, R.; Console, L.; Galluccio, M.; Pochini, L.; Indiveri, C. Chemical Approaches for Studying the Biology and Pharmacology of Membrane Transporters: The Histidine/Large Amino Acid Transporter SLC7A5 as a Benchmark. Molecules 2021, 26, 6562. [Google Scholar] [CrossRef] [PubMed]
- del Amo, E.M.; Urtti, A.; Yliperttula, M. Pharmacokinetic role of L-type amino acid transporters LAT1 and LAT2. Eur. J. Pharm. Sci. 2008, 35, 161–174. [Google Scholar] [CrossRef]
- Okano, N.; Hana, K.; Naruge, D.; Kawai, K.; Kobayashi, T.; Nagashima, F.; Endou, H.; Furuse, J. Biomarker Analyses in Patients With Advanced Solid Tumors Treated With the LAT1 Inhibitor JPH203. Vivo 2020, 34, 2595–2606. [Google Scholar] [CrossRef]
- Bahrami, K.; Jarvinen, J.; Laitinen, T.; Reinisalo, M.; Honkakoski, P.; Poso, A.; Huttunen, K.M.; Rautio, J. Structural Features Affecting the Interactions and Transportability of LAT1-Targeted Phenylalanine Drug Conjugates. Mol. Pharm. 2022, 20, 206–218. [Google Scholar] [CrossRef]
- Shi, Y. Common folds and transport mechanisms of secondary active transporters. Annu. Rev. Biophys. 2013, 42, 51–72. [Google Scholar] [CrossRef] [PubMed]
- Forrest, L.R.; Kramer, R.; Ziegler, C. The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta 2011, 1807, 167–188. [Google Scholar] [CrossRef] [Green Version]
- Shaffer, P.L.; Goehring, A.; Shankaranarayanan, A.; Gouaux, E. Structure and mechanism of a Na+-independent amino acid transporter. Science 2009, 325, 1010–1014. [Google Scholar] [CrossRef] [Green Version]
- Yan, R.; Zhao, X.; Lei, J.; Zhou, Q. Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex. Nature 2019, 568, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Scalise, M.; Pappacoda, G.; Mazza, T.; Console, L.; Pochini, L.; Indiveri, C. Cysteine 467 of the ASCT2 Amino Acid Transporter Is a Molecular Determinant of the Antiport Mechanism. Int. J. Mol. Sci. 2022, 23, 1127. [Google Scholar] [CrossRef] [PubMed]
- Krammer, E.M.; Ghaddar, K.; Andre, B.; Prevost, M. Unveiling the Mechanism of Arginine Transport through AdiC with Molecular Dynamics Simulations: The Guiding Role of Aromatic Residues. PLoS ONE 2016, 11, e0160219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Napolitano, L.; Galluccio, M.; Scalise, M.; Parravicini, C.; Palazzolo, L.; Eberini, I.; Indiveri, C. Novel insights into the transport mechanism of the human amino acid transporter LAT1 (SLC7A5). Probing critical residues for substrate translocation. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 727–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quan, L.; Ohgaki, R.; Hara, S.; Okuda, S.; Wei, L.; Okanishi, H.; Nagamori, S.; Endou, H.; Kanai, Y. Amino acid transporter LAT1 in tumor-associated vascular endothelium promotes angiogenesis by regulating cell proliferation and VEGF-A-dependent mTORC1 activation. J. Exp. Clin. Cancer Res. 2020, 39, 266. [Google Scholar] [CrossRef] [PubMed]
- Bo, T.; Kobayashi, S.; Inanami, O.; Fujii, J.; Nakajima, O.; Ito, T.; Yasui, H. LAT1 inhibitor JPH203 sensitizes cancer cells to radiation by enhancing radiation-induced cellular senescence. Transl. Oncol. 2021, 14, 101212. [Google Scholar] [CrossRef] [PubMed]
- Ylikangas, H.; Malmioja, K.; Peura, L.; Gynther, M.; Nwachukwu, E.O.; Leppanen, J.; Laine, K.; Rautio, J.; Lahtela-Kakkonen, M.; Huttunen, K.M.; et al. Quantitative insight into the design of compounds recognized by the L-type amino acid transporter 1 (LAT1). ChemMedChem 2014, 9, 2699–2707. [Google Scholar] [CrossRef]
- Augustyn, E.; Finke, K.; Zur, A.A.; Hansen, L.; Heeren, N.; Chien, H.C.; Lin, L.; Giacomini, K.M.; Colas, C.; Schlessinger, A.; et al. LAT-1 activity of meta-substituted phenylalanine and tyrosine analogs. Bioorg. Med. Chem. Lett. 2016, 26, 2616–2621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geier, E.G.; Schlessinger, A.; Fan, H.; Gable, J.E.; Irwin, J.J.; Sali, A.; Giacomini, K.M. Structure-based ligand discovery for the Large-neutral Amino Acid Transporter 1, LAT-1. Proc. Natl. Acad. Sci. USA 2013, 110, 5480–5485. [Google Scholar] [CrossRef] [Green Version]
- Hutchinson, K.; Silva, D.B.; Bohlke, J.; Clausen, C.; Thomas, A.A.; Bonomi, M.; Schlessinger, A. Describing inhibitor specificity for the amino acid transporter LAT1 from metainference simulations. Biophys. J. 2022, 121, 4476–4491. [Google Scholar] [CrossRef] [PubMed]
- Kongpracha, P.; Nagamori, S.; Wiriyasermkul, P.; Tanaka, Y.; Kaneda, K.; Okuda, S.; Ohgaki, R.; Kanai, Y. Structure-activity relationship of a novel series of inhibitors for cancer type transporter L-type amino acid transporter 1 (LAT1). J. Pharmacol. Sci. 2017, 133, 96–102. [Google Scholar] [CrossRef]
- Zur, A.A.; Chien, H.C.; Augustyn, E.; Flint, A.; Heeren, N.; Finke, K.; Hernandez, C.; Hansen, L.; Miller, S.; Lin, L.; et al. LAT1 activity of carboxylic acid bioisosteres: Evaluation of hydroxamic acids as substrates. Bioorg. Med. Chem. Lett. 2016, 26, 5000–5006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oda, K.; Hosoda, N.; Endo, H.; Saito, K.; Tsujihara, K.; Yamamura, M.; Sakata, T.; Anzai, N.; Wempe, M.F.; Kanai, Y.; et al. L-type amino acid transporter 1 inhibitors inhibit tumor cell growth. Cancer Sci. 2010, 101, 173–179. [Google Scholar] [CrossRef]
- Wempe, M.F.; Rice, P.J.; Lightner, J.W.; Jutabha, P.; Hayashi, M.; Anzai, N.; Wakui, S.; Kusuhara, H.; Sugiyama, Y.; Endou, H. Metabolism and pharmacokinetic studies of JPH203, an L-amino acid transporter 1 (LAT1) selective compound. Drug Metab. Pharmacokinet. 2012, 27, 155–161. [Google Scholar] [CrossRef] [Green Version]
- Toyoshima, J.; Kusuhara, H.; Wempe, M.F.; Endou, H.; Sugiyama, Y. Investigation of the role of transporters on the hepatic elimination of an LAT1 selective inhibitor JPH203. J. Pharm. Sci. 2013, 102, 3228–3238. [Google Scholar] [CrossRef] [PubMed]
- Rosilio, C.; Nebout, M.; Imbert, V.; Griessinger, E.; Neffati, Z.; Benadiba, J.; Hagenbeek, T.; Spits, H.; Reverso, J.; Ambrosetti, D.; et al. L-type amino-acid transporter 1 (LAT1): A therapeutic target supporting growth and survival of T-cell lymphoblastic lymphoma/T-cell acute lymphoblastic leukemia. Leukemia 2015, 29, 1253–1266. [Google Scholar] [CrossRef]
- Hayashi, K.; Jutabha, P.; Maeda, S.; Supak, Y.; Ouchi, M.; Endou, H.; Fujita, T.; Chida, M.; Anzai, N. LAT1 acts as a crucial transporter of amino acids in human thymic carcinoma cells. J. Pharmacol. Sci. 2016, 132, 201–204. [Google Scholar] [CrossRef] [Green Version]
- Choi, D.W.; Kim, D.K.; Kanai, Y.; Wempe, M.F.; Endou, H.; Kim, J.K. JPH203, a selective L-type amino acid transporter 1 inhibitor, induces mitochondria-dependent apoptosis in Saos2 human osteosarcoma cells. Korean J. Physiol. Pharmacol. 2017, 21, 599–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yothaisong, S.; Dokduang, H.; Anzai, N.; Hayashi, K.; Namwat, N.; Yongvanit, P.; Sangkhamanon, S.; Jutabha, P.; Endou, H.; Loilome, W. Inhibition of l-type amino acid transporter 1 activity as a new therapeutic target for cholangiocarcinoma treatment. Tumour. Biol. 2017, 39, 1010428317694545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueno, S.; Kimura, T.; Yamaga, T.; Kawada, A.; Ochiai, T.; Endou, H.; Sakurai, H. Metformin enhances anti-tumor effect of L-type amino acid transporter 1 (LAT1) inhibitor. J. Pharmacol. Sci. 2016, 131, 110–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enomoto, K.; Sato, F.; Tamagawa, S.; Gunduz, M.; Onoda, N.; Uchino, S.; Muragaki, Y.; Hotomi, M. A novel therapeutic approach for anaplastic thyroid cancer through inhibition of LAT1. Sci. Rep. 2019, 9, 14616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, C.; Wolfe, H.; Wells, A.; Chien, H.C.; Colas, C.; Schlessinger, A.; Giacomini, K.M.; Thomas, A.A. l-Type amino acid transporter 1 activity of 1,2,3-triazolyl analogs of l-histidine and l-tryptophan. Bioorg. Med. Chem. Lett. 2019, 29, 2254–2258. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Singh, S.K.; Kawate, T.; Jin, Y.; Gouaux, E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature 2005, 437, 215–223. [Google Scholar] [CrossRef]
- Krishnamurthy, H.; Piscitelli, C.L.; Gouaux, E. Unlocking the molecular secrets of sodium-coupled transporters. Nature 2009, 459, 347–355. [Google Scholar] [CrossRef]
- Jungnickel, K.E.J.; Parker, J.L.; Newstead, S. Structural basis for amino acid transport by the CAT family of SLC7 transporters. Nat. Commun. 2018, 9, 550. [Google Scholar] [CrossRef] [Green Version]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Soding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Lu, F.; Zhou, L.; Dang, S.; Sun, L.; Li, X.; Wang, J.; Shi, Y. Structure and mechanism of an amino acid antiporter. Science 2009, 324, 1565–1568. [Google Scholar] [CrossRef]
- Fang, Y.; Jayaram, H.; Shane, T.; Kolmakova-Partensky, L.; Wu, F.; Williams, C.; Xiong, Y.; Miller, C. Structure of a prokaryotic virtual proton pump at 3.2 A resolution. Nature 2009, 460, 1040–1043. [Google Scholar] [CrossRef] [Green Version]
- Ilgu, H.; Jeckelmann, J.M.; Gapsys, V.; Ucurum, Z.; de Groot, B.L.; Fotiadis, D. Insights into the molecular basis for substrate binding and specificity of the wild-type L-arginine/agmatine antiporter AdiC. Proc. Natl. Acad. Sci. USA 2016, 113, 10358–10363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilgu, H.; Jeckelmann, J.M.; Kalbermatter, D.; Ucurum, Z.; Lemmin, T.; Fotiadis, D. High-resolution structure of the amino acid transporter AdiC reveals insights into the role of water molecules and networks in oligomerization and substrate binding. BMC Biol. 2021, 19, 179. [Google Scholar] [CrossRef]
- Kowalczyk, L.; Ratera, M.; Paladino, A.; Bartoccioni, P.; Errasti-Murugarren, E.; Valencia, E.; Portella, G.; Bial, S.; Zorzano, A.; Fita, I.; et al. Molecular basis of substrate-induced permeation by an amino acid antiporter. Proc. Natl. Acad. Sci. USA. 2011, 108, 3935–3940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, J.; Kim, B.H.; Grishin, N.V. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008, 36, 2295–2300. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. Methods Mol. Biol. 2021, 2199, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Henin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Halgren, T. New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des. 2007, 69, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Achmad, A.; Lestari, S.; Holik, H.A.; Rahayu, D.; Bashari, M.H.; Faried, A.; Kartamihardja, A.H.S. Highly Specific L-Type Amino Acid Transporter 1 Inhibition by JPH203 as a Potential Pan-Cancer Treatment. Processes 2021, 9, 1170. [Google Scholar] [CrossRef]

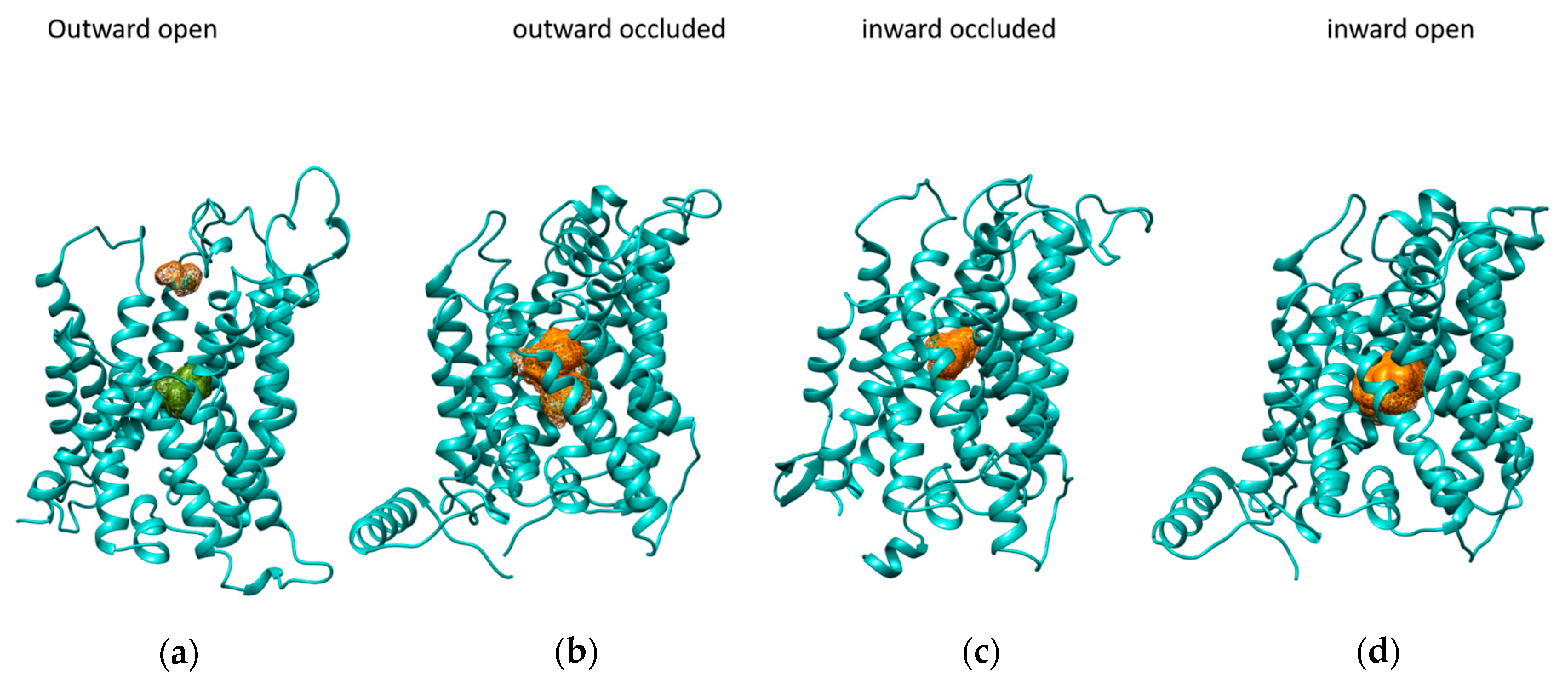
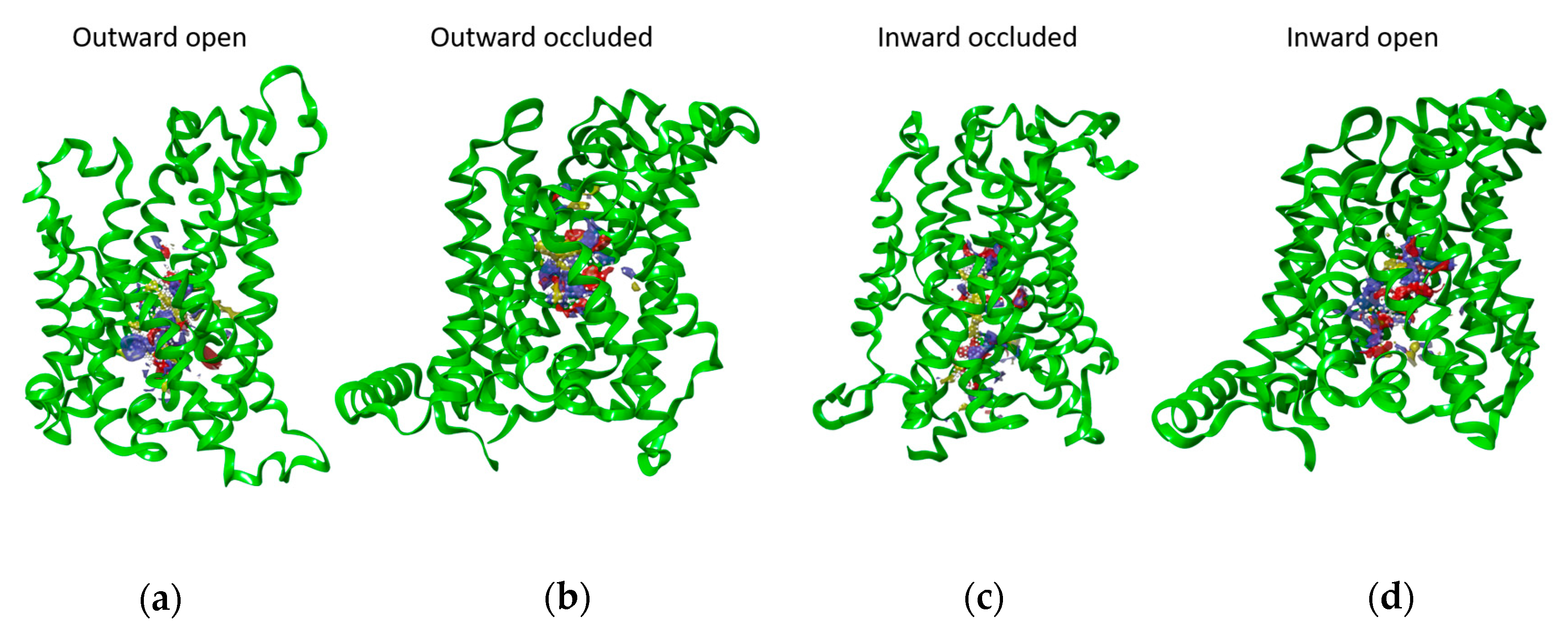
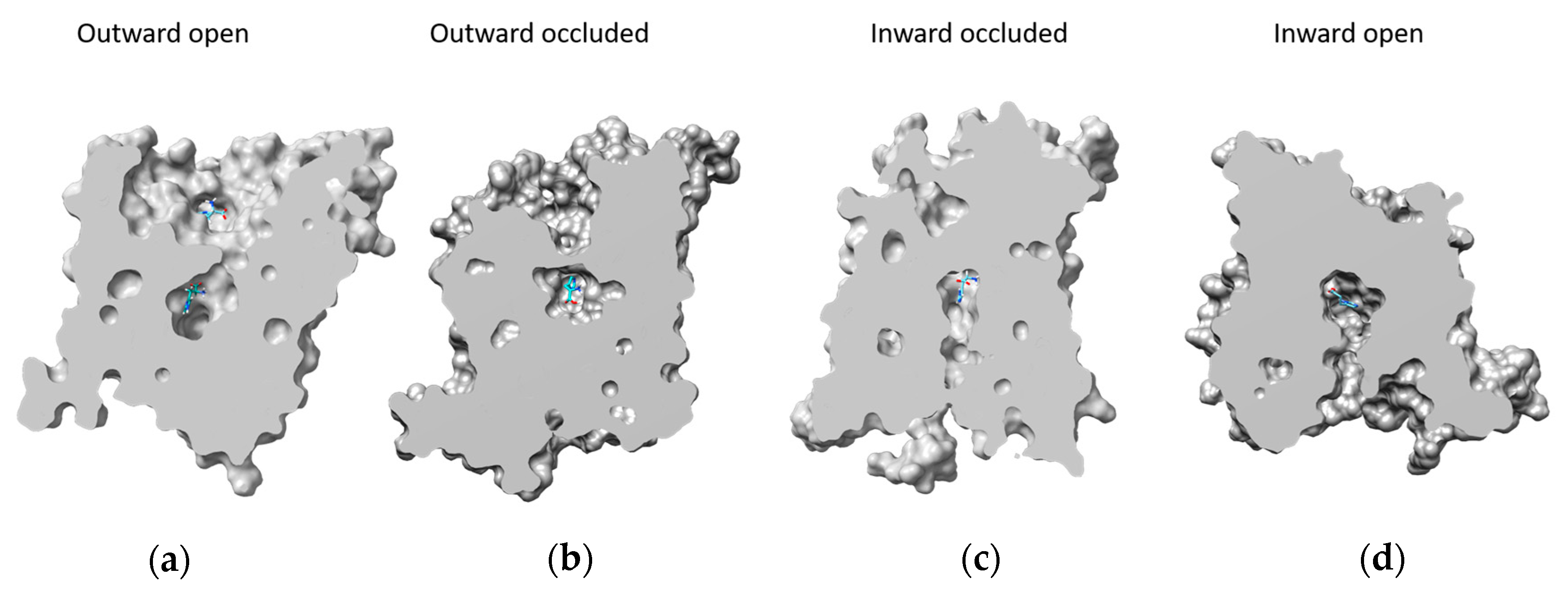

| Amino Acid | Outward Open | Outward Occluded | Inward Occluded | Inward Open |
|---|---|---|---|---|
| His | −5.72 | −6.3 | −5.31 | −4.93 |
| Ile | −3.56 | −5.15 | −4.11 | −3.8 |
| Val | −3.77 | −5.2 | −4.39 | −4.14 |
| Leu | −3.59 | −5.05 | −3.86 | −3.79 |
| Phe | −4.57 | −5.53 | −5.22 | −4.85 |
| Cys | −3.85 | −4.91 | −4.08 | −4.01 |
| Met | −3.76 | −4.98 | −3.79 | −3.02 |
| Ala | −2.33 | −3.61 | −2.56 | −2.41 |
| Gly | −1.73 | −4.42 | −1.94 | −1.67 |
| Thr | −3.82 | −5.19 | −4.78 | −3.44 |
| Ser | −3.30 | −6.85 | −4.16 | −3.25 |
| Trp | −4.86 | −7.31 | −5.86 | −5.87 |
| Tyr | −4.07 | −5.64 | −5.21 | −5.49 |
| Pro | −4.35 | −5.79 | −3.73 | −4.75 |
| Glu | −3.84 | −5.02 | −3.66 | −3.48 |
| Gln | −4.09 | −5.53 | −4.17 | −4.08 |
| Asp | −3.99 | −5.07 | −3.74 | −3.77 |
| Asn | −4.44 | −5.37 | −5.24 | −4.24 |
| Lys | −3.79 | −4.91 | −3.69 | −3.74 |
| Arg | −3.05 | −4.40 | −3.01 | −2.63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brunocilla, C.; Console, L.; Rovella, F.; Indiveri, C. Insights into the Transport Cycle of LAT1 and Interaction with the Inhibitor JPH203. Int. J. Mol. Sci. 2023, 24, 4042. https://doi.org/10.3390/ijms24044042
Brunocilla C, Console L, Rovella F, Indiveri C. Insights into the Transport Cycle of LAT1 and Interaction with the Inhibitor JPH203. International Journal of Molecular Sciences. 2023; 24(4):4042. https://doi.org/10.3390/ijms24044042
Chicago/Turabian StyleBrunocilla, Chiara, Lara Console, Filomena Rovella, and Cesare Indiveri. 2023. "Insights into the Transport Cycle of LAT1 and Interaction with the Inhibitor JPH203" International Journal of Molecular Sciences 24, no. 4: 4042. https://doi.org/10.3390/ijms24044042
APA StyleBrunocilla, C., Console, L., Rovella, F., & Indiveri, C. (2023). Insights into the Transport Cycle of LAT1 and Interaction with the Inhibitor JPH203. International Journal of Molecular Sciences, 24(4), 4042. https://doi.org/10.3390/ijms24044042








