The Gut–Organ-Axis Concept: Advances the Application of Gut-on-Chip Technology
Abstract
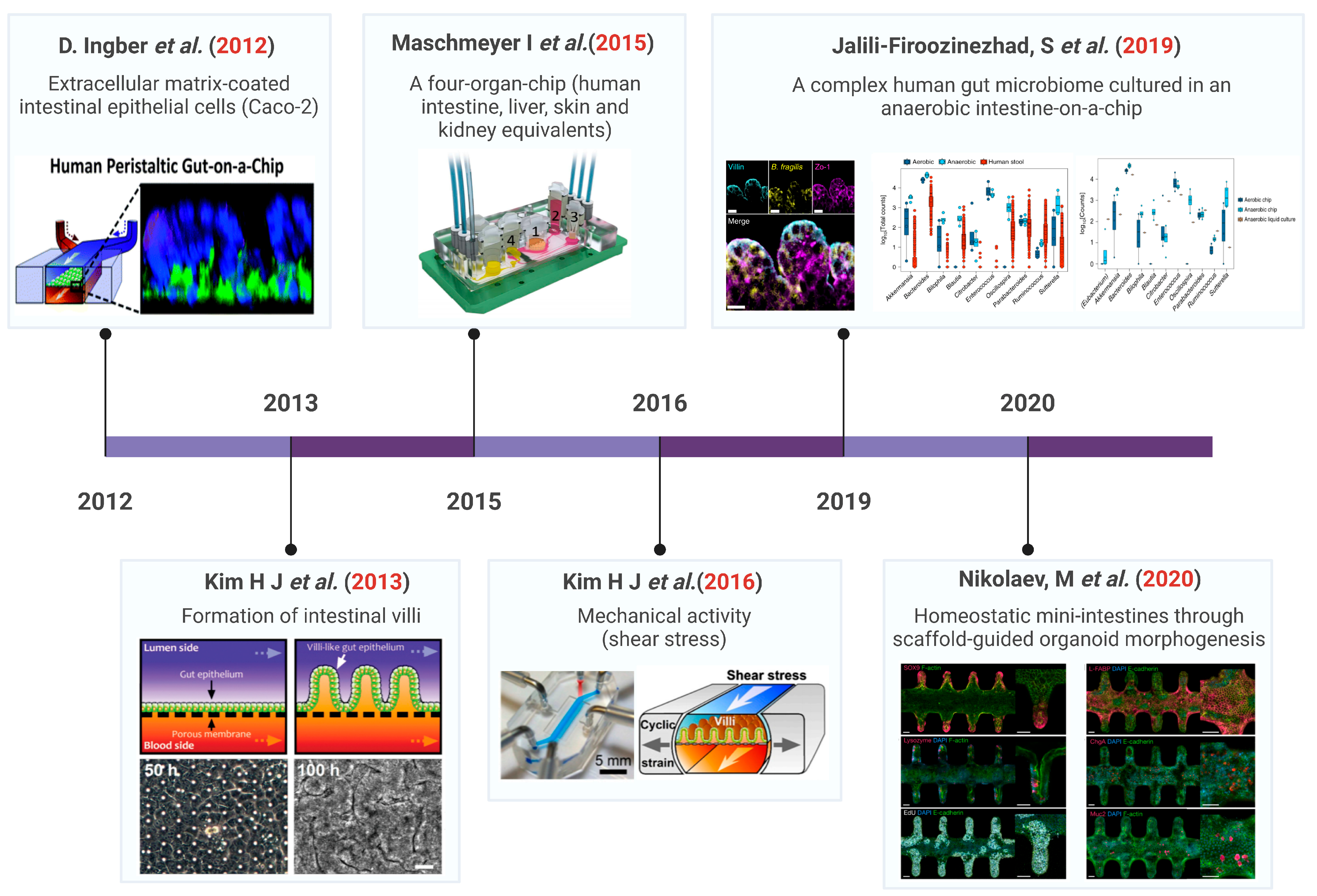
1. Introduction
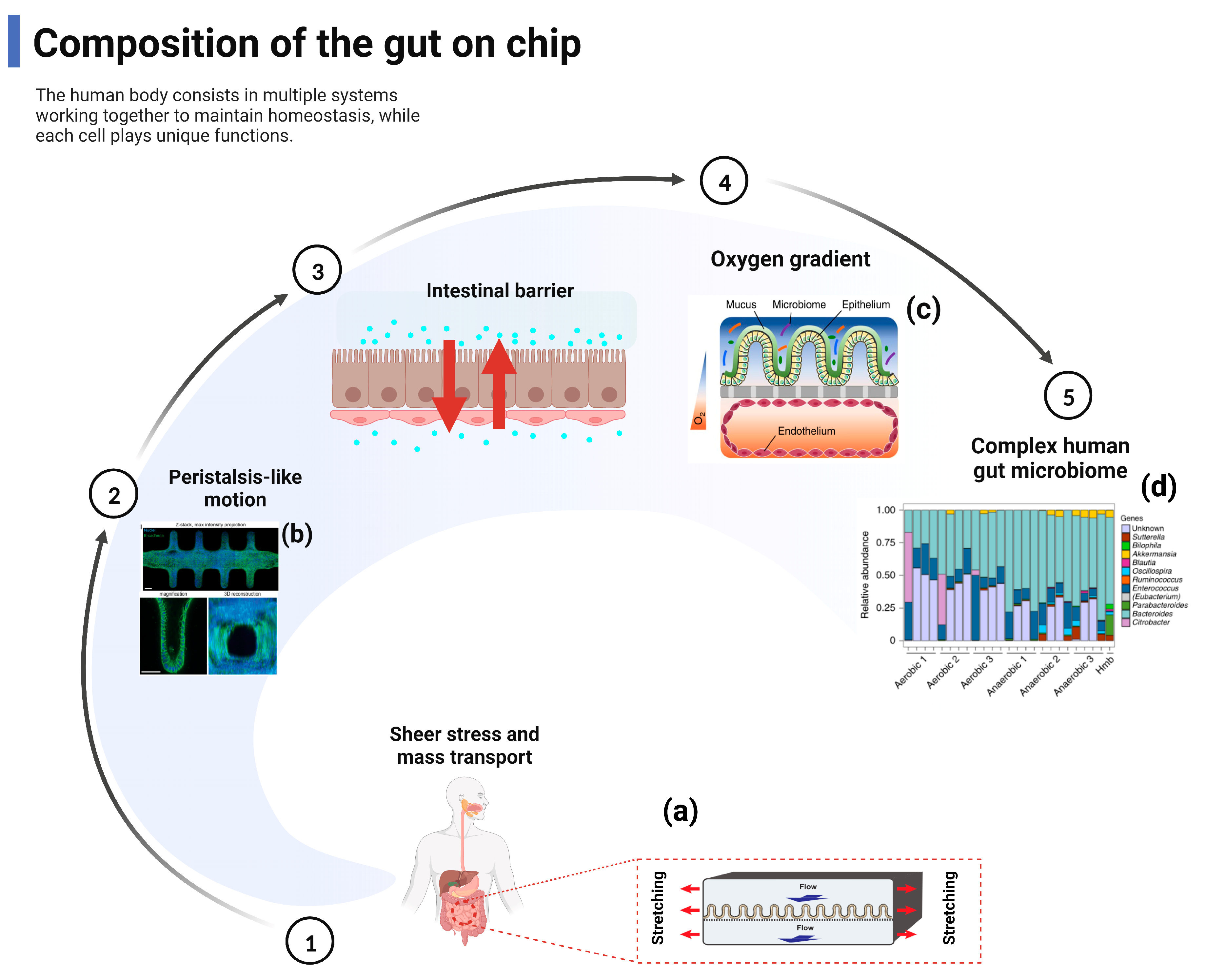
2. Composition of the Gut–Organ-on-Chip
3. Gut–Organ-Axis on Chip
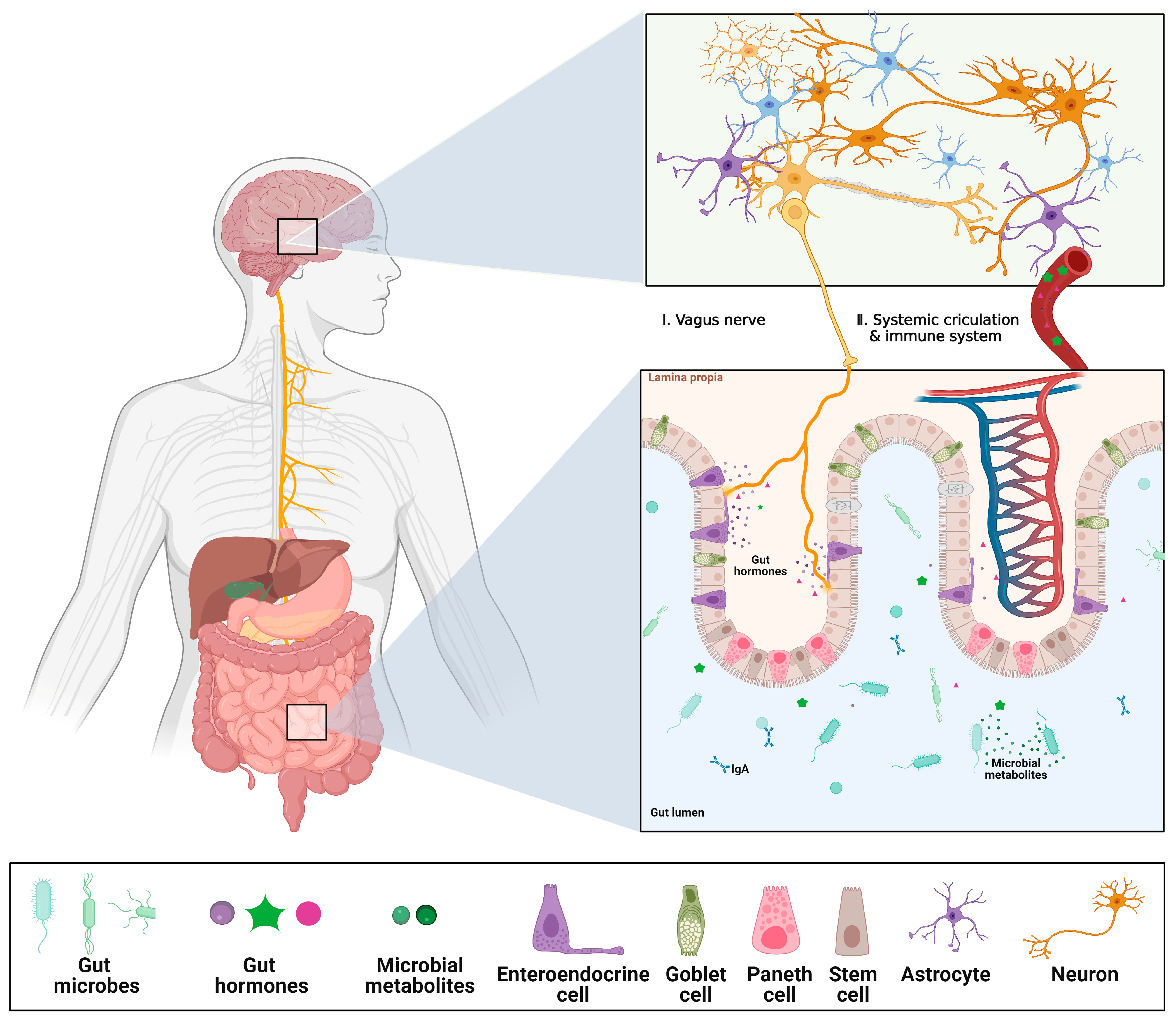
3.1. Gut–Brain Axis (GBA) on Chip
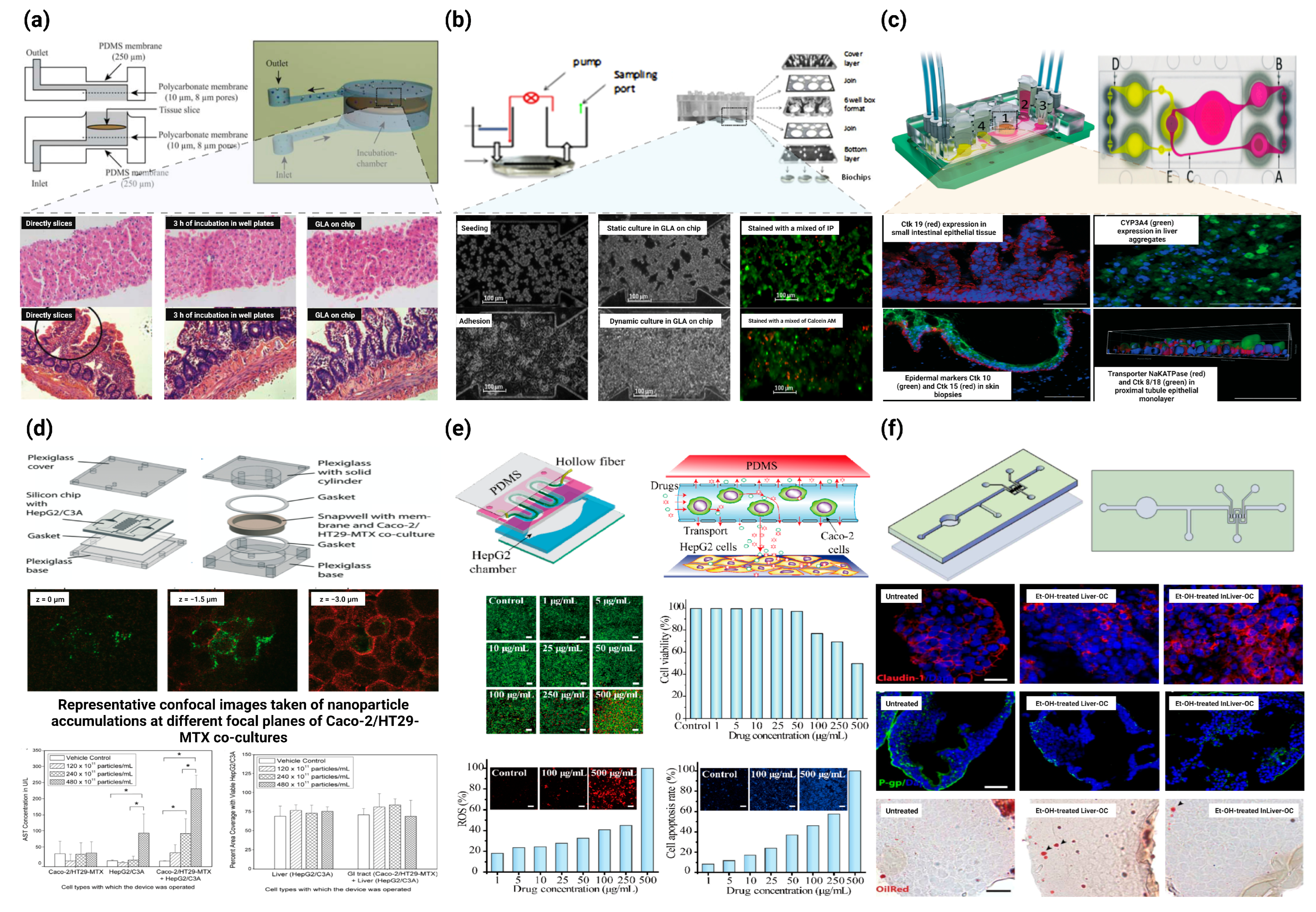
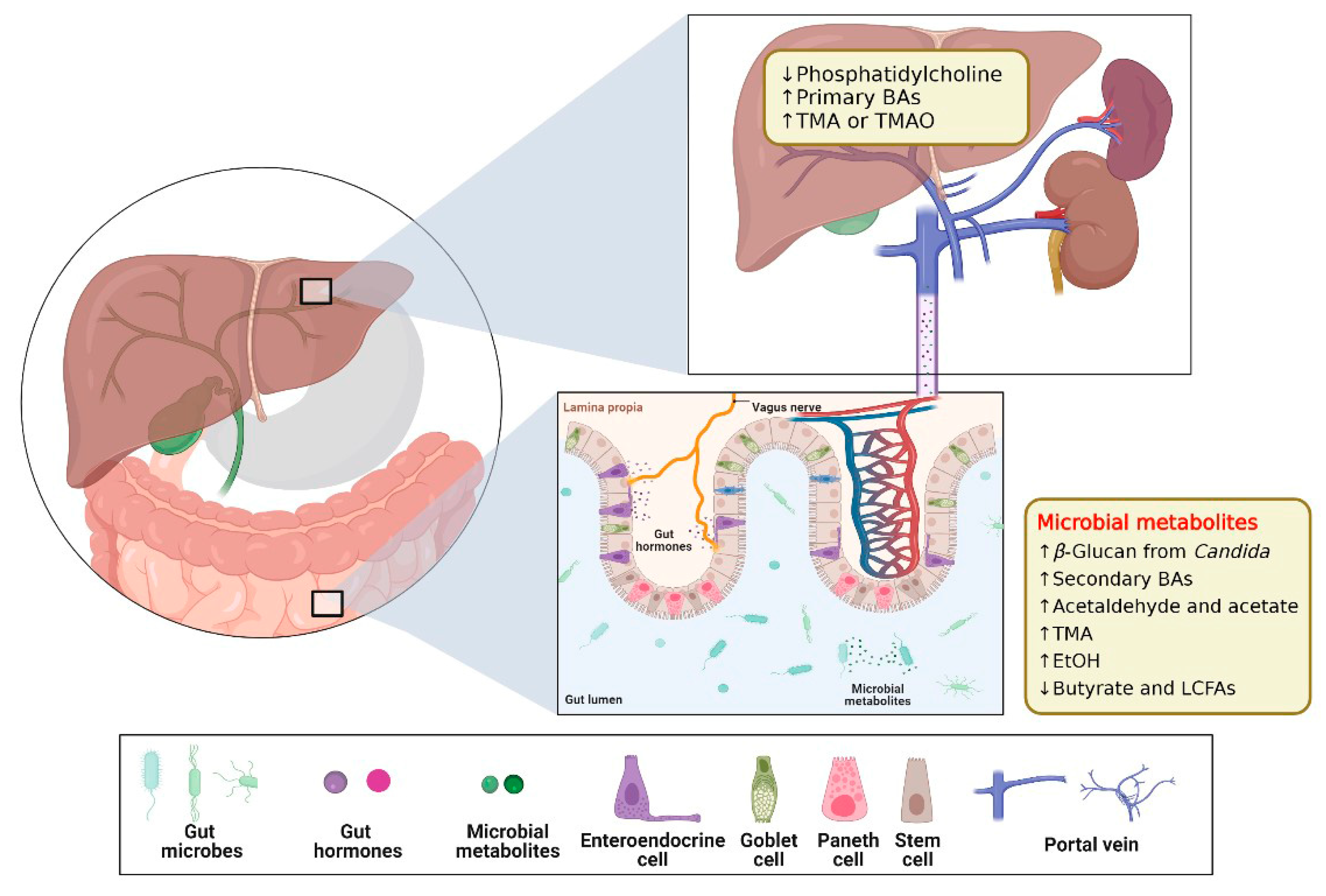
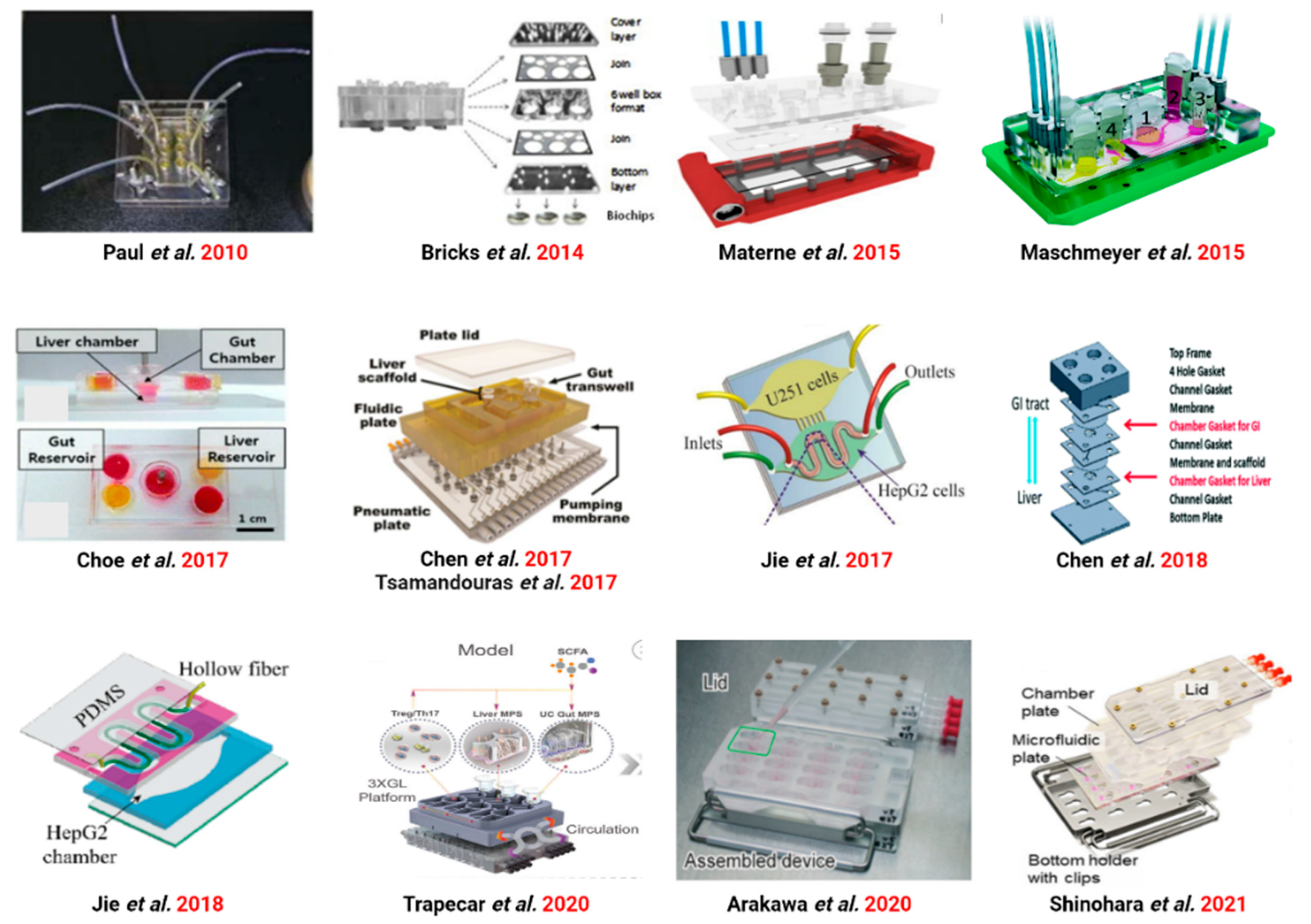
3.2. Gut–Liver Axis (GLA) on Chip
3.2.1. GLA Chip of Physiological Mechanisms Studies
3.2.2. GLA Chip for Drug Metabolism Studies
3.2.3. GLA Chip for Drug Toxicity Studies
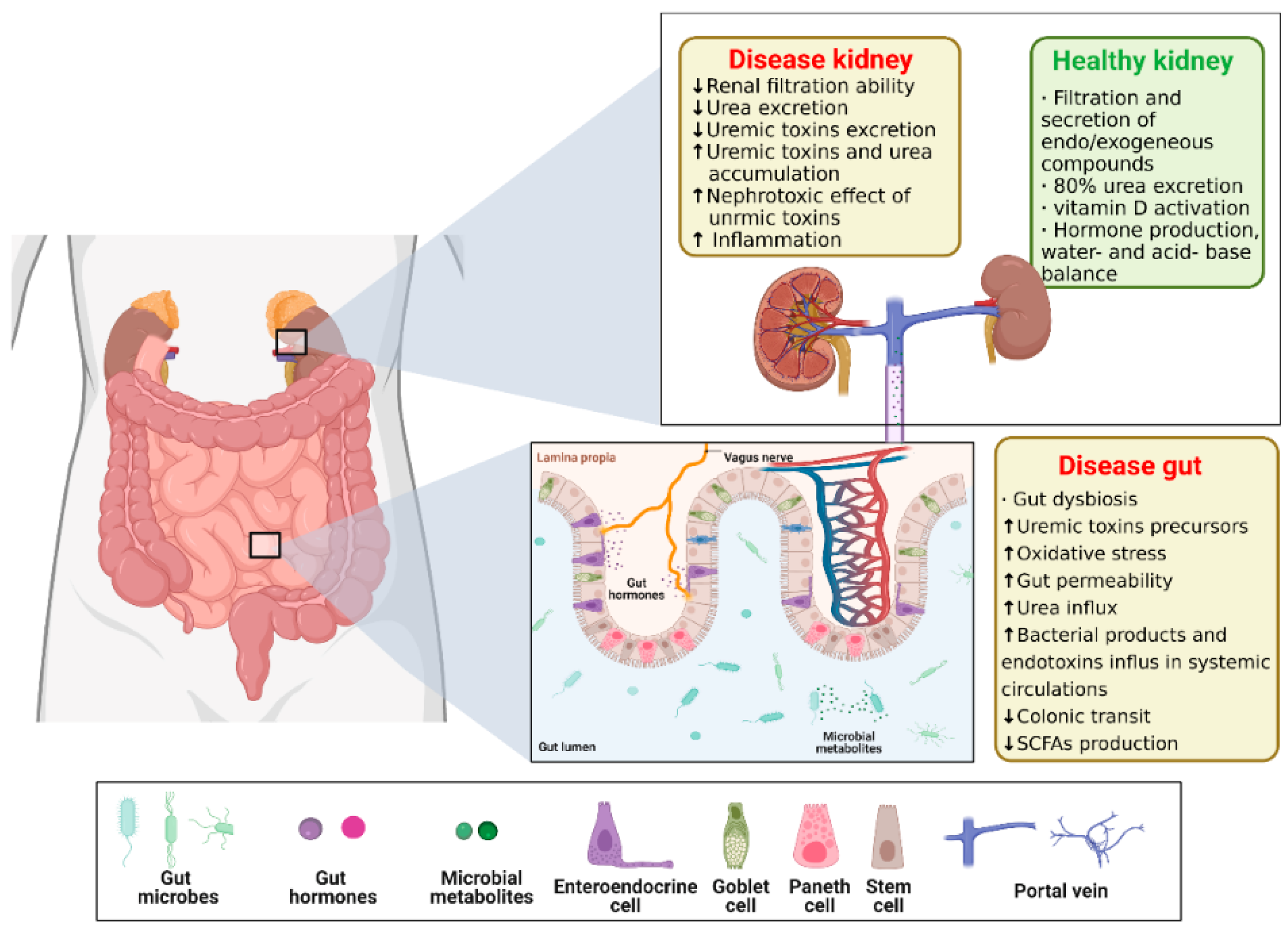
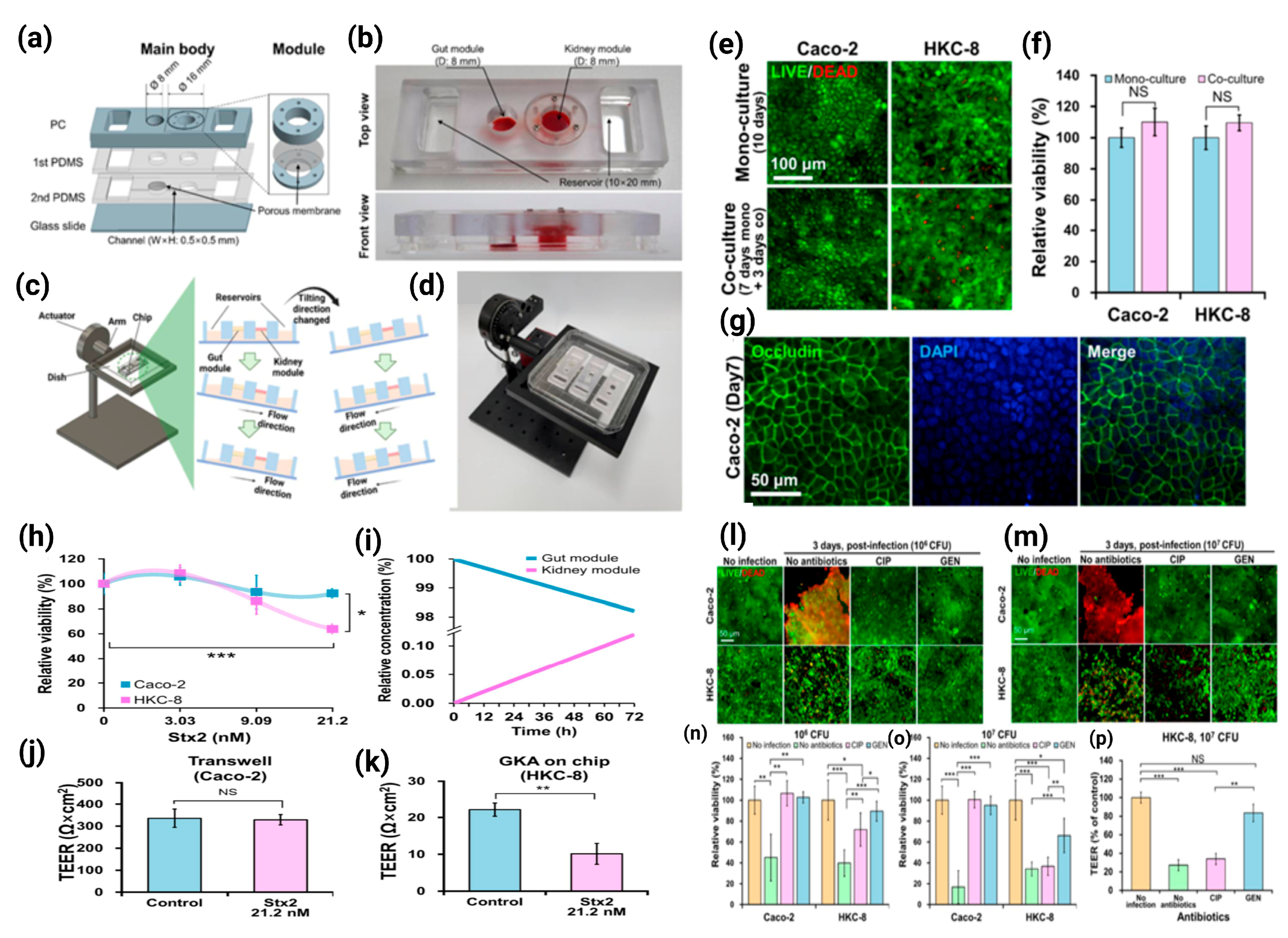
3.3. Gut–Kidney Axis (GKA) on Chip
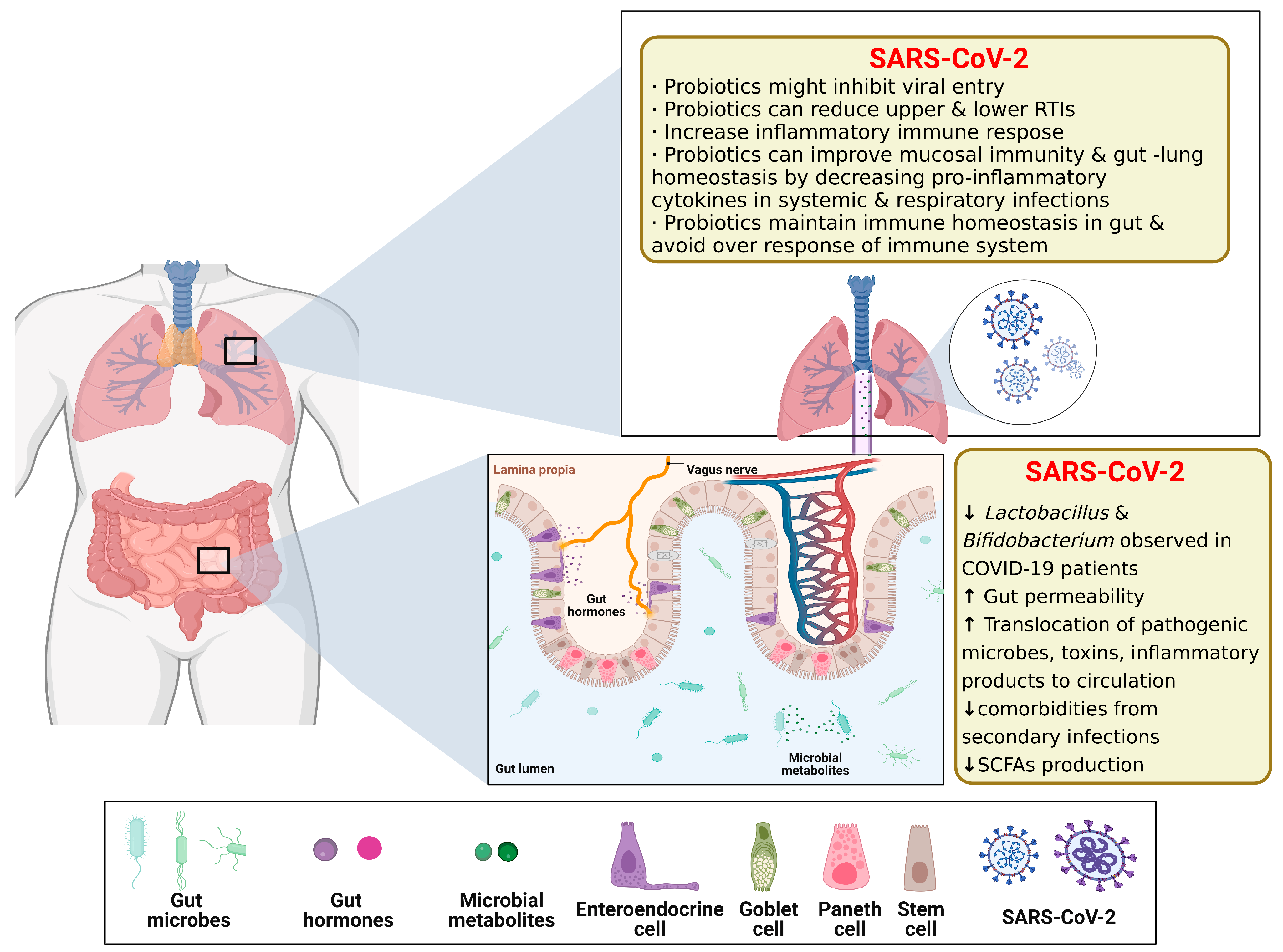
3.4. Gut–Lung Axis (GLAx) on Chip
3.5. Others
4. Future Advances in Gut–Organ-Axis-on-a-Chip: Challenges and Opportunities
- (1)
- Although some studies have been able to construct gut microarrays suitable for the survival of anaerobic bacteria, only specific types or classes of anaerobic bacteria have been tested, and there are still technical bottlenecks for testing the function of complex gut flora on the microarrays, which need to be addressed by further research.
- (2)
- The human body is a complex whole, and although existing studies have proposed a variety of interoperable models such as the brain–intestinal axis, the liver–intestinal axis, and the kidney–intestinal axis, how to consider the crosstalk between multiple models is a hot issue that should be of concern in subsequent studies of disease occurrence and drug action. Organ-on-a-chip systems are still marginal in the pharmaceutical industry, and pharmacokinetic simulation studies, toxicology studies, pharmacodynamic studies, and the reduction of each organ corresponding to the physiological environment and physiological effects in vivo are also important in drug development.
- (3)
- Although many researchers have sought to reduce the cost of using organ chips by finding more suitable materials and more convenient models, the specialized and cumbersome nature of chip design and manufacture also limits their large-scale use, so how can we improve their applicability by reducing the constraints such as the difficulty and cost of use from the perspective of raw material selection, template design, and chip manufacture? In addition to the finished chip and its suitability for use, the stability and repeatability of the chip quality is also a factor to be examined.
- (4)
- In the future development of gut–organ-axis chips, the signal molecules observed in the “mini-organ and mini-gut” can be used to infer the in vivo regulation of the gut–organ-axis, thus suggesting further possibilities for the future application of gut–brain microarrays in gut–organ-axis-related diseases, the exploration of disease mechanisms and the development of new drugs.
- (5)
- In terms of detection, the technological capabilities of the future intestinal axis microarray platform will allow real-time, in situ, and dynamic maintenance and monitoring of a large number of biological parameters such as shear stress, pH, oxygen, cytokines, as well as the use of methods such as electrochemical or optical- and fluorescence-based methods in conjunction with such organ systems on a chip. It is also combined with ELISA, PCR, and single-cell mRNA sequencing to correlate biomarker, molecular characterization, cell physiology, and histopathology of pathology.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cryan, J.F.; O’Riordan, K.J.; Sandhu, K.; Peterson, V.; Dinan, T.G. The gut microbiome in neurological disorders. Lancet Neurol. 2020, 19, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; Backhed, F. From association to causality: The role of the gut microbiota and its functional products on host metabolism. Mol. Cell 2020, 78, 584–596. [Google Scholar] [CrossRef] [PubMed]
- Compare, D.; Coccoli, P.; Rocco, A.; Nardone, O.M.; De Maria, S.; Carteni, M.; Nardone, G. Gut-liver axis: The impact of gut microbiota on non alcoholic fatty liver disease. Nutr. Metab. Cardiovasc. Dis. 2012, 22, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Lyte, M. Microbial endocrinology in the microbiome-gut-brain axis: How bacterial production and utilization of neurochemicals influence behavior. PLoS Pathog. 2013, 9, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Moser, G.; Fournier, C.; Peter, J. Intestinal microbiome-gut-brain axis and irritable bowel syndrome. Wien. Med. Wochenschr. 2018, 168, 62–66. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef]
- Raimondi, M.T.; Albani, D.; Giordano, C. An organ-on-a-chip engineered platform to study the microbiota-gut-brain axis in neurodegeneration. Trends Mol. Med. 2019, 25, 737–740. [Google Scholar] [CrossRef]
- Van Schaik, W. The human gut resistome. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140087. [Google Scholar] [CrossRef]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Loomba, R.; Schnabl, B.; Knight, R. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef]
- Margolis, K.G.; Cryan, J.F.; Mayer, E.A. The microbiota-gut-brain axis: From motility to mood. Gastroenterology 2021, 160, 1486–1501. [Google Scholar] [CrossRef]
- Yang, T.; Richards, E.M.; Pepine, C.J.; Raizada, M.K. The gut microbiota and the brain-gut-kidney axis in hypertension and chronic kidney disease. Nat. Rev. Nephrol. 2018, 14, 442–456. [Google Scholar] [CrossRef]
- Martin, C.R.; Osadchiy, V.; Kalani, A.; Mayer, E.A. The brain-gut-microbiome axis. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 133–148. [Google Scholar] [CrossRef]
- Marsland, B.J.; Trompette, A.; Gollwitzer, E.S. The gut-lung axis in respiratory disease. Ann. Am. Thorac. Soc. 2015, 12, S150–S156. [Google Scholar] [CrossRef]
- Zhang, J.H.; Fan, J.J.; Zeng, X.; Nie, M.M.; Luan, J.Y.; Wang, Y.C.; Ju, D.W.; Yin, K. Hedgehog signaling in gastrointestinal carcinogenesis and the gastrointestinal tumor microenvironment. Acta Pharm. Sin. B 2021, 11, 609–620. [Google Scholar] [CrossRef]
- Lee, M.; Chang, E.B. Inflammatory bowel diseases (IBD) and the microbiome-searching the crime scene for clues. Gastroenterology 2021, 160, 524–537. [Google Scholar] [CrossRef]
- Collins, S.L.; Patterson, A.D. The gut microbiome: An orchestrator of xenobiotic metabolism. Acta Pharm. Sin. B 2020, 10, 19–32. [Google Scholar] [CrossRef]
- Zhang, B.; Korolj, A.; Lai, B.F.L.; Radisic, M. Advances in organ-on-a-chip engineering. Nat. Rev. Mater. 2018, 3, 257–278. [Google Scholar] [CrossRef]
- Odijk, M.; van der Meer, A.D.; Levner, D.; Kim, H.J.; van der Helm, M.W.; Segerink, L.I.; Frimat, J.-P.; Hamilton, G.A.; Ingber, D.E.; van den Berg, A. Measuring direct current trans-epithelial electrical resistance in organ-on-a-chip microsystems. Lab A Chip 2015, 15, 745–752. [Google Scholar] [CrossRef]
- Ehrenfellner, B.; Zissler, A.; Steinbacher, P.; Monticelli, F.C.; Pittner, S. Are animal models predictive for human postmortem muscle protein degradation? Int. J. Leg. Med. 2017, 131, 1615–1621. [Google Scholar] [CrossRef]
- Van der Worp, H.B.; Howells, D.W.; Sena, E.S.; Porritt, M.J.; Rewell, S.; O’Collins, V.; Macleod, M.R. Can animal models of disease reliably inform human studies? PLoS Med. 2010, 7, e1000245. [Google Scholar] [CrossRef] [PubMed]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Park, M.H.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Kim, H.J.; Ingber, D.E. Microfluidic organ-on-a-chip models of human intestine. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Ahadian, S.; Civitarese, R.; Bannerman, D.; Mohammadi, M.H.; Lu, R.; Wang, E.; Davenport-Huyer, L.; Lai, B.; Zhang, B.; Zhao, Y.; et al. Organ-on-a-chip platforms: A convergence of advanced materials, cells, and microscale technologies. Adv. Healthc. Mater. 2018, 7, 1700506. [Google Scholar] [CrossRef] [PubMed]
- Hartung, T. Toxicology for the twenty-first century. Nature 2009, 460, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Peng, Y.; Li, H.; Chen, W. Organ-on-a-Chip: A new paradigm for drug development. Trends Pharmacol. Sci. 2021, 42, 119–133. [Google Scholar] [CrossRef]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2021, 20, 345–361. [Google Scholar] [CrossRef]
- Kim, M.-H.; van Noort, D.; Sung, J.H.; Park, S. Organ-on-a-chip for studying gut-brain interaction mediated by extracellular vesicles in the gut microenvironment. Int. J. Mol. Sci. 2021, 22, 3513. [Google Scholar] [CrossRef]
- Dickson, I. Anaerobic intestine-on-a-chip system enables complex microbiota co-culture. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 390. [Google Scholar] [CrossRef]
- Nikolaev, M.; Mitrofanova, O.; Broguiere, N.; Geraldo, S.; Dutta, D.; Tabata, Y.; Elci, B.; Brandenberg, N.; Kolotuev, I.; Gjorevski, N.; et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 2020, 585, 574–583. [Google Scholar] [CrossRef]
- Kim, M.-H.; Kim, D.; Sung, J.H. A Gut-Brain Axis-on-a-Chip for studying transport across epithelial and endothelial barriers. J. Ind. Eng. Chem. 2021, 101, 126–134. [Google Scholar] [CrossRef]
- Guo, Y.; Luo, R.; Wang, Y.; Deng, P.; Song, T.; Zhang, M.; Wang, P.; Zhang, X.; Cui, K.; Tao, T.; et al. SARS-CoV-2 induced intestinal responses with a biomimetic human gut-on-chip. Sci. Bull. 2021, 66, 783–793. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.-H.; Alves, D.R.; Kim, S.; Lee, L.P.; Sung, J.H.; Park, S. Gut-kidney axis on chip for studying effects of antibiotics on risk of hemolytic uremic syndrome by shiga toxin-producing Escherichia coli. Toxins 2021, 13, 775. [Google Scholar] [CrossRef]
- Chen, W.L.K.; Edington, C.; Suter, E.; Yu, J.; Velazquez, J.J.; Velazquez, J.G.; Shockley, M.; Large, E.M.; Venkataramanan, R.; Hughes, D.J.; et al. Integrated gut/liver microphysiological systems elucidates inflammatory inter-tissue crosstalk. Biotechnol. Bioeng. 2017, 114, 2648–2659. [Google Scholar] [CrossRef]
- Shinohara, M.; Arakawa, H.; Oda, Y.; Shiraki, N.; Sugiura, S.; Nishiuchi, T.; Satoh, T.; Iino, K.; Leo, S.; Kato, Y.; et al. Coculture with hiPS-derived intestinal cells enhanced human hepatocyte functions in a pneumatic-pressure-driven two-organ microphysiological system. Sci. Rep. 2021, 11, 5437. [Google Scholar] [CrossRef]
- Arakawa, H.; Sugiura, S.; Kawanishi, T.; Shin, K.; Toyoda, H.; Satoh, T.; Sakai, Y.; Kanamori, T.; Kato, Y. Kinetic analysis of sequential metabolism of triazolam and its extrapolation to humans using an entero-hepatic two-organ microphysiological system. Lab A Chip 2020, 20, 537–547. [Google Scholar] [CrossRef]
- Marrero, D.; Pujol-Vila, F.; Vera, D.; Gabriel, G.; Illa, X.; Elizalde-Torrent, A.; Alvarez, M.; Villa, R. Gut-on-a-chip: Mimicking and monitoring the human intestine. Biosens. Bioelectron. 2021, 181, 113156. [Google Scholar] [CrossRef]
- Garcia-Gutierrez, E.; Cotter, P.D. Relevance of organ(s)-on-a-chip systems to the investigation of food-gut microbiota-host interactions. Crit. Rev. Microbiol. 2021, 48, 463–488. [Google Scholar] [CrossRef]
- Rahman, S.; Ghiboub, M.; Donkers, J.M.; van de Steeg, E.; van Tol, E.A.F.; Hakvoort, T.B.M.; de Jonge, W.J. The progress of intestinal epithelial models from cell lines to gut-on-chip. Int. J. Mol. Sci. 2021, 22, 3472. [Google Scholar] [CrossRef]
- Signore, M.A.; De Pascali, C.; Giampetruzzi, L.; Siciliano, P.A.; Francioso, L. Gut-on-Chip microphysiological systems: Latest advances in the integration of sensing strategies and adoption of mature detection mechanisms. Sens. Bio-Sens. Res. 2021, 33, 100443. [Google Scholar] [CrossRef]
- Kim, H.J.; Li, H.; Collins, J.J.; Ingber, D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. USA 2016, 113, E7–E15. [Google Scholar] [CrossRef]
- Beaurivage, C.; Naumovska, E.; Chang, Y.X.; Elstak, E.D.; Nicolas, A.; Wouters, H.; van Moolenbroek, G.; Lanz, H.L.; Trietsch, S.J.; Joore, J.; et al. Development of a gut-on-a-chip model for high throughput disease modeling and drug discovery. Int. J. Mol. Sci. 2019, 20, 5661. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab A Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Hinman, S.S.; Wang, Y.; Allbritton, N.L. Photopatterned membranes and chemical gradients enable scalable phenotypic organization of primary human colon epithelial models. Anal. Chem. 2019, 91, 15240–15247. [Google Scholar] [CrossRef] [PubMed]
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Swenor, B.; Nestor, B.; Cronce, M.J.; Tovaglieri, A.; et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Naumovska, E.; Aalderink, G.; Valencia, C.W.; Kosim, K.; Nicolas, A.; Brown, S.; Vulto, P.; Erdmann, K.S.; Kurek, D. Direct on-chip differentiation of intestinal tubules from induced pluripotent stem cells. Int. J. Mol. Sci. 2020, 21, 4964. [Google Scholar] [CrossRef]
- Workman, M.J.; Gleeson, J.P.; Troisi, E.J.; Estrada, H.Q.; Kerns, S.J.; Hinojosa, C.D.; Hamilton, G.A.; Targan, S.R.; Svendsen, C.N.; Barrett, R.J. Enhanced utilization of induced pluripotent stem cell-derived human intestinal organoids using microengineered chips. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 669–675. [Google Scholar] [CrossRef]
- Kim, R.; Attayek, P.J.; Wang, Y.; Furtado, K.L.; Tamayo, R.; Sims, C.E.; Allbritton, N.L. An in vitro intestinal platform with a self-sustaining oxygen gradient to study the human gut/microbiome interface. Biofabrication 2020, 12, 015006. [Google Scholar] [CrossRef]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jaeger, C.; Seguin-Devaux, C.; et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef]
- Grassart, A.; Malarde, V.; Gobba, S.; Sartori-Rupp, A.; Kerns, J.; Karalis, K.; Marteyn, B.; Sansonetti, P.; Sauvonnet, N. Bioengineered Human Organ-on-Chip Reveals Intestinal Microenvironment and Mechanical Forces Impacting Shigella Infection. Cell Host Microbe 2019, 26, 435–444. [Google Scholar] [CrossRef]
- Kim, D.H.; Cheon, J.H. Pathogenesis of inflammatory bowel disease and recent advances in biologic therapies. Immune Netw. 2017, 17, 25–40. [Google Scholar] [CrossRef]
- Shin, M.K.; Kim, S.K.; Jung, H. Integration of intra- and extravasation in one cell-based microfluidic chip for the study of cancer metastasis. Lab A Chip 2011, 11, 3880–3887. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Aleman, J.; Shin, S.R.; Kilic, T.; Kim, D.; Shaegh, S.A.M.; Massa, S.; Riahi, R.; Chae, S.; Hu, N.; et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. USA 2017, 114, E2293–E2302. [Google Scholar] [CrossRef]
- Skardal, A.; Murphy, S.V.; Devarasetty, M.; Mead, I.; Kang, H.-W.; Seol, Y.-J.; Zhang, Y.S.; Shin, S.-R.; Zhao, L.; Aleman, J.; et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 2017, 7, 8837. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, Z.; Rahim, N.A.A.; van Noort, D.; Yu, H. Towards a human-on-chip: Culturing multiple cell types on a chip with compartmentalized microenvironments. Lab A Chip 2009, 9, 3185–3192. [Google Scholar] [CrossRef]
- Vernetti, L.; Gough, A.; Baetz, N.; Blutt, S.; Broughman, J.R.; Brown, J.A.; Foulke-Abel, J.; Hasan, N.; In, J.; Kelly, E.; et al. Functional coupling of human microphysiology systems: Intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci. Rep. 2017, 7, 42296. [Google Scholar] [CrossRef]
- Choe, A.; Ha, S.K.; Choi, I.; Choi, N.; Sung, J.H. Microfluidic Gut-liver chip for reproducing the first pass metabolism. Biomed. Microdevices 2017, 19, 4. [Google Scholar] [CrossRef]
- Singh, D.; Cho, W.C.; Upadhyay, G. Drug-induced liver toxicity and prevention by herbal antioxidants: An overview. Front. Physiol. 2016, 6, 363. [Google Scholar] [CrossRef]
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Huebner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A.; et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab A Chip 2015, 15, 2688–2699. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Ashammakhi, N.; Nasiri, R.; de Barros, N.R.; Tebon, P.; Thakor, J.; Goudie, M.; Shamloo, A.; Martin, M.G.; Khademhosseini, A. Gut-on-a-chip: Current progress and future opportunities. Biomaterials 2020, 255, 120196. [Google Scholar] [CrossRef]
- Fois, C.A.M.; Le, T.Y.L.; Schindeler, A.; Naficy, S.; McClure, D.D.; Read, M.N.; Valtchev, P.; Khademhosseini, A.; Dehghani, F. Models of the Gut for Analyzing the Impact of Food and Drugs. Adv. Healthc. Mater. 2019, 8, 1900968. [Google Scholar] [CrossRef] [PubMed]
- Hewes, S.A.; Wilson, R.L.; Estes, M.K.; Shroyer, N.F.; Blutt, S.E.; Grande-Allen, K.J. In Vitro Models of the Small Intestine: Engineering Challenges and Engineering Solutions. Tissue Eng. Part B Rev. 2020, 26, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ingber, D.E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 2013, 5, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A.; et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 2871. [Google Scholar] [CrossRef]
- Williamson, I.A.; Arnold, J.W.; Samsa, L.A.; Gaynor, L.; DiSalvo, M.; Cocchiaro, J.L.; Carroll, I.; Azcarate-Peril, M.A.; Rawls, J.F.; Allbritton, N.L.; et al. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 301–319. [Google Scholar] [CrossRef]
- Li, X.; Tian, T. Recent advances in an organ-on-a-chip: Biomarker analysis and applications. Anal. Methods 2018, 10, 3122–3130. [Google Scholar] [CrossRef]
- Soucy, J.R.; Bindas, A.J.; Koppes, A.N.; Koppes, R.A. Instrumented microphysiological systems for real-time measurement and manipulation of cellular electrochemical processes. Iscience 2019, 21, 521–548. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. Brain-gut-microbiota axis-mood, metabolism and behaviour. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 69–70. [Google Scholar] [CrossRef]
- Iannone, L.F.; Preda, A.; Blottiere, H.M.; Clarke, G.; Albani, D.; Belcastro, V.; Carotenuto, M.; Cattaneo, A.; Citraro, R.; Ferraris, C.; et al. Microbiota-gut brain axis involvement in neuropsychiatric disorders. Expert Rev. Neurother. 2019, 19, 1037–1050. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous Bacteria from the Gut Microbiota Regulate Host Serotonin Biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef]
- Samuel, B.S.; Shaito, A.; Motoike, T.; Rey, F.E.; Backhed, F.; Manchester, J.K.; Hammer, R.E.; Williams, S.C.; Crowley, J.; Yanagisawa, M.; et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. USA 2008, 105, 16767–16772. [Google Scholar] [CrossRef]
- Haghikia, A.; Joerg, S.; Duscha, A.; Berg, J.; Manzel, A.; Waschbisch, A.; Hammer, A.; Lee, D.-H.; May, C.; Wilck, N.; et al. Dietary fatty acids directly impact central nervous system autoimmunity via the small intestine. Immunity 2015, 43, 817–829. [Google Scholar] [CrossRef]
- Alkasir, R.; Li, J.; Li, X.; Jin, M.; Zhu, B. Human gut microbiota: The links with dementia development. Protein Cell 2017, 8, 90–102. [Google Scholar] [CrossRef]
- Shang, J.M.; Ma, S.R.; Zang, C.X.; Bao, X.Q.; Wang, Y.; Zhang, D. Gut microbiota mediates the absorption of FLZ, a new drug for Parkinson’s disease treatment. Acta Pharm. Sin. B 2021, 11, 1213–1226. [Google Scholar] [CrossRef]
- Raimondi, I.; Izzo, L.; Tunesi, M.; Comar, M.; Albania, D.; Giordano, C. Organ-on-a-chip in vitro models of the brain and the blood-brain barrier and their value to study the microbiota-gut-brain axis in neurodegeneration. Front. Bioeng. Biotechnol. 2020, 7, 435. [Google Scholar] [CrossRef]
- Gao, C.L.; Xu, Y.Z.; Liang, Z.Z.; Wang, Y.J.; Shang, Q.H.; Zhang, S.B.; Wang, C.F.; Ni, M.M.; Wu, D.L.; Huang, Z.J.; et al. A novel PGAM5 inhibitor LFHP-1c protects bloodebrain barrier integrity in ischemic stroke. Acta Pharm. Sin. B 2021, 11, 1867–1884. [Google Scholar] [CrossRef]
- Sharma, G.; Sharma, A.R.; Lee, S.-S.; Bhattacharya, M.; Nam, J.-S. Advances in nanocarriers enabled brain targeted drug delivery across blood brain barrier. Int. J. Pharm. 2019, 559, 360–372. [Google Scholar] [CrossRef]
- Jiang, L.; Li, S.; Zheng, J.; Li, Y.; Huang, H. Recent progress in microfluidic models of the blood-brain barrier. Micromachines 2019, 10, 375. [Google Scholar] [CrossRef]
- Han, L.; Jiang, C. Evolution of blood-brain barrier in brain diseases and related systemic nanoscale brain-targeting drug delivery strategies. Acta Pharm. Sin. B 2021, 11, 2306–2325. [Google Scholar] [CrossRef]
- Voelkl, B.; Altman, N.S.; Forsman, A.; Forstmeier, W.; Gurevitch, J.; Jaric, I.; Karp, N.A.; Kas, M.J.; Schielzeth, H.; Van de Casteele, T.; et al. Reproducibility of animal research in light of biological variation. Nat. Rev. Neurosci. 2020, 21, 384–393. [Google Scholar] [CrossRef]
- Wang, X.; Hou, Y.; Ai, X.; Sun, J.; Xu, B.; Meng, X.; Zhang, Y.; Zhang, S. Potential applications of microfluidics based blood brain barrier (BBB)-on-chips for in vitro drug development. Biomed. Pharmacother. 2020, 132, 110822. [Google Scholar] [CrossRef] [PubMed]
- Trapecar, M.; Wogram, E.; Svoboda, D.; Communal, C.; Omer, A.; Lungjangwa, T.; Sphabmixay, P.; Velazquez, J.; Schneider, K.; Wright, C.W.; et al. Human physiomimetic model integrating microphysiological systems of the gut, liver, and brain for studies of neurodegenerative diseases. Sci. Adv. 2021, 7, eabd1707. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Sung, J.H. Gut-liver on a chip toward an in vitro model of hepatic steatosis. Biotechnol. Bioeng. 2018, 115, 2817–2827. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Q.; Yu, D.K.; Xue, L.; Li, H.; Du, J.R. Probiotics modulate the microbiota-gut-brain axis and improve memory deficits in aged SAMP8 mice. Acta Pharm. Sin. B 2020, 10, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Osadchiy, V.; Martin, C.R.; Mayer, E.A. The gut-brain axis and the microbiome: Mechanisms and clinical implications. Clin. Gastroenterol. Hepatol. 2019, 17, 322–332. [Google Scholar] [CrossRef]
- Trapecar, M.; Communal, C.; Velazquez, J.; Maass, C.A.; Huang, Y.-J.; Schneider, K.; Wright, C.W.; Butty, V.; Eng, G.; Yilmaz, O.; et al. Gut-liver physiomimetics reveal paradoxical modulation of ibd-related inflammation by short-chain fatty acids. Cell Syst. 2020, 10, 223–231. [Google Scholar] [CrossRef]
- Starkel, P.; Schnabl, B. Bidirectional communication between liver and gut during alcoholic liver disease. Semin. Liver Dis. 2016, 36, 331–339. [Google Scholar] [CrossRef]
- Van Midwoud, P.M.; Merema, M.T.; Verpoorte, E.; Groothuis, G.M.M. A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices. Lab A Chip 2010, 10, 2778–2786. [Google Scholar] [CrossRef]
- Bricks, T.; Paullier, P.; Legendre, A.; Fleury, M.-J.; Zeller, P.; Merlier, F.; Anton, P.M.; Leclerc, E. Development of a new microfluidic platform integrating co-cultures of intestinal and liver cell lines. Toxicol. Vitr. 2014, 28, 885–895. [Google Scholar] [CrossRef]
- Duan, X.; Zheng, L.; Zhang, X.; Wang, B.; Xiao, M.; Zhao, W.; Liu, S.; Sui, G. A membrane-free liver-gut-on-chip platform for the assessment on dysregulated mechanisms of cholesterol and bile acid metabolism induced by PM2.5. Acs Sens. 2020, 5, 3483–3492. [Google Scholar] [CrossRef]
- Tsamandouras, N.; Chen, W.L.K.; Edington, C.D.; Stokes, C.L.; Griffith, L.G.; Cirit, M. Integrated gut and liver microphysiological systems for quantitative in vitro pharmacokinetic studies. Aaps J. 2017, 19, 1499–1512. [Google Scholar] [CrossRef] [PubMed]
- Jie, M.; Mao, S.; Liu, H.; He, Z.; Li, H.-F.; Lin, J.-M. Evaluation of drug combination for glioblastoma based on an intestine-liver metabolic model on microchip. Analyst 2017, 142, 3629–3638. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.J.; Miller, P.; Shuler, M.L. A pumpless body-on-a-chip model using a primary culture of human intestinal cells and a 3D culture of liver cells. Lab A Chip 2018, 18, 2036–2046. [Google Scholar] [CrossRef] [PubMed]
- Jie, M.; Lin, H.; He, Z.; Liu, H.; Li, H.; Lin, J.-M. An on-chip intestine-liver model for multiple drugs absorption and metabolism behavior simulation. Sci. China Chem. 2018, 61, 236–242. [Google Scholar] [CrossRef]
- Xu, L.N.; Yin, L.H.; Qi, Y.; Tan, X.M.; Gao, M.; Peng, J.Y. 3D disorganization and rearrangement of genome provide insights into pathogenesis of NAFLD by integrated Hi-C, Nanopore, and RNA sequencing. Acta Pharm. Sin. B 2021, 11, 3150–3164. [Google Scholar] [CrossRef]
- Skardal, A.; Devarasetty, M.; Forsythe, S.; Atala, A.; Soker, S. A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol. Bioeng. 2016, 113, 2020–2032. [Google Scholar] [CrossRef]
- Esch, M.B.; Mahler, G.J.; Stokor, T.; Shuler, M.L. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab A Chip 2014, 14, 3081–3092. [Google Scholar] [CrossRef]
- De Gregorio, V.; Telesco, M.; Corrado, B.; Rosiello, V.; Urciuolo, F.; Netti, P.A.; Imparato, G. Intestine-liver axis on-chip reveals the intestinal protective role on hepatic damage by emulating ethanol first-pass metabolism. Front. Bioeng. Biotechnol. 2020, 8, 163. [Google Scholar] [CrossRef]
- Jha, V.; Garcia-Garcia, G.; Iseki, K.; Li, Z.; Naicker, S.; Plattner, B.; Saran, R.; Wang, A.Y.M.; Yang, C.W. Chronic kidney disease: Global dimension and perspectives. Lancet 2013, 382, 260–272. [Google Scholar] [CrossRef]
- Pan, L.B.; Han, P.; Ma, S.R.; Peng, R.; Wang, C.; Kong, W.J.; Cong, L.; Fu, J.; Zhang, Z.W.; Yu, H.; et al. Abnormal metabolism of gut microbiota reveals the possible molecular mechanism of nephropathy induced by hyperuricemia. Acta Pharm. Sin. B 2020, 10, 249–261. [Google Scholar] [CrossRef]
- Rukavina Mikusic, N.L.; Martin Kouyoumdzian, N.; Roberto Choi, M. Gut microbiota and chronic kidney disease: Evidences and mechanisms that mediate a new communication in the gastrointestinal-renal axis. Pflug. Arch. Eur. J. Physiol. 2020, 472, 303–320. [Google Scholar] [CrossRef]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- Li, Z.; Su, W.; Zhu, Y.; Tao, T.; Li, D.; Peng, X.; Qin, J. Drug absorption related nephrotoxicity assessment on an intestine-kidney chip. Biomicrofluidics 2017, 11, 034114. [Google Scholar] [CrossRef]
- Ashammakhi, N.; Wesseling-Perry, K.; Hasan, A.; Elkhammas, E.; Zhang, Y.S. Kidney-on-a-chip: Untapped opportunities. Kidney Int. 2018, 94, 1073–1086. [Google Scholar] [CrossRef]
- Giordano, L.; Mihaila, S.M.; Amirabadi, H.E.; Masereeuw, R. Microphysiological systems to recapitulate the gut-kidney axis. Trends Biotechnol. 2021, 39, 811–823. [Google Scholar] [CrossRef]
- Sze, M.A.; Dimitriu, P.A.; Hayashi, S.; Elliott, W.M.; McDonough, J.E.; Gosselink, J.V.; Cooper, J.; Sin, D.D.; Mohn, W.W.; Hogg, J.C. The lung tissue microbiome in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2012, 185, 1073–1080. [Google Scholar] [CrossRef]
- Zhao, J.C.; Schloss, P.D.; Kalikin, L.M.; Carmody, L.A.; Foster, B.K.; Petrosino, J.F.; Cavalcoli, J.D.; VanDevanter, D.R.; Murray, S.; Li, J.Z.; et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc. Natl. Acad. Sci. USA 2012, 109, 5809–5814. [Google Scholar] [CrossRef]
- Li, Q.J.; Yi, D.R.; Lei, X.B.; Zhao, J.Y.; Zhang, Y.X.; Cui, X.L.; Xiao, X.; Jiao, T.; Dong, X.J.; Zhao, X.S.; et al. Corilagin inhibits SARS-CoV-2 replication by targeting viral RNA-dependent RNA polymerase. Acta Pharm. Sin. B 2021, 11, 1555–1567. [Google Scholar] [CrossRef]
- Lyu, M.; Fan, G.W.; Xiao, G.X.; Wang, T.Y.; Xu, D.; Gao, J.; Ge, S.Q.; Li, Q.L.; Ma, Y.L.; Zhang, H.; et al. Traditional Chinese medicine in COVID-19. Acta Pharm. Sin. B 2021, 11, 3337–3363. [Google Scholar] [CrossRef]
- Anh Thu, D.; Marsland, B.J. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef]
- Adhikary, P.P.; Ul Ain, Q.; Hocke, A.C.; Hedtrich, S. COVID-19 highlights the model dilemma in biomedical research. Nat. Rev. Mater. 2021, 6, 374–376. [Google Scholar] [CrossRef] [PubMed]
- Oleaga, C.; Bernabini, C.; Smith, A.S.T.; Srinivasan, B.; Jackson, M.; McLamb, W.; Platt, V.; Bridges, R.; Cai, Y.; Santhanam, N.; et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci. Rep. 2016, 6, 20030. [Google Scholar] [CrossRef] [PubMed]
- Herland, A.; Maoz, B.M.; Das, D.; Somayaji, M.R.; Prantil-Baun, R.; Novak, R.; Cronce, M.; Huffstater, T.; Jeanty, S.S.F.; Ingram, M.; et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat. Biomed. Eng. 2020, 4, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cirit, M.; Wishnok, J.S.; Griffith, L.G.; Tannenbaum, S.R. Analysis of an integrated human multiorgan microphysiological system for combined tolcapone metabolism and brain metabolomics. Anal. Chem. 2019, 91, 8667–8675. [Google Scholar] [CrossRef]




| Gut Axis on Chips | Cell/Organ Type | Targeted Application and Major Results | Ref. |
|---|---|---|---|
| GBA on chip | -Gut epithelial cells -Brain endothelial cells | -Co-culture creates a positive cell barrier -Reflects interactions between microbial by-products, intestinal epithelium and BBB -Transport of fluorescently labeled exosomes across the intestinal barrier to the blood–brain barrier can be observed | [30] |
| -Human monocyte-derived dendritic cells and macrophages -Hepatocytes and Kupffer cells -Neurons, astrocytes, and microglia -Treg/TH 17 cells | -A model of excellence for investigating neurodegenerative diseases -Systemic interactions enhance the in vivo-like behavior characteristics of brain micro-physiological systems -Microbial-associated SCFAs increase the expression of pathology-related pathways in Parkinson’s disease | [82] | |
| GLA on chip | -Hepatocytes and Kupffer cells -Enterocyte, goblet cells, and dendritic cells | -Reveals the regulation of bile acid metabolism -Provides evidence for physiologically relevant intestine–hepatic crosstalk -Significant non-linear regulation of cytokine responses observed in inflammatory intestine–hepatic interactions | [33] |
| -Human-induced pluripotent stem cell-derived intestinal cells and fresh human hepatocytes | -Exploring unknown physiological mechanisms of in vitro organ–organ interactions | [34] | |
| -Caco-2 cells -Hepatic HepaRG cells | -Exploration of the pharmacokinetic mechanism model of triazolam (TRZ) and its metabolites in GLA on chip -TRZ is metabolized to α- and 4-hydroxytriazolam and their respective glucuronides | [35] | |
| -Caco-2 cells -HepG2 cells | - Assessment of the metabolism of the flavonoid apigenin -The co-culture of intestinal and liver cells on the microarray led to a metabolic profile that was stronger than the monoculture effect | [56] | |
| -Caco-2 cells -Hepatic cells -Treg/TH 17 cells | -Used as an in vitro model of ulcerative colitis (UC) -SCFAs can ameliorate or worsen the severity of UC, depending on the involvement of effector CD4+ T cells -SCFA increased ketone body, glycolysis, and lipogenesis production, while it significantly reduced innate immune activation in the UC gut | [86] | |
| -Caco-2 TC7 cells -HepG2 C3A | - Dynamic intestine liver coculture model -First pass metabolism of phenacetin investigation -Higher metabolic performance of the bioreactor when compared the Petri coculture | [89] | |
| -Human intestinal myofibroblasts -Caco-2 cells | -Simulates the first-pass mechanism that occurs in vivo -Emphasis on ethanol-induced 3D-him hyperpermeability and interstitial injury, and the preventive role of the intestine on liver injury -Simulation of metabolic enzyme release following high-dose ethanol administration | [98] | |
| -Caco-2 cells -HepG2 cells | -Evaluation of drug combinations for the treatment of glioblastoma -Continuous infusion of irinotecan (CPT-11), temozolomide (TMZ), cyclophosphamide (CP), and other drugs dynamically stimulate cells as an interventional drug model -After intestinal absorption and hepatic metabolism, the prodrugs are converted into active metabolites and induce apoptosis in glioblastoma cells -Enables long-term cell co-culture, drug delivery, metabolism and real-time analysis of drug effects | [92] | |
| -Caco-2 cells -HepG2 cells | -Investigating the effects of dynein and dacarbazine on cell viability, hepatotoxicity, and cell cycle arrest during combination drug therapy -Served as a platform model for long-term observation of the uptake, transport, and metabolism of the combined drugs | [94] | |
| CKA on chip | -Caco-2 cells -HKC-8 cells | -As a model for investigating STEC O157:H7 (O157) infection and Shiga toxin 2 toxicity in intestinal and renal cells | [32] |
| GLAx on chip | -Caco-2 cells -HUVECs cells | -Creation of an intestinal infection model that allows reproduction of SARSCoV-2-induced human-associated intestinal pathophysiology at the organ level | [31] |
| multi-organ on chip | Intestine, liver, kidney proximal tubule, blood–brain barrier, and skeletal muscle | -Evaluated the pharmacokinetics (PK) and toxicity of terfenadine, trimethoprim, and vitamin D3 -Demonstrated the potential of multi-organ co-chips for multi-organ toxicity and absorption, distribution, metabolism, and excretion (ADME) | [55] |
| Intestine, liver, kidney, and coupled bone marrow | -Exploration of the first physiological pharmacokinetic model of absorption, metabolism, and excretion of drugs through humans -Predicted pharmacodynamic parameters of oral nicotine and intravenous cisplatin | [113] | |
| Brain, pancreas, liver, lungs, heart, intestines, endometrium | -Evaluation of mephedrone metabolite profiles and metabolomics | [114] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Chen, X.; Gong, P.; Li, G.; Yao, W.; Yang, W. The Gut–Organ-Axis Concept: Advances the Application of Gut-on-Chip Technology. Int. J. Mol. Sci. 2023, 24, 4089. https://doi.org/10.3390/ijms24044089
Guo Y, Chen X, Gong P, Li G, Yao W, Yang W. The Gut–Organ-Axis Concept: Advances the Application of Gut-on-Chip Technology. International Journal of Molecular Sciences. 2023; 24(4):4089. https://doi.org/10.3390/ijms24044089
Chicago/Turabian StyleGuo, Yuxi, Xuefeng Chen, Pin Gong, Guoliang Li, Wenbo Yao, and Wenjuan Yang. 2023. "The Gut–Organ-Axis Concept: Advances the Application of Gut-on-Chip Technology" International Journal of Molecular Sciences 24, no. 4: 4089. https://doi.org/10.3390/ijms24044089
APA StyleGuo, Y., Chen, X., Gong, P., Li, G., Yao, W., & Yang, W. (2023). The Gut–Organ-Axis Concept: Advances the Application of Gut-on-Chip Technology. International Journal of Molecular Sciences, 24(4), 4089. https://doi.org/10.3390/ijms24044089







