Metabolic Background, Not Photosynthetic Physiology, Determines Drought and Drought Recovery Responses in C3 and C2 Moricandias
Abstract
1. Introduction
2. Results
2.1. Cross-Species Comparison under Control Conditions
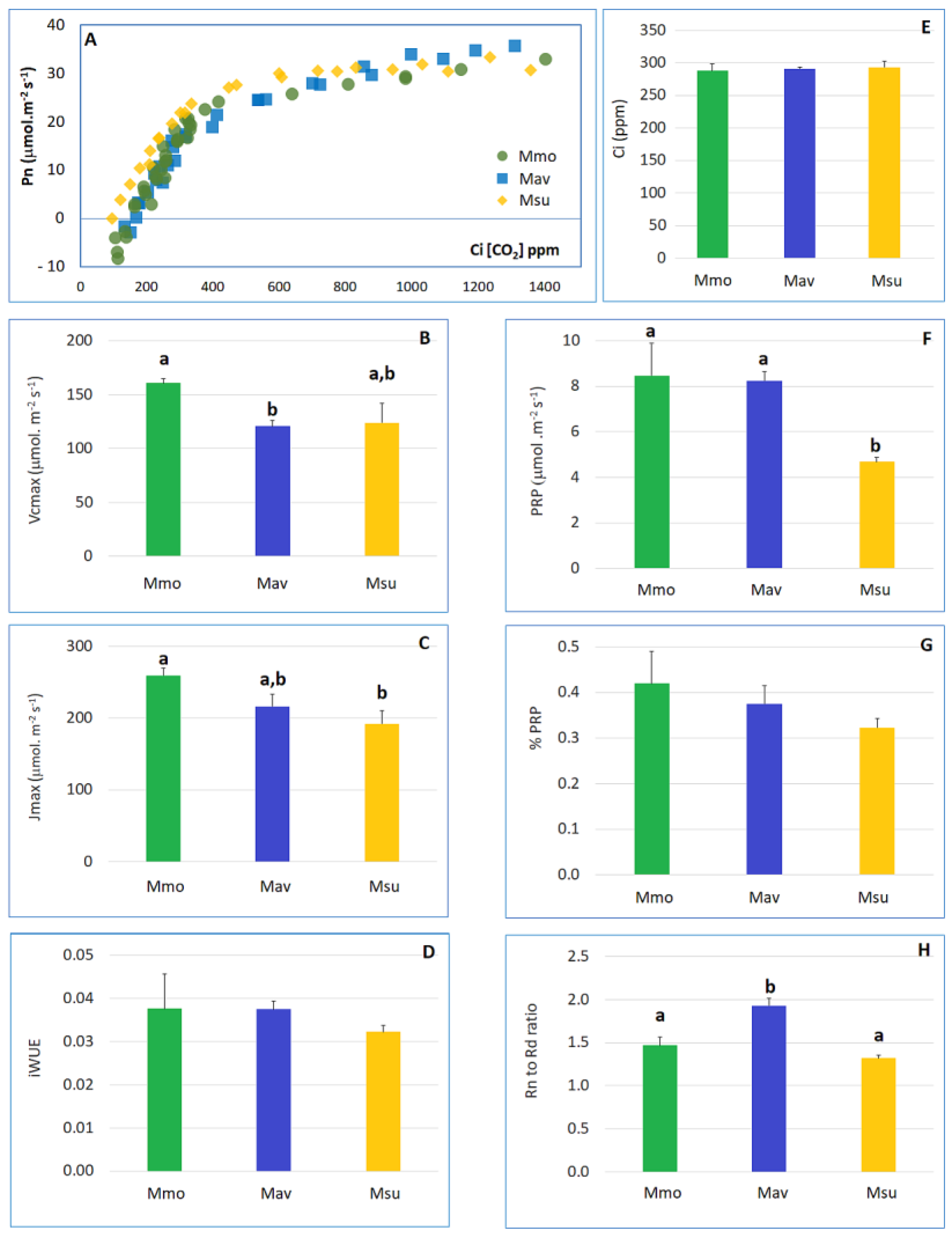
2.1.1. Physiology
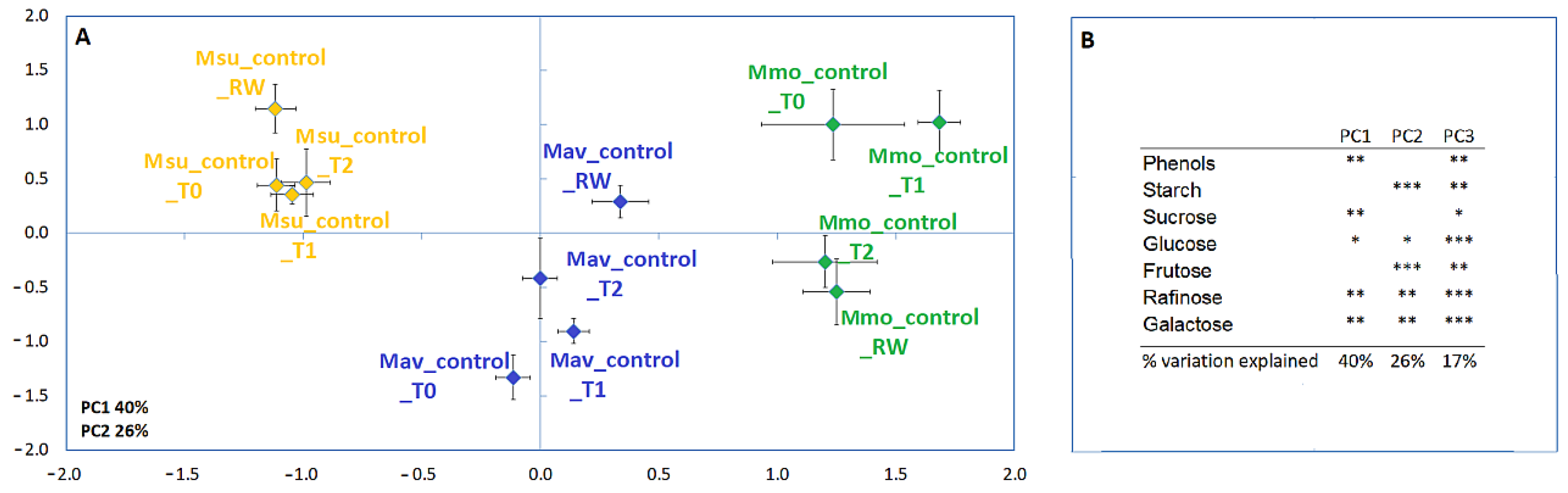
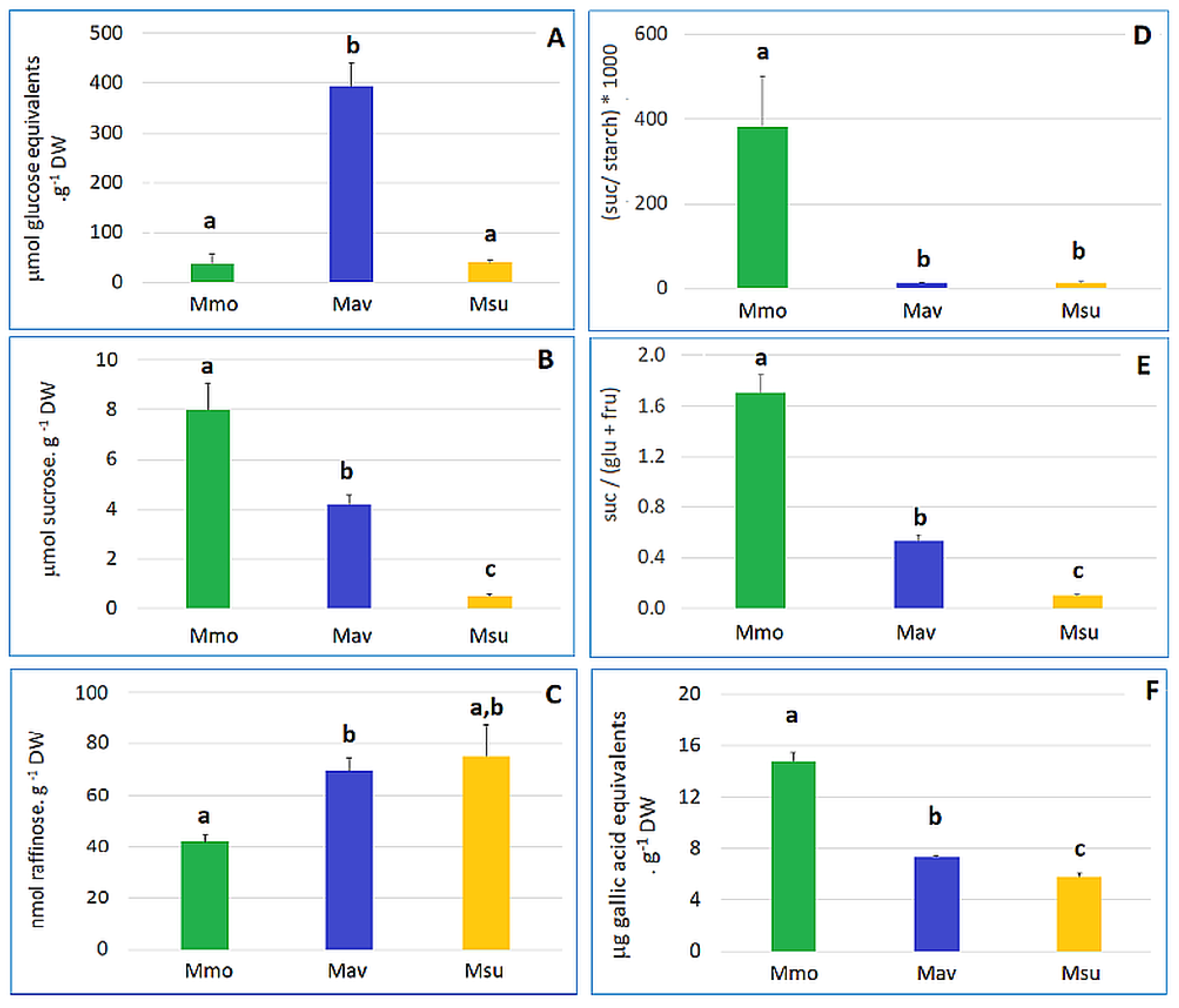
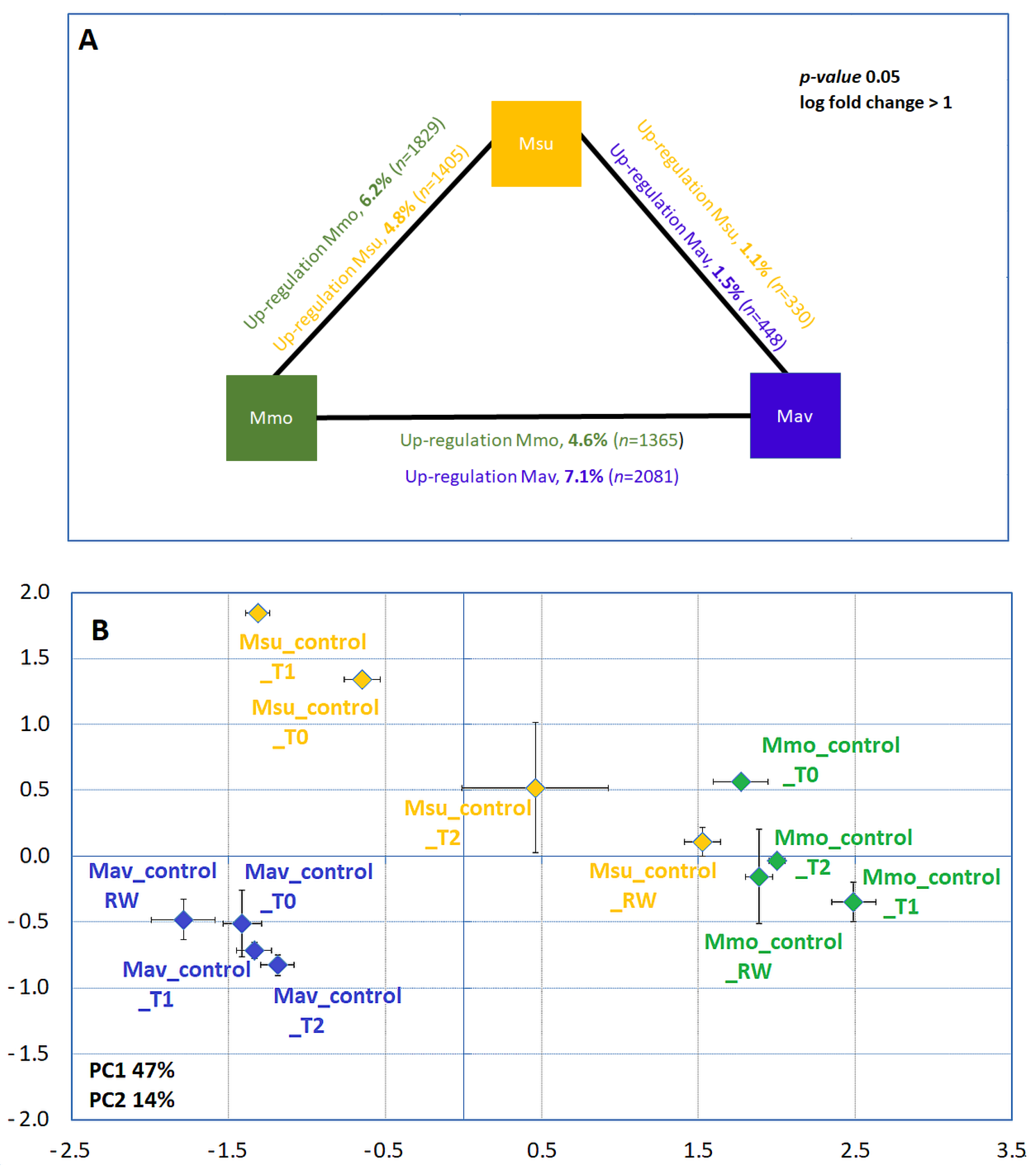
2.1.2. Biochemistry and Transcriptome Profiles
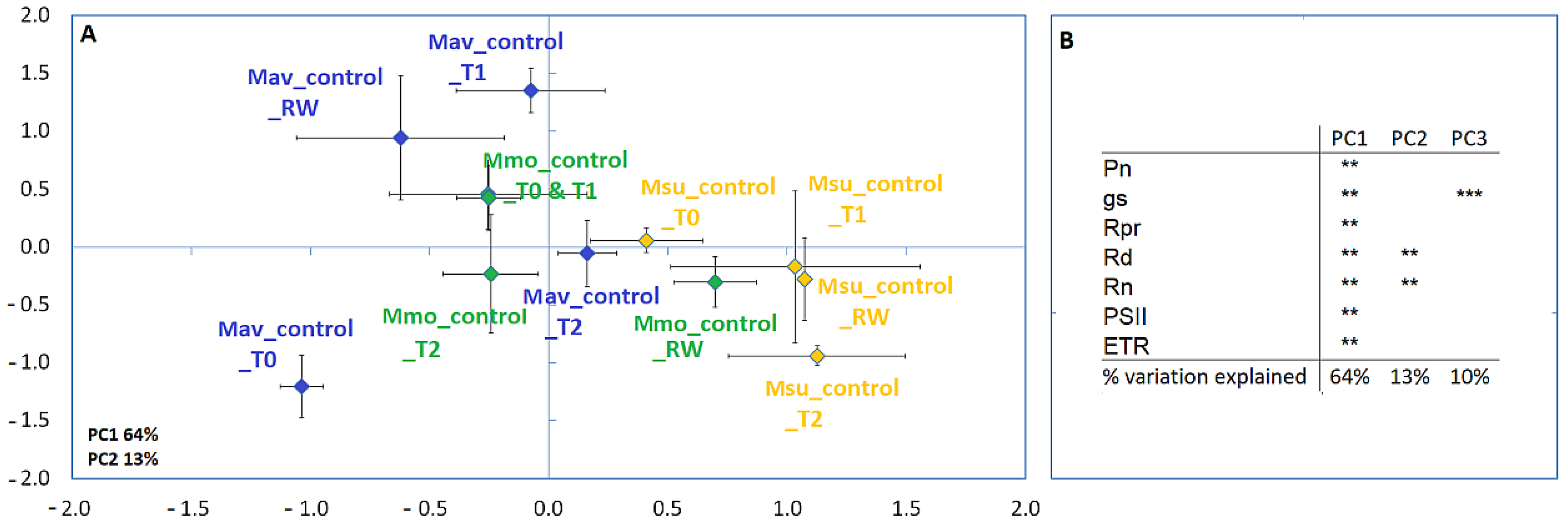
2.2. Progressive Soil Water Deficit and Rewatering
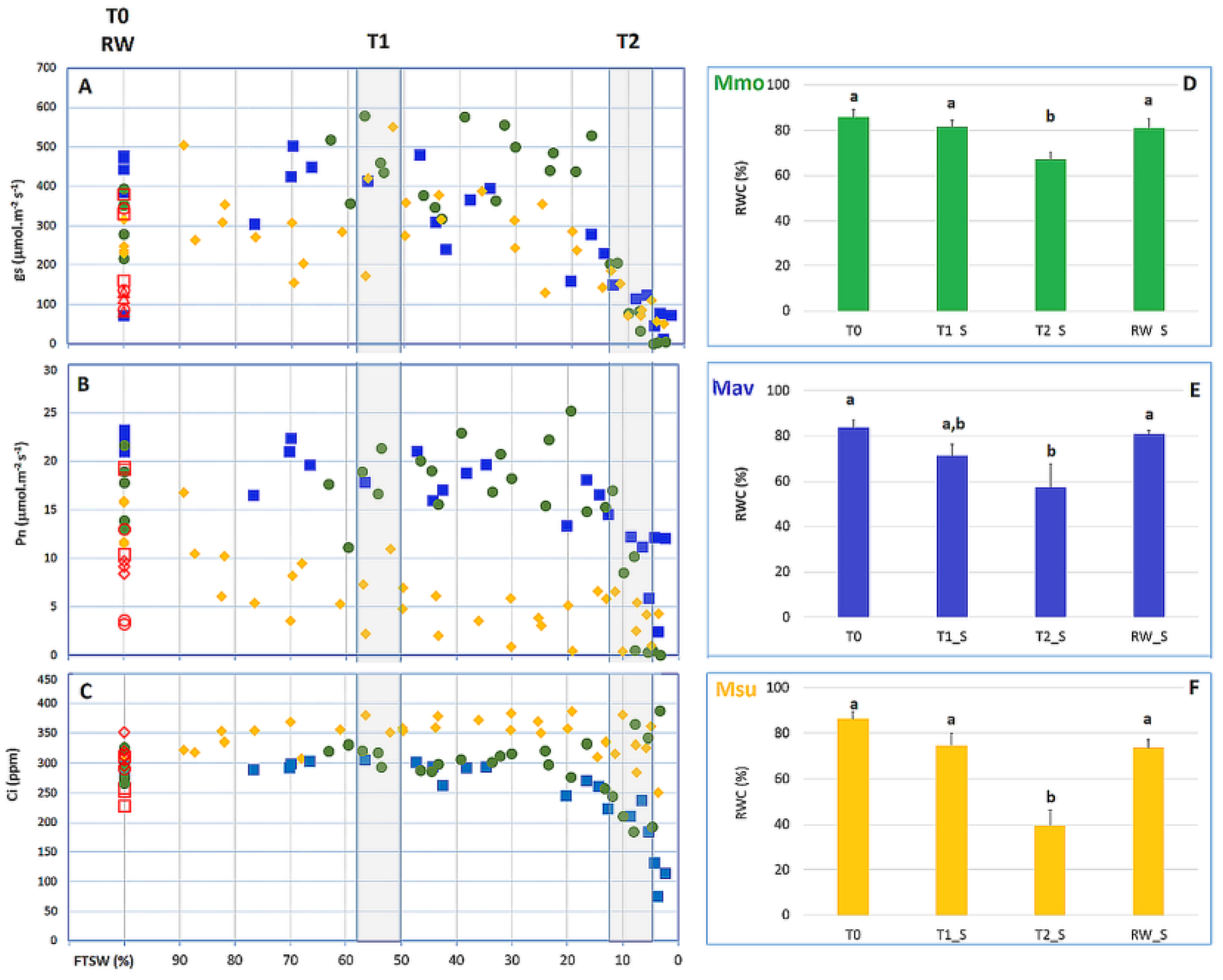
2.2.1. Physiological Evaluation
2.2.2. Cross-Species Comparison at the Physiological and Biochemical Levels
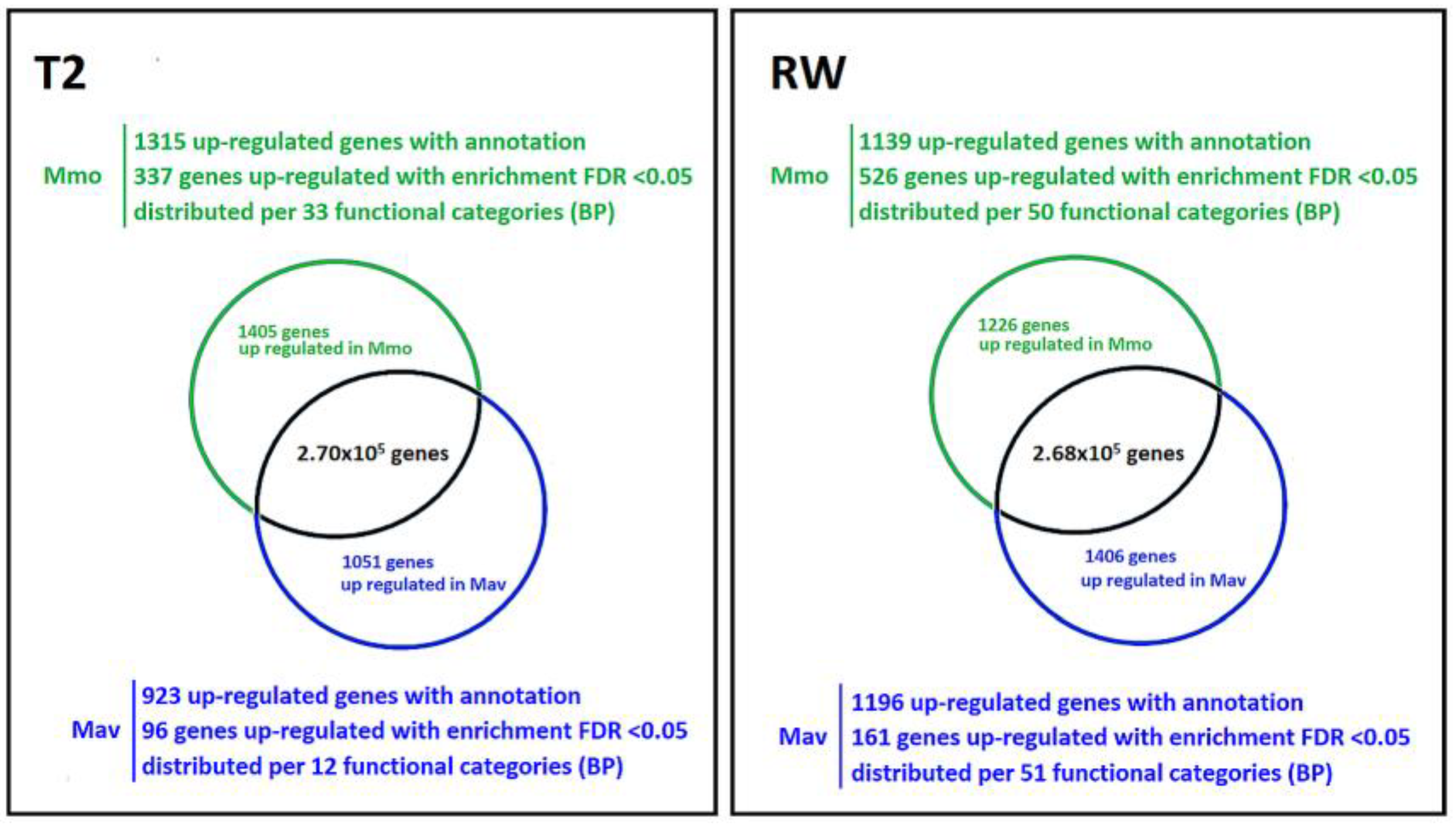
2.2.3. Cross-Species Transcriptome Comparison at T2 and RW
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Plant Growth Conditions
4.3. Drought Treatment and Leaf Sampling
4.4. Plant Gas Exchange
4.5. Biochemical Analysis
4.6. Estimation of the Reducing Capacity by the Folin–Ciocalteau Method
4.7. Extraction and Quantification of Water-Soluble Carbohydrates and Starch
4.8. RNA Isolation, RNA-Seq and Bioinformatic Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinheiro, C.; David, T.S.; Baptista, P.; Guerra-Guimarães, L. Mediterranean Woody Agroecosystems in a Warming and Drier Climate: The Importance of Knowledge-Based Management. Flora 2022, 291, 152070. [Google Scholar] [CrossRef]
- Costa, J.M.; Marques da Silva, J.; Pinheiro, C.; Barón, M.; Mylona, P.; Centritto, M.; Haworth, M.; Loreto, F.; Uzilday, B.; Turkan, I.; et al. Opportunities and Limitations of Crop Phenotyping in Southern European Countries. Front. Plant Sci. 2019, 10, 1125. [Google Scholar] [CrossRef] [PubMed]
- Tramblay, Y.; Koutroulis, A.; Samaniego, L.; Vicente-Serrano, S.M.; Volaire, F.; Boone, A.; Le Page, M.; Llasat, M.C.; Albergel, C.; Burak, S.; et al. Challenges for Drought Assessment in the Mediterranean Region under Future Climate Scenarios. Earth Sci. Rev. 2020, 210, 103348. [Google Scholar] [CrossRef]
- Bloom, A.J. Photorespiration and Nitrate Assimilation: A Major Intersection between Plant Carbon and Nitrogen. Photosynth. Res. 2015, 123, 117–128. [Google Scholar] [CrossRef]
- Osborne, C.P.; Sack, L. Evolution of C 4 Plants: A New Hypothesis for an Interaction of CO2 and Water Relations Mediated by Plant Hydraulics. Philos. Trans. R. Soc. B 2012, 367, 583–600. [Google Scholar] [CrossRef]
- Schlüter, U.; Weber, A.P.M. Regulation and Evolution of C4 Photosynthesis. Annu. Rev. Plant Biol. 2020, 71, 183–215. [Google Scholar] [CrossRef]
- Haworth, M.; Marino, G.; Centritto, M. The Impact of Atmospheric Composition on the Evolutionary Development of Stomatal Control and Biochemistry of Photosynthesis over the Past 450 Ma. In Evolutionary Developmental Biology; de la Rosa, L.N., Müller, G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–12. ISBN 978-3-319-33038-9. [Google Scholar]
- Sage, R.F.; Christin, P.-A.; Edwards, E.J. The C4 Plant Lineages of Planet Earth. J. Exp. Bot. 2011, 62, 3155–3169. [Google Scholar] [CrossRef]
- Lundgren, M.R. C2 Photosynthesis: A Promising Route towards Crop Improvement? New Phytol. 2020, 228, 1734–1740. [Google Scholar] [CrossRef]
- Osborne, C.P.; Beerling, D.J. Nature’s Green Revolution: The Remarkable Evolutionary Rise of C₄ Plants. Philos. Trans. R. Soc. B 2006, 361, 173–194. [Google Scholar] [CrossRef]
- Sage, R.F.; Sage, T.L.; Kocacinar, F. Photorespiration and the Evolution of C4 Photosynthesis. Annu. Rev. Plant Biol. 2012, 63, 19–47. [Google Scholar] [CrossRef]
- Perfectti, F.; Gómez, J.M.; González-Megías, A.; Abdelaziz, M.; Lorite, J. Molecular Phylogeny and Evolutionary History of Moricandia DC (Brassicaceae). PeerJ 2017, 5, e3964. [Google Scholar] [CrossRef] [PubMed]
- Jobe, T.O.; Karvansara, P.R.; Zenzen, I.; Kopriva, S. Ensuring Nutritious Food under Elevated CO2 Conditions: A Case for Improved C4 Crops. Front. Plant Sci. 2020, 11, 1267. [Google Scholar] [CrossRef] [PubMed]
- Apel, P.; Horstmann, C.; Pfeffer, M. The Moricandia Syndrome in Species of the Brassicaceae—Evolutionary Aspects. Photosynthetica 1997, 33, 205–215. [Google Scholar] [CrossRef]
- Mallmann, J.; Heckmann, D.; Bräutigam, A.; Lercher, M.J.; Weber, A.P.; Westhoff, P.; Gowik, U. The Role of Photorespiration during the Evolution of C4 Photosynthesis in the Genus Flaveria. Elife 2014, 3, e02478. [Google Scholar] [CrossRef] [PubMed]
- Borghi, G.L.; Arrivault, S.; Günther, M.; Barbosa Medeiros, D.; Dell’Aversana, E.; Fusco, G.M.; Carillo, P.; Ludwig, M.; Fernie, A.R.; Lunn, J.E.; et al. Metabolic Profiles in C3, C3-C4 Intermediate, C4-like, and C4 Species in the Genus Flaveria. J. Exp. Bot. 2022, 73, 1581–1601. [Google Scholar] [CrossRef] [PubMed]
- Schlüter, U.; Bräutigam, A.; Gowik, U.; Melzer, M.; Christin, P.-A.; Kurz, S.; Mettler-Altmann, T.; Weber, A.P. Photosynthesis in C3-C4 Intermediate Moricandia Species. J. Exp. Bot. 2017, 68, 191–206. [Google Scholar] [CrossRef]
- Bellasio, C.; Farquhar, G.D. A Leaf-Level Biochemical Model Simulating the Introduction of C2 and C4 Photosynthesis in C3 Rice: Gains, Losses and Metabolite Fluxes. New Phytol. 2019, 223, 150–166. [Google Scholar] [CrossRef]
- Chaves, M.M.; Costa, J.M.; Zarrouk, O.; Pinheiro, C.; Lopes, C.M.; Pereira, J.S. Controlling Stomatal Aperture in Semi-Arid Regions—The Dilemma of Saving Water or Being Cool? Plant Sci. 2016, 251, 54–64. [Google Scholar] [CrossRef]
- Peterhansel, C.; Blume, C.; Offermann, S. Photorespiratory Bypasses: How Can They Work? J. Exp. Bot. 2013, 64, 709–715. [Google Scholar] [CrossRef]
- Haworth, M.; Marino, G.; Loreto, F.; Centritto, M. The Evolution of Diffusive and Biochemical Capacities for Photosynthesis was Predominantly Shaped by [CO2] with a Smaller Contribution from [O2]. Sci. Total Environ. 2022, 840, 156606. [Google Scholar] [CrossRef]
- Westhoff, P.; Gowik, U. Evolution of C4 Photosynthesis—Looking for the Master Switch. Plant Physiol. 2010, 154, 598–601. [Google Scholar] [CrossRef] [PubMed]
- Ethier, G.J.; Livingston, N.J. On the Need to Incorporate Sensitivity to CO2 Transfer Conductance into the Farquhar-von Caemmerer-Berry Leaf Photosynthesis Model. Plant Cell Environ. 2004, 27, 137–153. [Google Scholar] [CrossRef]
- Lin, M.-Y.; Koppers, N.; Denton, A.; Schlüter, U.; Weber, A.P.M. Whole Genome Sequencing and Assembly Data of Moricandia moricandioides and M. arvensis. Data Brief 2021, 35, 106922. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.; Watts, R. Moricandia. In Wild Crop Relatives: Genomic and Breeding Resources: Oilseeds; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 191–198. ISBN 978-3-642-14871-2. [Google Scholar]
- Voshall, A.; Moriyama, E.N. Next-Generation Transcriptome Assembly and Analysis: Impact of Ploidy. Methods 2020, 176, 14–24. [Google Scholar] [CrossRef]
- 3TO4 Final Report of the Project 3to4: Converting C3 to C4 Photosynthesis for Sustainable Agriculture. 2017. Available online: https://cordis.europa.eu/project/id/289582/reporting (accessed on 18 October 2022).
- Xu, Z.; Escamilla-Treviño, L.; Zeng, L.; Lalgondar, M.; Bevan, D.; Winkel, B.; Mohamed, A.; Cheng, C.-L.; Shih, M.-C.; Poulton, J.; et al. Functional Genomic Analysis of Arabidopsis thaliana Glycoside Hydrolase Family 1. Plant Mol. Biol. 2004, 55, 343–367. [Google Scholar] [CrossRef]
- Sinclair, T.R.; Ludlow, M.M. Influence of Soil Water Supply on the Plant Water Balance of Four Tropical Grain Legumes. Funct. Plant Biol. 1986, 13, 329–341. [Google Scholar] [CrossRef]
- Centritto, M.; Brilli, F.; Fodale, R.; Loreto, F. Different Sensitivity of Isoprene Emission, Respiration and Photosynthesis to High Growth Temperature Coupled with Drought Stress in Black Poplar (Populus nigra) Saplings. Tree Physiol. 2011, 31, 275–286. [Google Scholar] [CrossRef]
- Killi, D.; Bussotti, F.; Raschi, A.; Haworth, M. Adaptation to High Temperature Mitigates the Impact of Water Deficit during Combined Heat and Drought Stress in C3 Sunflower and C4 Maize Varieties with Contrasting Drought Tolerance. Physiol. Plant. 2017, 159, 130–147. [Google Scholar] [CrossRef]
- Marino, G.; Haworth, M.; Scartazza, A.; Tognetti, R.; Centritto, M. A Comparison of the Variable J and Carbon-Isotopic Composition of Sugars Methods to Assess Mesophyll Conductance from the Leaf to the Canopy Scale in Drought-Stressed Cherry. Int. J. Mol. Sci. 2020, 21, 1222. [Google Scholar] [CrossRef]
- Körner, C.; Scheel, J.A.; Bauer, H. Maximum Leaf Diffusive Conductance in Vascular Plants. Photosynthetica 1979, 13, 45–82. [Google Scholar]
- Pinheiro, C.; Dickinson, E.; Marriott, A.; Ribeiro, I.C.; Pintó-Marijuan, M.; António, C.; Zarrouk, O.; Chaves, M.M.; Dodd, I.C.; Munné-Bosch, S.; et al. Distinctive Phytohormonal and Metabolic Profiles of Arabidopsis thaliana and Eutrema salsugineum under Similar Soil Drying. Planta 2019, 249, 1417–1433. [Google Scholar] [CrossRef] [PubMed]
- Lüttge, U. Ability of Crassulacean Acid Metabolism Plants to Overcome Interacting Stresses in Tropical Environments. AoB Plants 2010, 2010, plq005. [Google Scholar] [CrossRef] [PubMed]
- Pikart, F.C.; Marabesi, M.A.; Mioto, P.T.; Gonçalves, A.Z.; Matiz, A.; Alves, F.R.R.; Mercier, H.; Aidar, M.P.M. The Contribution of Weak CAM to the Photosynthetic Metabolic Activities of a Bromeliad Species under Water Deficit. Plant Physiol. Biochem. 2018, 123, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Arantes, M.K.; da Silva Filho, M.P.; Pennacchi, J.P.; das Chagas Mendonça, A.M.; Barbosa, J.P.R.A.D. Phenotypic Plasticity of Leaf Anatomical Traits Helps to Explain Gas-Exchange Response to Water Shortage in Grasses of Different Photosynthetic Types. Theor. Exp. Plant Physiol. 2020, 32, 341–356. [Google Scholar] [CrossRef]
- Winter, K.; Usuda, H.; Tsuzuki, M.; Schmitt, M.; Edwards, G.E.; Thomas, R.J.; Evert, R.F. Influence of Nitrate and Ammonia on Photosynthetic Characteristics and Leaf Anatomy of Moricandia arvensis. Plant Physiol. 1982, 70, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Mercier, H.; Rodrigues, M.A.; Andrade, S.; Coutinho, L.L.; Gobara, B.N.K.; Matiz, A.; Mioto, P.T.; Gonçalves, A.Z. Transcriptional Foliar Profile of the C3-CAM Bromeliad Guzmania monostachia. PLoS ONE 2019, 14, e0224429. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.N.; Gaspar, M.; Smith, J.A.C.; Mercier, H. Ammonium Intensifies CAM Photosynthesis and Counteracts Drought Effects by Increasing Malate Transport and Antioxidant Capacity in Guzmania monostachia. J. Exp. Bot. 2018, 69, 1993–2003. [Google Scholar] [CrossRef]
- Stein, O.; Granot, D. An Overview of Sucrose Synthases in Plants. Front. Plant Sci. 2019, 10, 95. [Google Scholar] [CrossRef]
- Fünfgeld, M.M.F.F.; Wang, W.; Ishihara, H.; Arrivault, S.; Feil, R.; Smith, A.M.; Stitt, M.; Lunn, J.E.; Niittylä, T. Sucrose Synthases Are Not Involved in Starch Synthesis in Arabidopsis Leaves. Nat. Plants 2022, 8, 574–582. [Google Scholar] [CrossRef]
- Baroja-Fernández, E.; Muñoz, F.J.; Li, J.; Bahaji, A.; Almagro, G.; Montero, M.; Etxeberria, E.; Hidalgo, M.; Sesma, M.T.; Pozueta-Romero, J. Sucrose Synthase Activity in the sus1/sus2/sus3/sus4 Arabidopsis Mutant Is Sufficient to Support Normal Cellulose and Starch Production. Proc. Natl. Acad. Sci. USA 2012, 109, 321–326. [Google Scholar] [CrossRef]
- Figueroa, C.M.; Lunn, J.E. A Tale of Two Sugars: Trehalose 6-Phosphate and Sucrose. Plant Physiol. 2016, 172, 7–27. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, B.B.; Gruissem, W.; Jones, R.L. (Eds.) Biochemistry and Molecular Biology of Plants, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2015; p. 1280. ISBN 978-0-470-71421-8. [Google Scholar]
- Fichtner, F.; Olas, J.J.; Feil, R.; Watanabe, M.; Krause, U.; Hoefgen, R.; Stitt, M.; Lunn, J.E. Functional Features of TREHALOSE-6-PHOSPHATE SYNTHASE1, an Essential Enzyme in Arabidopsis. Plant Cell 2020, 32, 1949–1972. [Google Scholar] [CrossRef] [PubMed]
- Vandesteene, L.; López-Galvis, L.; Vanneste, K.; Feil, R.; Maere, S.; Lammens, W.; Rolland, F.; Lunn, J.E.; Avonce, N.; Beeckman, T.; et al. Expansive Evolution of the TREHALOSE-6-PHOSPHATE PHOSPHATASE Gene Family in Arabidopsis. Plant Physiol. 2012, 160, 884–896. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Yang, J.; Wang, Q.; Zhu, H.; Chen, Z.; Dao, Y.; Wang, K. Overexpression of the Trehalose-6-Phosphate Phosphatase Family Gene AtTPPF Improves the Drought Tolerance of Arabidopsis thaliana. BMC Plant Biol. 2019, 19, 381. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, Q.; Xu, S.; Liu, W.-C.; Zhu, X.; Song, C.-P. Trehalose-6-Phosphate Phosphatase E Modulates ABA-Controlled Root Growth and Stomatal Movement in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 1518–1534. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, F.; Lunn, J.E. The Role of Trehalose 6-Phosphate (Tre6P) in Plant Metabolism and Development. Annu. Rev. Plant Biol. 2021, 72, 737–760. [Google Scholar] [CrossRef]
- Wei, J.; Chen, Q.; Lin, J.; Chen, F.; Chen, R.; Liu, H.; Chu, P.; Lu, Z.; Li, S.; Yu, G. Genome-Wide Identification and Expression Analysis of Tomato Glycoside Hydrolase Family 1 β-Glucosidase Genes in Response to Abiotic Stresses. Biotechnol. Biotechnol. Equip. 2022, 36, 268–280. [Google Scholar] [CrossRef]
- Dong, X.; Jiang, Y.; Hur, Y. Genome-Wide Analysis of Glycoside Hydrolase Family 1 β-Glucosidase Genes in Brassica rapa and their Potential Role in Pollen Development. Int. J. Mol. Sci. 2019, 20, 1663. [Google Scholar] [CrossRef]
- Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. BRENDA, the ELIXIR Core Data Resource in 2021: New Developments and Updates. Nucleic Acids Res. 2021, 49, D498–D508. [Google Scholar] [CrossRef]
- Xu, Z.-Y.; Lee, K.H.; Dong, T.; Jeong, J.C.; Jin, J.B.; Kanno, Y.; Kim, D.H.; Kim, S.Y.; Seo, M.; Bressan, R.A.; et al. A Vacuolar β-Glucosidase Homolog That Possesses Glucose-Conjugated Abscisic Acid Hydrolyzing Activity Plays an Important Role in Osmotic Stress Responses in Arabidopsis. Plant Cell 2012, 24, 2184–2199. [Google Scholar] [CrossRef]
- Lee, K.H.; Piao, H.L.; Kim, H.-Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.-J.; Hwang, I. Activation of Glucosidase via Stress-Induced Polymerization Rapidly Increases Active Pools of Abscisic Acid. Cell 2006, 126, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Watanabe, S.; Shimada, H.; Sakamoto, A. Dynamics of the Leaf Endoplasmic Reticulum Modulate β-Glucosidase-Mediated Stress-Activated ABA Production from its Glucosyl Ester. J. Exp. Bot 2020, 71, 2058–2071. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, R.; Hirai, M.Y. Atypical Myrosinase as a Mediator of Glucosinolate Functions in Plants. Front. Plant Sci. 2019, 10, 1008. [Google Scholar] [CrossRef] [PubMed]
- Escamilla-Treviño, L.L.; Chen, W.; Card, M.L.; Shih, M.-C.; Cheng, C.-L.; Poulton, J.E. Arabidopsis thaliana β-Glucosidases BGLU45 and BGLU46 Hydrolyse Monolignol Glucosides. Phytochemistry 2006, 67, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Cosio, C.; Penel, C.; Dunand, C. Peroxidases Have More Functions than a Swiss Army Knife. Plant Cell Rep. 2005, 24, 255–265. [Google Scholar] [CrossRef]
- Nianiou-Obeidat, I.; Madesis, P.; Kissoudis, C.; Voulgari, G.; Chronopoulou, E.; Tsaftaris, A.; Labrou, N.E. Plant Glutathione Transferase-Mediated Stress Tolerance: Functions and Biotechnological Applications. Plant Cell Rep. 2017, 36, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Vaish, S.; Gupta, D.; Mehrotra, R.; Mehrotra, S.; Basantani, M.K. Glutathione S-Transferase: A Versatile Protein Family. 3 Biotech 2020, 10, 321. [Google Scholar] [CrossRef]
- Monticolo, F.; Colantuono, C.; Chiusano, M.L. Shaping the Evolutionary Tree of Green Plants: Evidence from the GST Family. Sci. Rep. 2017, 7, 14363. [Google Scholar] [CrossRef]
- Wagner, U.; Edwards, R.; Dixon, D.P.; Mauch, F. Probing the Diversity of the Arabidopsis Glutathione S-Transferase Gene Family. Plant Mol. Biol. 2002, 49, 515–532. [Google Scholar] [CrossRef]
- Ioannou, E.; Papageorgiou, A.C.; Labrou, N.E. Directed Evolution of Phi Class Glutathione Transferases Involved in Multiple-Herbicide Resistance of Grass Weeds and Crops. Int. J. Mol. Sci. 2022, 23, 7469. [Google Scholar] [CrossRef]
- Diaz-Pérez, J.C.; Shackel, K.A.; Sutter, E.G. Relative Water Content and Water Potential of Tissue 1. J. Exp. Bot. 1995, 46, 111–118. [Google Scholar] [CrossRef]
- Haworth, M.; Marino, G.; Centritto, M. An Introductory Guide to Gas Exchange Analysis of Photosynthesis and its Application to Plant Phenotyping and Precision Irrigation to Enhance Water Use Efficiency. J. Water Clim. Change 2018, 9, 786–808. [Google Scholar] [CrossRef]
- Haworth, M.; Scutt, C.P.; Douthe, C.; Marino, G.; Gomes, M.T.G.; Loreto, F.; Flexas, J.; Centritto, M. Allocation of the Epidermis to Stomata Relates to Stomatal Physiological Control: Stomatal Factors Involved in the Evolutionary Diversification of the Angiosperms and Development of Amphistomaty. Environ. Exp. Bot. 2018, 151, 55–63. [Google Scholar] [CrossRef]
- Centritto, M.; Loreto, F.; Chartzoulakis, K. The Use of Low [CO2] to Estimate Diffusional and Non-Diffusional Limitations of Photosynthetic Capacity of Salt-Stressed Olive Saplings. Plant Cell Environ. 2003, 26, 585–594. [Google Scholar] [CrossRef]
- Genty, B.; Briantais, J.-M.; Baker, N.R. The Relationship between the Quantum Yield of Photosynthetic Electron Transport and Quenching of Chlorophyll Fluorescence. Biochim. Biophys. Acta 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Rodeghiero, M.; Niinemets, Ü.; Cescatti, A. Major Diffusion Leaks of Clamp-on Leaf Cuvettes Still Unaccounted: How Erroneous are the Estimates of Farquhar et al. Model Parameters?: Diffusion Leaks and Plant Gas Exchange. Plant Cell Environ. 2007, 30, 1006–1022. [Google Scholar] [CrossRef]
- Loriaux, S.D.; Avenson, T.J.; Welles, J.M.; Mcdermitt, D.K.; Eckles, R.D.; Riensche, B.; Genty, B. Closing in on Maximum Yield of Chlorophyll Fluorescence Using a Single Multiphase Flash of Sub-Saturating Intensity. Plant Cell Environ. 2013, 36, 1755–1770. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, T.D. Estimating the Rate of Photorespiration in Leaves. Physiol. Plant. 1988, 73, 147–152. [Google Scholar] [CrossRef]
- Laisk, A. Kinetics of Photosynthesis and Photorespiration in C3 Plants. Nauka: Moscow, Russia, 1977. (In Russian) [Google Scholar]
- Di Marco, G.; Manes, F.; Tricoli, D.; Vitale, E. Fluorescence Parameters Measured Concurrently with Net Photosynthesis to Investigate Chloroplastic CO2 Concentration in Leaves of Quercus ilex L. J. Plant Physiol. 1990, 136, 538–543. [Google Scholar] [CrossRef]
- Loreto, F.; Velikova, V.; Marco, G.D. Respiration in the Light Measured by 12CO2 Emission in 13CO2 Atmosphere in Maize Leaves. Funct. Plant Biol. 2001, 28, 1103–1108. [Google Scholar] [CrossRef]
- Vassey, T.L.; Sharkey, T.D. Mild Water Stress of Phaseolus vulgaris Plants Leads to Reduced Starch Synthesis and Extractable Sucrose Phosphate Synthase Activity. Plant Physiol. 1989, 89, 1066–1070. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, C.; Wienkoop, S.; de Almeida, J.F.; Brunetti, C.; Zarrouk, O.; Planchon, S.; Gori, A.; Tattini, M.; Ricardo, C.P.; Renaut, J.; et al. Phellem Cell-Wall Components Are Discriminants of Cork Quality in Quercus suber. Front. Plant Sci. 2019, 10, 944. [Google Scholar] [CrossRef] [PubMed]
- Boss, P.K.; Davies, C.; Robinson, S.P. Analysis of the Expression of Anthocyanin Pathway Genes in Developing Vitis vinifera L. Cv Shiraz Grape Berries and the Implications for Pathway Regulation. Plant Physiol. 1996, 111, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Rossi, J.A. Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents. Am. J. Enol Vitic. 1965, 16, 144–158. [Google Scholar]
- Antonio, C.; Pinheiro, C.; Chaves, M.M.; Ricardo, C.P.; Ortuño, M.F.; Thomas-Oates, J. Analysis of Carbohydrates in Lupinus albus Stems on Imposition of Water Deficit, Using Porous Graphitic Carbon Liquid Chromatography-Electrospray Ionization Mass Spectrometry. J. Chromatogr. A 2008, 1187, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Hatterscheid, G.W.J. Mikoplattenleser Zur Enzymatischen Zuckerbestimmung. BioTec Analytik 1991, 4, 46–48. [Google Scholar]
- Kent, W.J. BLAT—The BLAST-like Alignment Tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.J.; Jiao, Q.X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG Mapping Tools for Uncovering Hidden Features in Biological Data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Thioulouse, J.; Dray, S. Interactive multivariate data analysis in R with the ade4 and ade4TkGUI packages. J. Stat. Softw. 2007, 22, 1–14. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 14 September 2022).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinheiro, C.; Emiliani, G.; Marino, G.; Fortunato, A.S.; Haworth, M.; De Carlo, A.; Chaves, M.M.; Loreto, F.; Centritto, M. Metabolic Background, Not Photosynthetic Physiology, Determines Drought and Drought Recovery Responses in C3 and C2 Moricandias. Int. J. Mol. Sci. 2023, 24, 4094. https://doi.org/10.3390/ijms24044094
Pinheiro C, Emiliani G, Marino G, Fortunato AS, Haworth M, De Carlo A, Chaves MM, Loreto F, Centritto M. Metabolic Background, Not Photosynthetic Physiology, Determines Drought and Drought Recovery Responses in C3 and C2 Moricandias. International Journal of Molecular Sciences. 2023; 24(4):4094. https://doi.org/10.3390/ijms24044094
Chicago/Turabian StylePinheiro, Carla, Giovanni Emiliani, Giovanni Marino, Ana S. Fortunato, Matthew Haworth, Anna De Carlo, Maria Manuela Chaves, Francesco Loreto, and Mauro Centritto. 2023. "Metabolic Background, Not Photosynthetic Physiology, Determines Drought and Drought Recovery Responses in C3 and C2 Moricandias" International Journal of Molecular Sciences 24, no. 4: 4094. https://doi.org/10.3390/ijms24044094
APA StylePinheiro, C., Emiliani, G., Marino, G., Fortunato, A. S., Haworth, M., De Carlo, A., Chaves, M. M., Loreto, F., & Centritto, M. (2023). Metabolic Background, Not Photosynthetic Physiology, Determines Drought and Drought Recovery Responses in C3 and C2 Moricandias. International Journal of Molecular Sciences, 24(4), 4094. https://doi.org/10.3390/ijms24044094









