N-Glycan on the Non-Consensus N-X-C Glycosylation Site Impacts Activity, Stability, and Localization of the Sda Synthase B4GALNT2
Abstract
:1. Introduction
2. Results
2.1. Vertebrate B4GALNT2 Sequences Contain Unusual and Conserved N-Glycosylation Site(s)
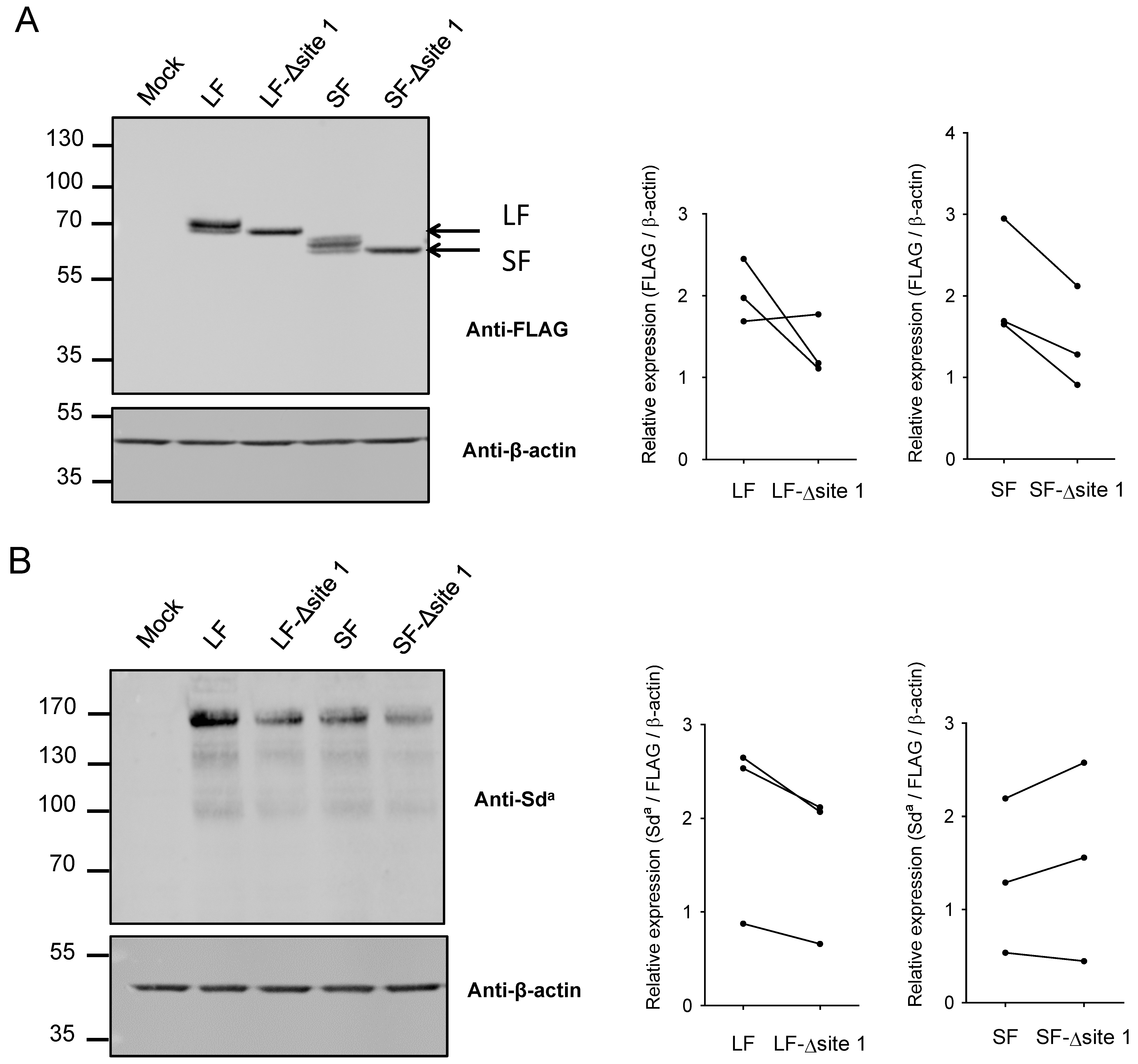
2.2. N-Glycosylation of B4GALNT2 in Mammals
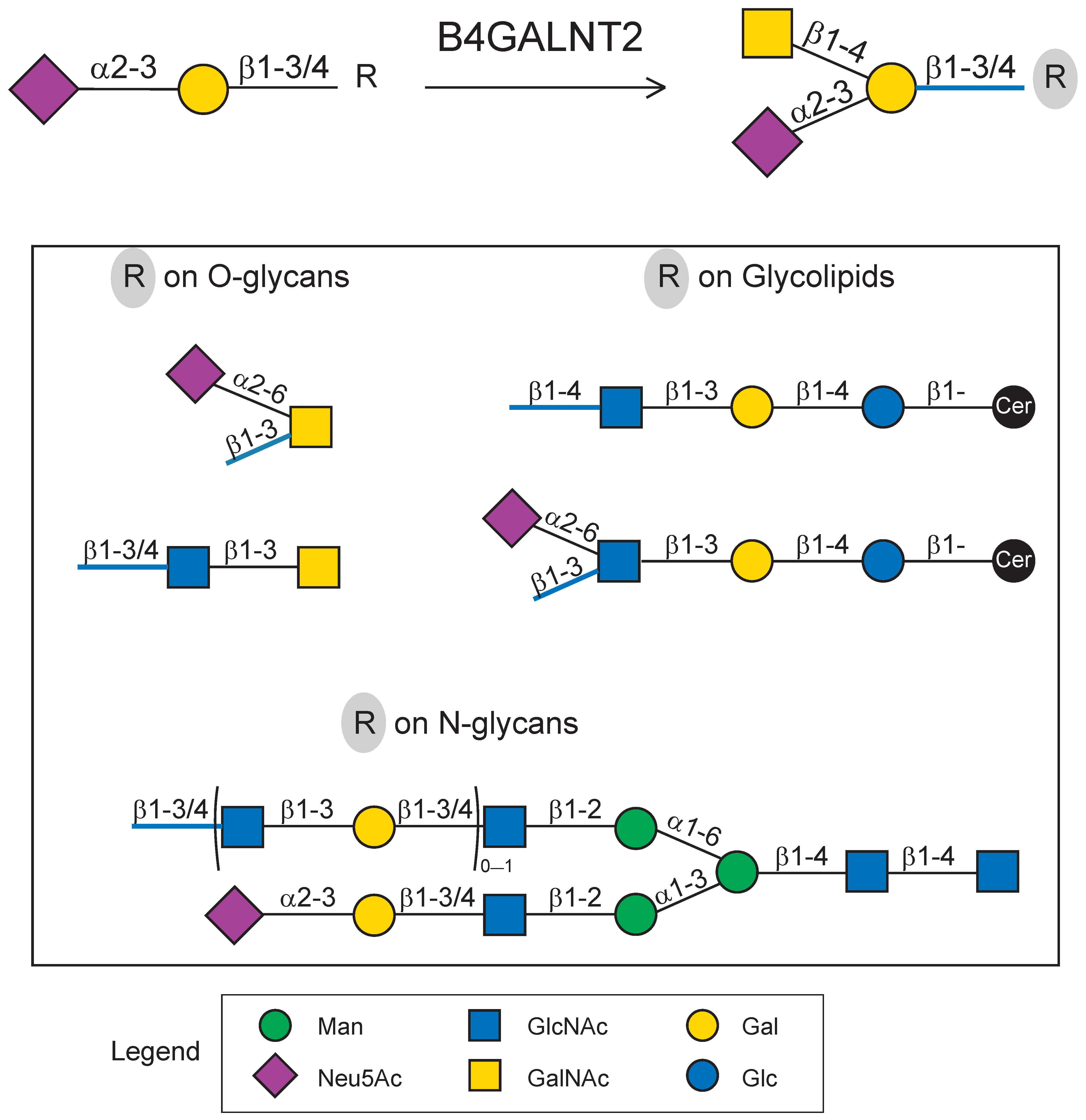
2.3. Impact of N-Glycan on B4GALNT2 Activity and Sda Synthesis
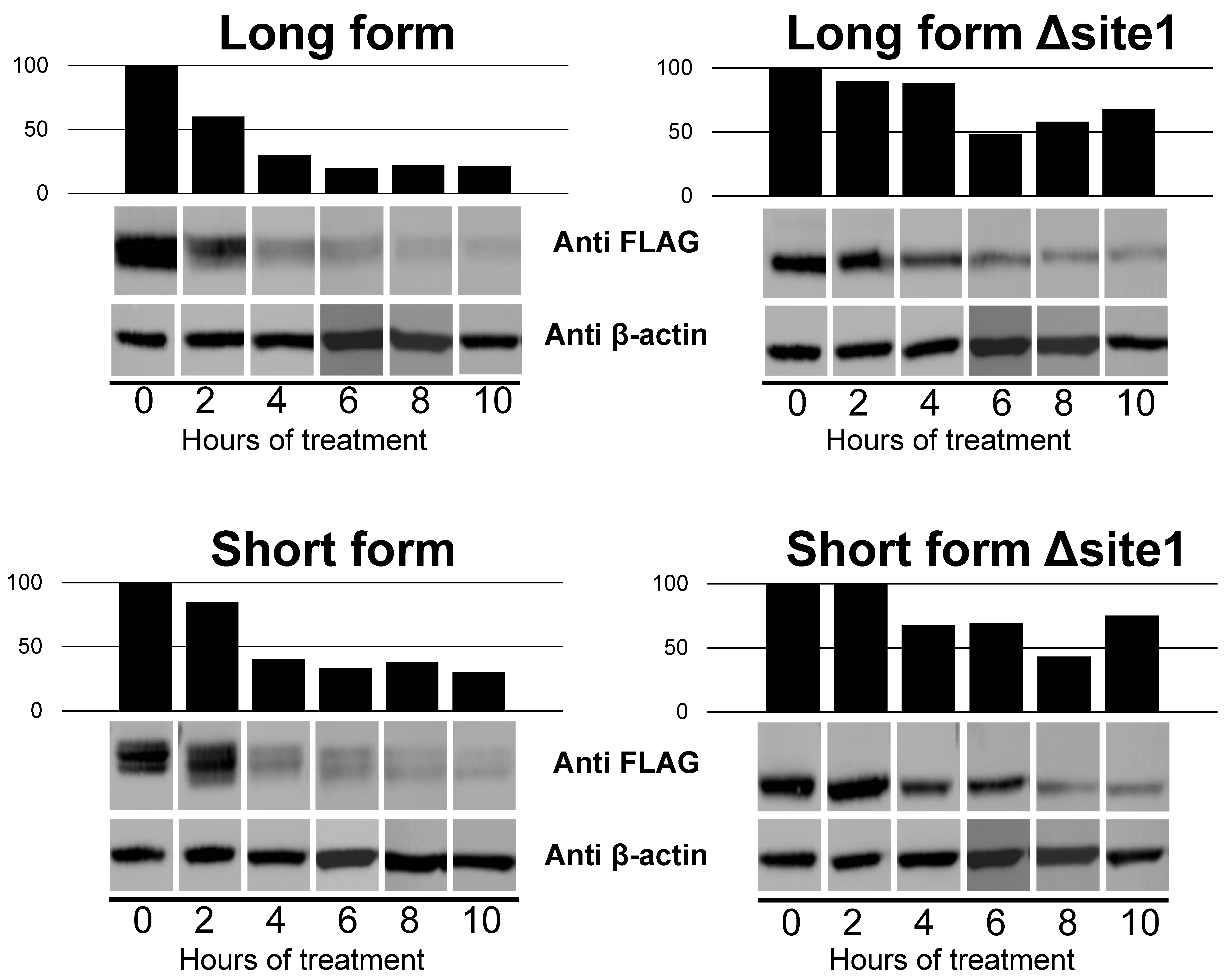
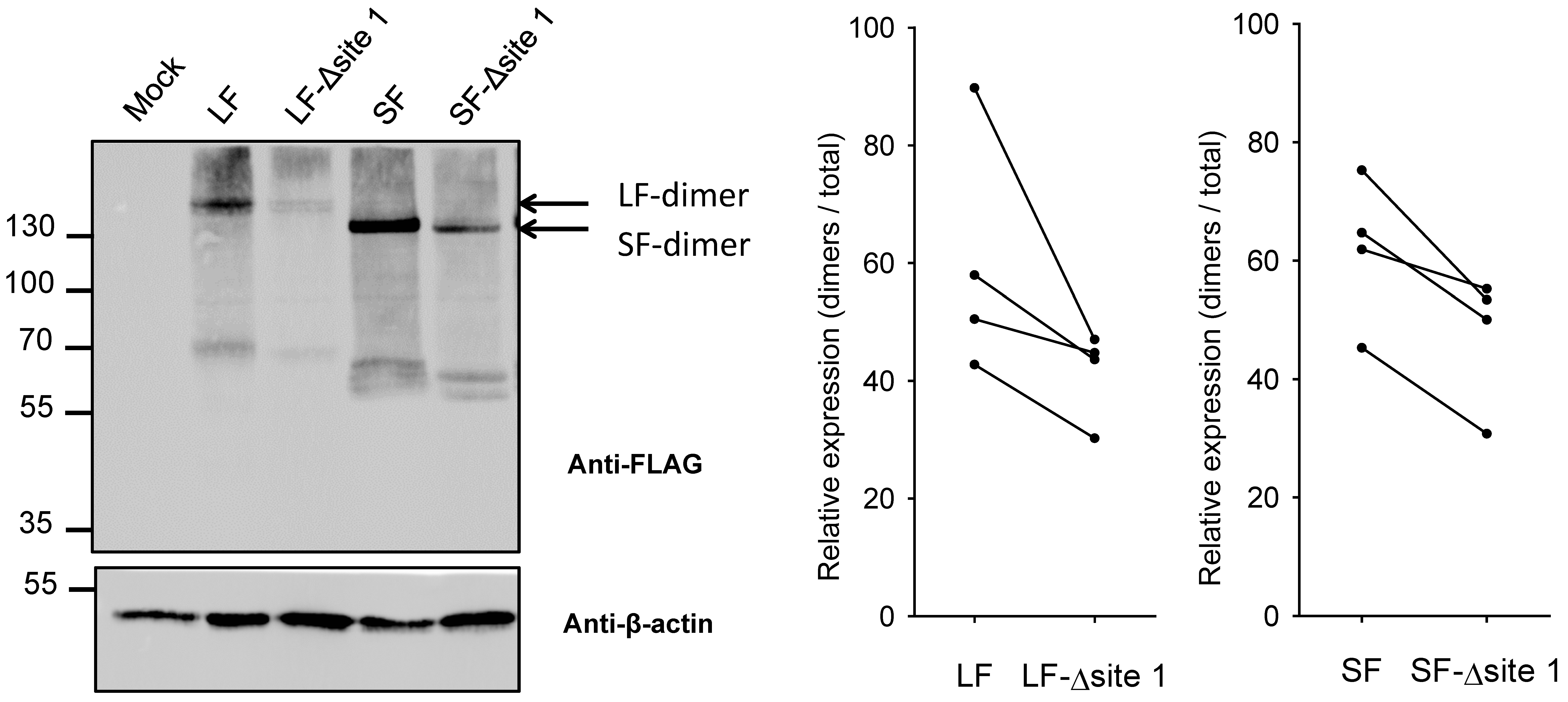
2.4. Impact of N-Glycan on Dimerization of B4GALNT2 Isoforms
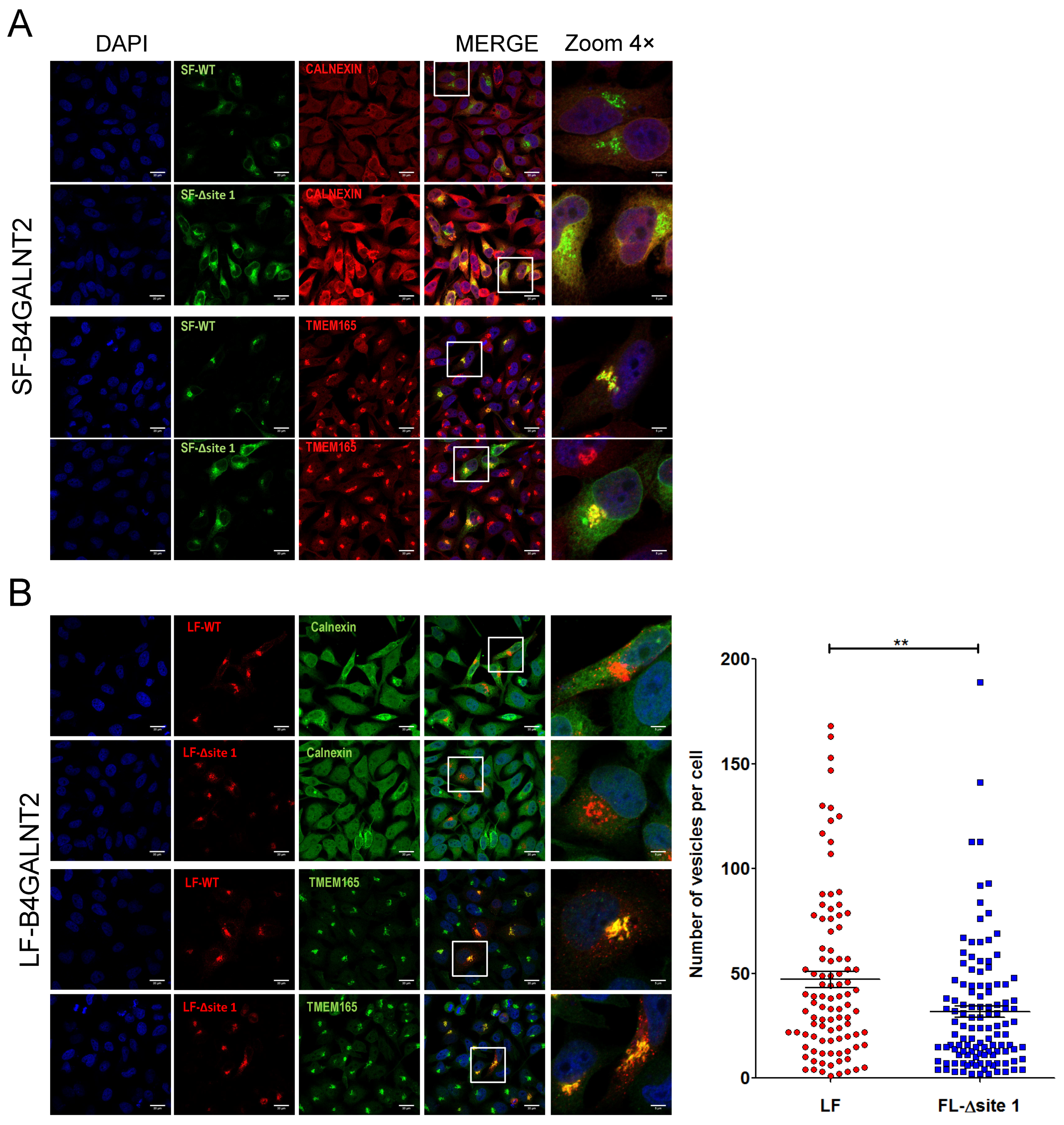
2.5. Subcellular Localization of B4GALNT2 Isoforms
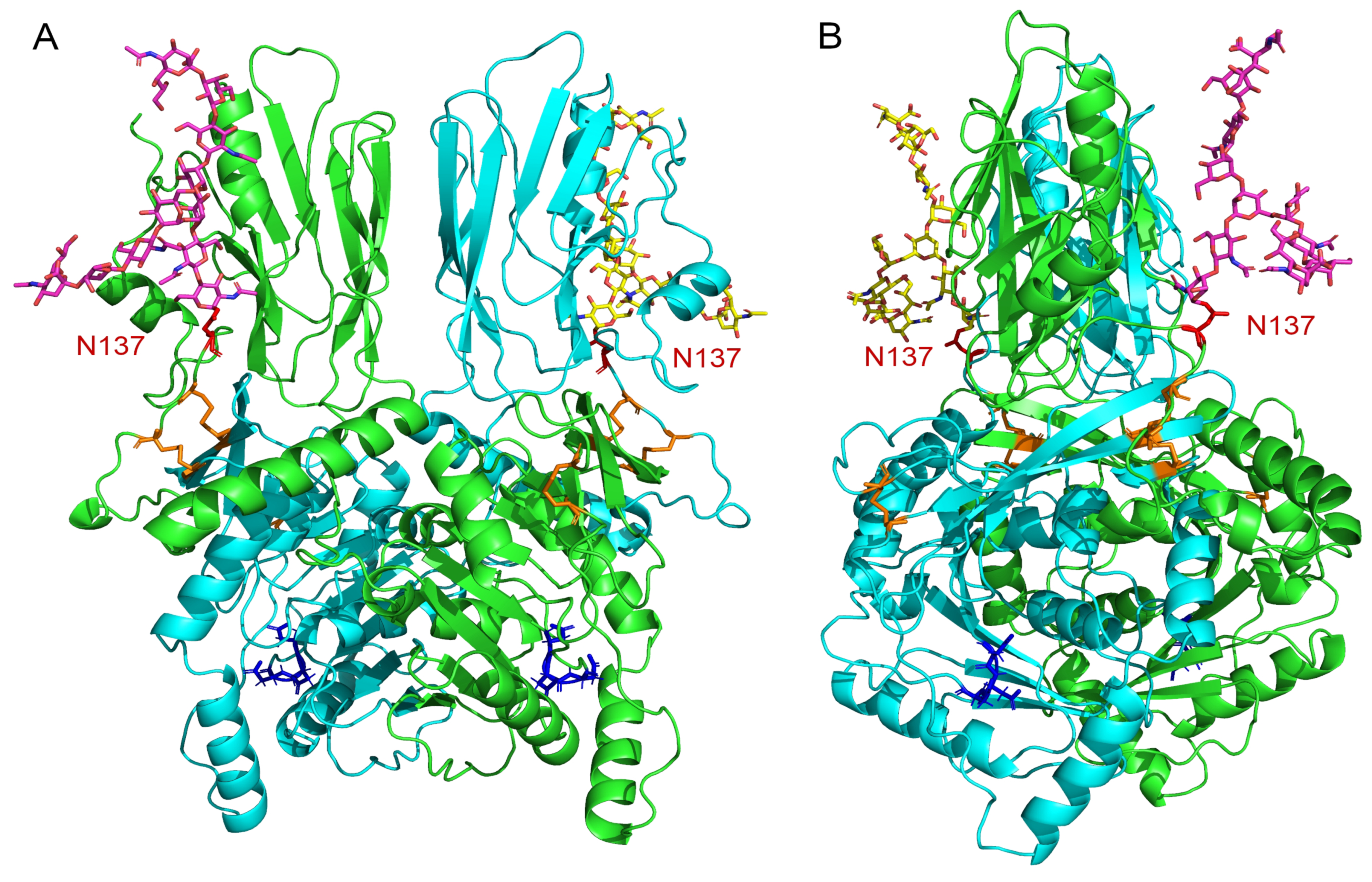
2.6. Model of the B4GALNT2 Structure
3. Discussion
4. Materials and Methods
4.1. Bioinformatic Analysis of B4GALNT Protein Sequences
4.2. Antibodies and Reagents
4.3. Plasmid Constructions of Human Long and Short B4GALNT2 Isoforms and Mutants
4.4. Cell Culture and Transfections
4.5. Western Blot Analysis
4.6. Total Proteins Preparation from Various Mammalian Gastrointestinal Tissues and Immunochemical Analyses
4.7. PNGase F and Endo H Treatments
4.8. B4GALNT2 Enzyme Activity
4.9. Flexibility Prediction
4.10. Modeling of the B4GALNT2 Dimeric Structure
- --is_prokaryote_list=false
- --max_template_date=2021-11-17
- --model_preset=multimer
- --db_preset=full_dbs
4.11. Statistical Methodology
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macvie, S.I.; Morton, J.A.; Pickles, M.M. The reactions and inheritance of a new blood group antigen, Sda. Vox Sang. 1967, 13, 485–492. [Google Scholar] [CrossRef]
- Renton, P.H.; Howell, P.; IKIN, E.W.; Giles, C.M.; Goldsmith, K.L.G. Anti-Sda, a New Blood Group Antibody. Vox Sang. 1967, 13, 493–501. [Google Scholar] [CrossRef]
- Groux-Degroote, S.; Vicogne, D.; Cogez, V.; Schulz, C.; Harduin-Lepers, A. B4GALNT2 Controls Sda and SLex Antigen Biosynthesis in Healthy and Cancer Human Colon. Chembiochem 2021, 22, 3381–3390. [Google Scholar] [CrossRef]
- Stenfelt, L.; Hellberg, Å.; Möller, M.; Thornton, N.; Larson, G.; Olsson, M.L. Missense mutations in the C-terminal portion of the B4GALNT2-encoded glycosyltransferase underlying the Sd(a−) phenotype. Biochem. Biophys. Rep. 2019, 19, 100659. [Google Scholar] [CrossRef]
- Dall’Olio, F.; Malagolini, N.; Chiricolo, M.; Trinchera, M.; Harduin-Lepers, A. The expanding roles of the Sda/Cad carbohydrate antigen and its cognate glycosyltransferase B4GALNT2. Biochim. Biophys. Acta 2014, 1840, 443–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groux-Degroote, S.; Wavelet, C.; Krzewinski-Recchi, M.A.; Portier, L.; Mortuaire, M.; Mihalache, A.; Trinchera, M.; Delannoy, P.; Malagolini, N.; Chiricolo, M.; et al. B4GALNT2 gene expression controls the biosynthesis of Sda and sialyl Lewis X antigens in healthy and cancer human gastrointestinal tract. Int. J. Biochem. Cell Biol. 2014, 53, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Levery, S.B.; Saito, S.; Satoh, M.; Hakomori, S. A novel ganglioside isolated from renal cell carcinoma. J. Biol. Chem. 2001, 276, 16695–16703. [Google Scholar] [CrossRef] [Green Version]
- Tsuchida, A.; Senda, M.; Ito, A.; Saito, S.; Kiso, M.; Ando, T.; Harduin-Lepers, A.; Matsuda, A.; Furukawa, K.; Furukawa, K. Roles of GalNAc-disialyl Lactotetraosyl Antigens in Renal Cancer Cells. Sci. Rep. 2018, 8, 7017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawamura, Y.I.; Kawashima, R.; Fukunaga, R.; Hirai, K.; Toyama-Sorimachi, N.; Tokuhara, M.; Shimizu, T.; Dohi, T. Introduction of Sda carbohydrate antigen in gastrointestinal cancer cells eliminates selectin ligands and inhibits metastasis. Cancer Res. 2005, 65, 6220–6227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pucci, M.; Gomes Ferreira, I.; Malagolini, N.; Ferracin, M.; Dall’Olio, F. The Sda Synthase B4GALNT2 Reduces Malignancy and Stemness in Colon Cancer Cell Lines Independently of Sialyl Lewis X Inhibition. Int. J. Mol. Sci. 2020, 21, 6558. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.T.; Xu, R.; Rodino-Klapac, L.R.; Oglesbay, E.; Camboni, M.; Montgomery, C.L.; Shontz, K.; Chicoine, L.G.; Clark, K.R.; Sahenk, Z.; et al. Overexpression of Galgt2 in skeletal muscle prevents injury resulting from eccentric contractions in both mdx and wild-type mice. Am. J. Physiol. Cell Physiol. 2009, 296, C476–C488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.; DeVries, S.; Camboni, M.; Martin, P.T. Overexpression of Galgt2 reduces dystrophic pathology in the skeletal muscles of alpha sarcoglycan-deficient mice. Am. J. Pathol. 2009, 175, 235–247. [Google Scholar] [CrossRef] [Green Version]
- Mohlke, K.L.; Purkayastha, A.A.; Westrick, R.J.; Smith, P.L.; Petryniak, B.; Lowe, J.B.; Ginsburg, D. Mvwf, a dominant modifier of murine von Willebrand factor, results from altered lineage-specific expression of a glycosyltransferase. Cell 1999, 96, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Lefrancois, L.; Puddington, L.; Machamer, C.E.; Bevan, M.J. Acquisition of cytotoxic T lymphocyte-specific carbohydrate differentiation antigens. J. Exp. Med. 1985, 162, 1275–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staubach, F.; Kunzel, S.; Baines, A.C.; Yee, A.; McGee, B.M.; Backhed, F.; Baines, J.F.; Johnsen, J.M. Expression of the blood-group-related glycosyltransferase B4galnt2 influences the intestinal microbiota in mice. ISME J. 2012, 6, 1345–1355. [Google Scholar] [CrossRef] [Green Version]
- Li, P.T.; Liao, C.J.; Yu, L.C.; Wu, W.G.; Chu, S.T. Localization of B4GALNT2 and its role in mouse embryo attachment. Fertil. Steril. 2012, 97, 1206–1212.e1201–1203. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Cummings, R.D.; Aebi, M.; Packer, N.H.; Seeberger, P.H.; Esko, J.D.; Stanley, P.; Hart, G.; Darvill, A.; Kinoshita, T.; et al. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology 2015, 25, 1323–1324. [Google Scholar] [CrossRef] [Green Version]
- Cheng, K.; Zhou, Y.; Neelamegham, S. DrawGlycan-SNFG: A robust tool to render glycans and glycopeptides with fragmentation information. Glycobiology 2017, 27, 200–205. [Google Scholar] [CrossRef] [Green Version]
- Lo Presti, L.; Cabuy, E.; Chiricolo, M.; Dall′Olio, F. Molecular cloning of the human beta1,4 N-acetylgalactosaminyltransferase responsible for the biosynthesis of the Sda histo-blood group antigen: The sequence predicts a very long cytoplasmic domain. J. Biochem. 2003, 134, 675–682. [Google Scholar] [CrossRef]
- Montiel, M.D.; Krzewinski-Recchi, M.A.; Delannoy, P.; Harduin-Lepers, A. Molecular cloning, gene organization and expression of the human UDP-GalNAc:Neu5Acα2-3Galβ-Rβ1,4-N-acetylgalactosaminyltransferase responsible for the biosynthesis of the blood group Sda/Cad antigen: Evidence for an unusual extended cytoplasmic domain. Biochem. J. 2003, 373, 369–379. [Google Scholar] [CrossRef] [Green Version]
- Pucci, M.; Malagolini, N.; Dall’Olio, F. Glycosyltransferase B4GALNT2 as a Predictor of Good Prognosis in Colon Cancer: Lessons from Databases. Int. J. Mol. Sci. 2021, 22, 4331. [Google Scholar] [CrossRef]
- Wavelet-Vermuse, C.; Groux-Degroote, S.; Vicogne, D.; Cogez, V.; Venturi, G.; Trinchera, M.; Brysbaert, G.; Krzewinski-Recchi, M.A.; Hadj Bachir, E.; Schulz, C.; et al. Analysis of the proximal promoter of the human colon-specific B4GALNT2 (Sda synthase) gene: B4GALNT2 is transcriptionally regulated by ETS1. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194747. [Google Scholar] [CrossRef] [PubMed]
- Malagolini, N.; Santini, D.; Chiricolo, M.; Dall’Olio, F. Biosynthesis and expression of the Sda and sialyl Lewis x antigens in normal and cancer colon. Glycobiology 2007, 17, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Groux-Degroote, S.; Schulz, C.; Cogez, V.; Noel, M.; Portier, L.; Vicogne, D.; Solorzano, C.; Dall’Olio, F.; Steenackers, A.; Mortuaire, M.; et al. The extended cytoplasmic tail of the human B4GALNT2 is critical for its Golgi targeting and post-Golgi sorting. FEBS J. 2018, 285, 3442–3463. [Google Scholar] [CrossRef] [Green Version]
- Rosnoblet, C.; Peanne, R.; Legrand, D.; Foulquier, F. Glycosylation disorders of membrane trafficking. Glycoconj. J. 2013, 30, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Mikolajczyk, K.; Kaczmarek, R.; Czerwinski, M. How glycosylation affects glycosylation: The role of N-glycans in glycosyltransferase activity. Glycobiology 2020, 30, 941–969. [Google Scholar] [CrossRef]
- Smith, P.L.; Lowe, J.B. Molecular cloning of a murine N-acetylgalactosamine transferase cDNA that determines expression of the T lymphocyte-specific CT oligosaccharide differentiation antigen. J. Biol. Chem. 1994, 269, 15162–15171. [Google Scholar] [CrossRef]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. In Proceedings of the Pacific Symposium on Biocomputing 2002, Kauai, HI, USA, 3–7 January 2002; pp. 310–322. [Google Scholar]
- Haraguchi, M.; Yamashiro, S.; Furukawa, K.; Takamiya, K.; Shiku, H.; Furukawa, K. The effects of the site-directed removal of N-glycosylation sites from β-1,4-N-acetylgalactosaminyltransferase on its function. Biochem. J. 1995, 312, 273–280. [Google Scholar] [CrossRef]
- Faid, V.; Denguir, N.; Chapuis, V.; Bihoreau, N.; Chevreux, G. Site-specific N-glycosylation analysis of human factor XI: Identification of a noncanonical NXC glycosite. Proteomics 2014, 14, 2460–2470. [Google Scholar] [CrossRef]
- Lowenthal, M.S.; Davis, K.S.; Formolo, T.; Kilpatrick, L.E.; Phinney, K.W. Identification of Novel N-Glycosylation Sites at Noncanonical Protein Consensus Motifs. J. Proteome Res. 2016, 15, 2087–2101. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Zhang, H. Identification and validation of atypical N-glycosylation sites. Anal. Chem. 2015, 87, 11948–11951. [Google Scholar] [CrossRef] [Green Version]
- Zielinska, D.F.; Gnad, F.; Wisniewski, J.R.; Mann, M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 2010, 141, 897–907. [Google Scholar] [CrossRef] [Green Version]
- Foulquier, F.; Amyere, M.; Jaeken, J.; Zeevaert, R.; Schollen, E.; Race, V.; Bammens, R.; Morelle, W.; Rosnoblet, C.; Legrand, D.; et al. TMEM165 deficiency causes a congenital disorder of glycosylation. Am. J. Hum. Genet. 2012, 91, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic. Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Park, C.; Zhang, J. Genome-wide evolutionary conservation of N-glycosylation sites. Mol. Biol. Evol. 2011, 28, 2351–2357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Colley, K.J. Minimal structural and glycosylation requirements for ST6Gal I activity and trafficking. Glycobiology 2000, 10, 531–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeanneau, C.; Chazalet, V.; Auge, C.; Soumpasis, D.M.; Harduin-Lepers, A.; Delannoy, P.; Imberty, A.; Breton, C. Structure-function analysis of the human sialyltransferase ST3Gal I: Role of N-glycosylation and a novel conserved sialylmotif. J. Biol. Chem. 2004, 279, 13461–13468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallejo-Ruiz, V.; Haque, R.; Mir, A.M.; Schwientek, T.; Mandel, U.; Cacan, R.; Delannoy, P.; Harduin-Lepers, A. Delineation of the minimal catalytic domain of human Galβ1-3GalNAc α2,3-sialyltransferase (hST3Gal I). Biochim. Biophys. Acta 2001, 1549, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, F.M.; Vilcaes, A.A.; Iglesias-Bartolome, R.; Daniotti, J.L. Critical role of evolutionarily conserved glycosylation at Asn211 in the intracellular trafficking and activity of sialyltransferase ST3Gal-II. Biochem. J. 2015, 469, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Bieberich, E.; Tencomnao, T.; Kapitonov, D.; Yu, R.K. Effect of N-glycosylation on turnover and subcellular distribution of N-acetylgalactosaminyltransferase I and sialyltransferase II in neuroblastoma cells. J. Neurochem. 2000, 74, 2359–2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martina, J.A.; Daniotti, J.L.; Maccioni, H.J. Influence of N-glycosylation and N-glycan trimming on the activity and intracellular traffic of GD3 synthase. J. Biol. Chem. 1998, 273, 3725–3731. [Google Scholar] [CrossRef] [Green Version]
- Uemura, S.; Kurose, T.; Suzuki, T.; Yoshida, S.; Ito, M.; Saito, M.; Horiuchi, M.; Inagaki, F.; Igarashi, Y.; Inokuchi, J. Substitution of the N-glycan function in glycosyltransferases by specific amino acids: ST3Gal-V as a model enzyme. Glycobiology 2006, 16, 258–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Battari, A.; Prorok, M.; Angata, K.; Mathieu, S.; Zerfaoui, M.; Ong, E.; Suzuki, M.; Lombardo, D.; Fukuda, M. Different glycosyltransferases are differentially processed for secretion, dimerization, and autoglycosylation. Glycobiology 2003, 13, 941–953. [Google Scholar] [CrossRef] [Green Version]
- Hassinen, A.; Kellokumpu, S. Organizational interplay of Golgi N-glycosyltransferases involves organelle microenvironment-dependent transitions between enzyme homo- and heteromers. J. Biol. Chem. 2014, 289, 26937–26948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassinen, A.; Rivinoja, A.; Kauppila, A.; Kellokumpu, S. Golgi N-glycosyltransferases form both homo- and heterodimeric enzyme complexes in live cells. J. Biol. Chem. 2010, 285, 17771–17777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoder-Agha, F.; Harrus, D.; Brysbaert, G.; Lensink, M.F.; Harduin-Lepers, A.; Glumoff, T.; Kellokumpu, S. Assembly of B4GALT1/ST6GAL1 heteromers in the Golgi membranes involves lateral interactions via highly charged surface domains. J. Biol. Chem. 2019, 294, 14383–14393. [Google Scholar] [CrossRef]
- Li, J.; Yen, T.Y.; Allende, M.L.; Joshi, R.K.; Cai, J.; Pierce, W.M.; Jaskiewicz, E.; Darling, D.S.; Macher, B.A.; Young, W.W., Jr. Disulfide bonds of GM2 synthase homodimers. Antiparallel orientation of the catalytic domains. J. Biol. Chem. 2000, 275, 41476–41486. [Google Scholar] [CrossRef] [Green Version]
- Cherepanova, N.A.; Shrimal, S.; Gilmore, R. Oxidoreductase activity is necessary for N-glycosylation of cysteine-proximal acceptor sites in glycoproteins. J. Cell Biol. 2014, 206, 525–539. [Google Scholar] [CrossRef] [Green Version]
- Cherepanova, N.A.; Venev, S.V.; Leszyk, J.D.; Shaffer, S.A.; Gilmore, R. Quantitative glycoproteomics reveals new classes of STT3A- and STT3B-dependent N-glycosylation sites. J. Cell Biol. 2019, 218, 2782–2796. [Google Scholar] [CrossRef] [Green Version]
- Blom, N.; Sicheritz-Ponten, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef]
- Colomb, F.; Krzewinski-Recchi, M.A.; El Machhour, F.; Mensier, E.; Jaillard, S.; Steenackers, A.; Harduin-Lepers, A.; Lafitte, J.J.; Delannoy, P.; Groux-Degroote, S. TNF regulates sialyl-Lewisx and 6-sulfo-sialyl-Lewisx expression in human lung through up-regulation of ST3GAL4 transcript isoform BX. Biochimie 2012, 94, 2045–2053. [Google Scholar] [CrossRef] [PubMed]
- Cilia, E.; Pancsa, R.; Tompa, P.; Lenaerts, T.; Vranken, W.F. From protein sequence to dynamics and disorder with DynaMine. Nat. Commun. 2013, 4, 2741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cilia, E.; Pancsa, R.; Tompa, P.; Lenaerts, T.; Vranken, W.F. The DynaMine webserver: Predicting protein dynamics from sequence. Nucleic. Acids Res. 2014, 42, W264–W270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Schrödinger, LLC. The PyMOL Molecular Graphics System, version 2.3.0; 2010. Available online: https://pymol.org/2/support.html?#citing (accessed on 23 December 2021).
- Hubbard, S.J.; Thornton, J.M. Naccess, Computer Program, version 2.1.1 ed; Department of Biochemistry and Molecular Biology, University College: London, UK, 1993. [Google Scholar]
- Woods Group, R. GLYCAM Web. Complex Carbohydrate Research Center, University of Georgia: Athens, GA, USA. Available online: http://legacy.glycam.org (accessed on 1 January 2020).
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger, LLC. The PyMOL Molecular Graphics System, version 1.8; 2015. Available online: https://pymol.org/2/ (accessed on 23 December 2021).


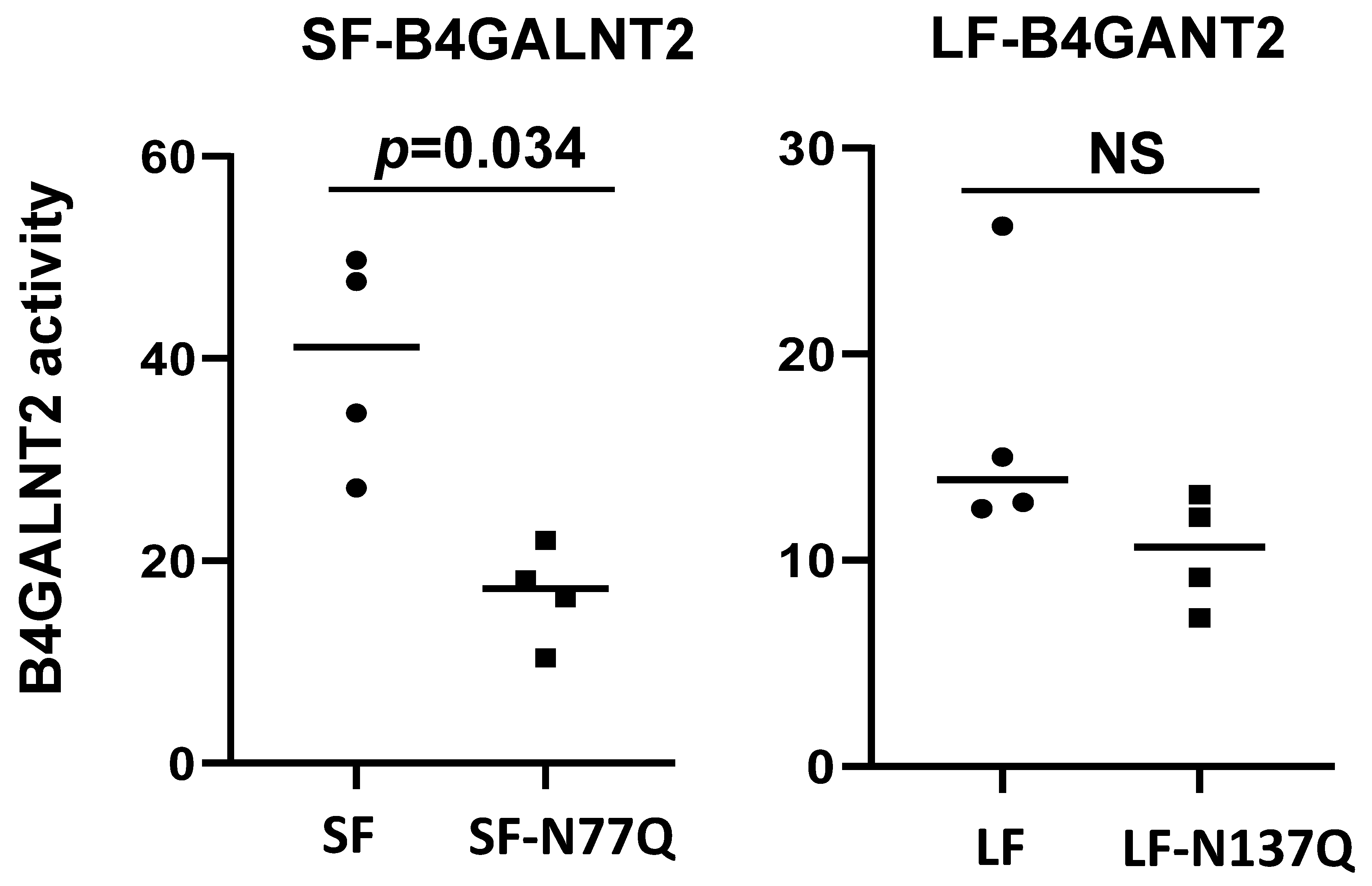
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cogez, V.; Vicogne, D.; Schulz, C.; Portier, L.; Venturi, G.; de Ruyck, J.; Decloquement, M.; Lensink, M.F.; Brysbaert, G.; Dall’Olio, F.; et al. N-Glycan on the Non-Consensus N-X-C Glycosylation Site Impacts Activity, Stability, and Localization of the Sda Synthase B4GALNT2. Int. J. Mol. Sci. 2023, 24, 4139. https://doi.org/10.3390/ijms24044139
Cogez V, Vicogne D, Schulz C, Portier L, Venturi G, de Ruyck J, Decloquement M, Lensink MF, Brysbaert G, Dall’Olio F, et al. N-Glycan on the Non-Consensus N-X-C Glycosylation Site Impacts Activity, Stability, and Localization of the Sda Synthase B4GALNT2. International Journal of Molecular Sciences. 2023; 24(4):4139. https://doi.org/10.3390/ijms24044139
Chicago/Turabian StyleCogez, Virginie, Dorothée Vicogne, Céline Schulz, Lucie Portier, Giulia Venturi, Jérôme de Ruyck, Mathieu Decloquement, Marc F. Lensink, Guillaume Brysbaert, Fabio Dall’Olio, and et al. 2023. "N-Glycan on the Non-Consensus N-X-C Glycosylation Site Impacts Activity, Stability, and Localization of the Sda Synthase B4GALNT2" International Journal of Molecular Sciences 24, no. 4: 4139. https://doi.org/10.3390/ijms24044139




