Evolution of Cytochrome P450 Enzymes and Their Redox Partners in Archaea
Abstract
1. Introduction
2. Results and Discussion
2.1. Archaea Has the Lowest P450 Diversity
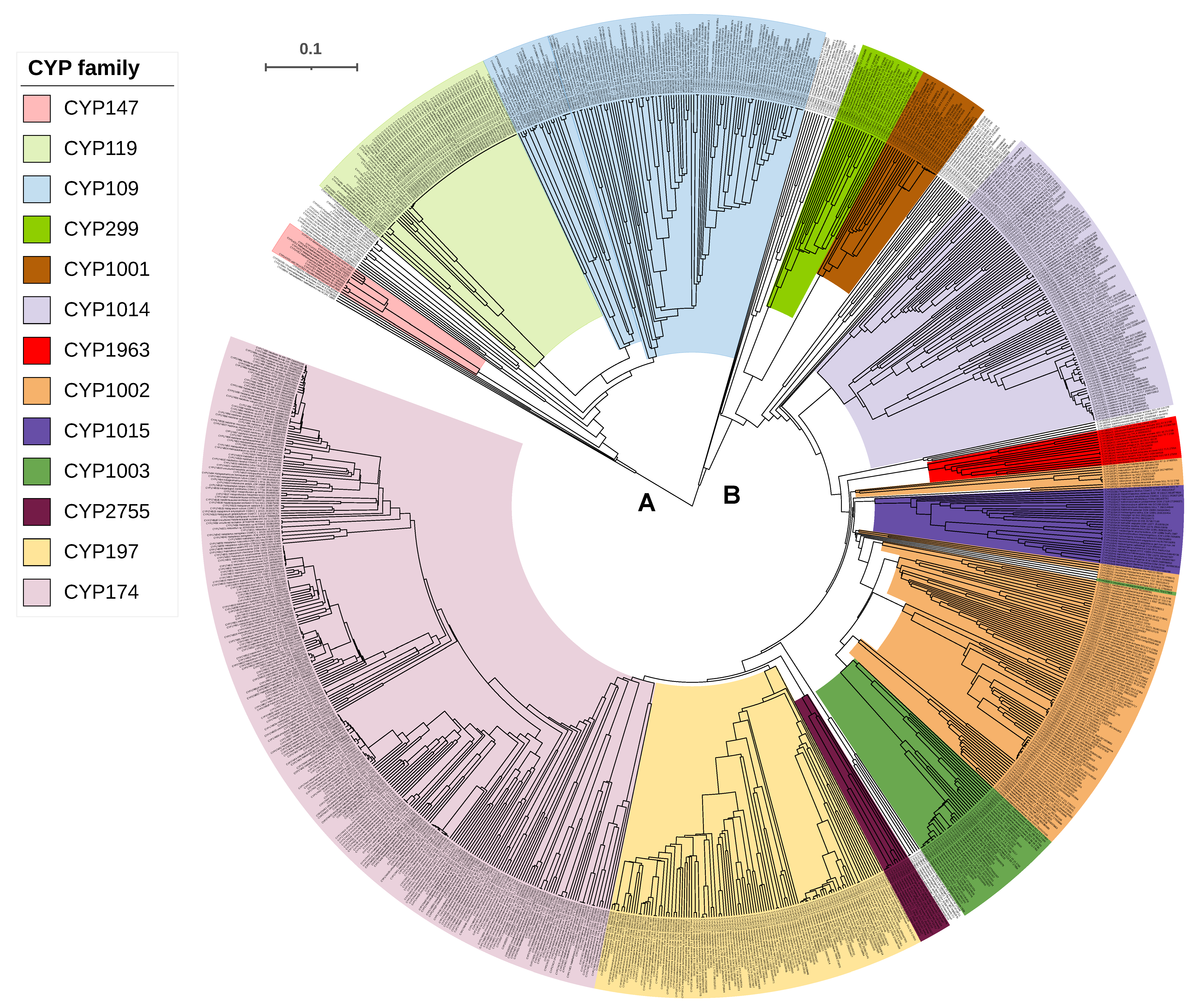
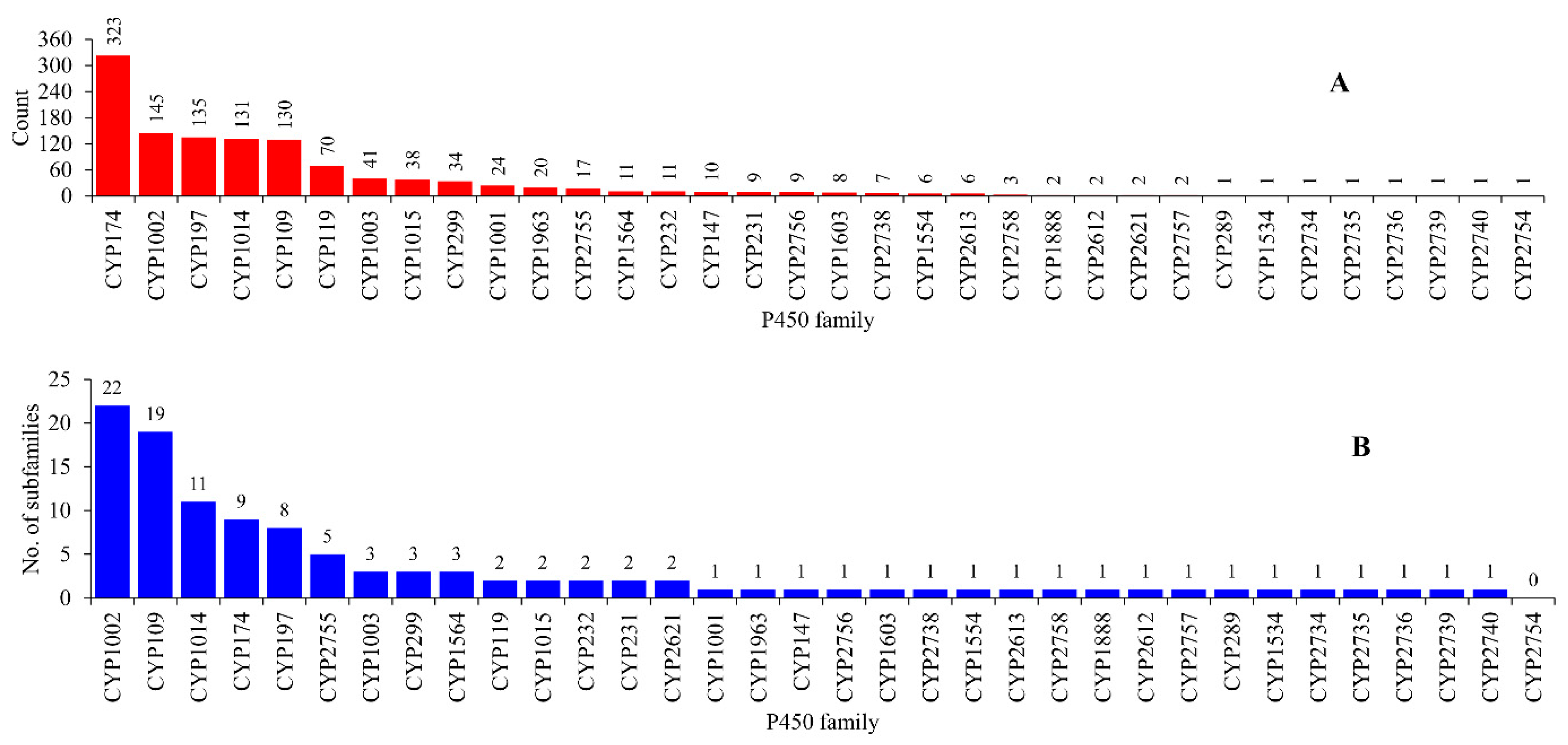
2.2. Some P450 Families and Subfamilies Are Expanded in Archaea
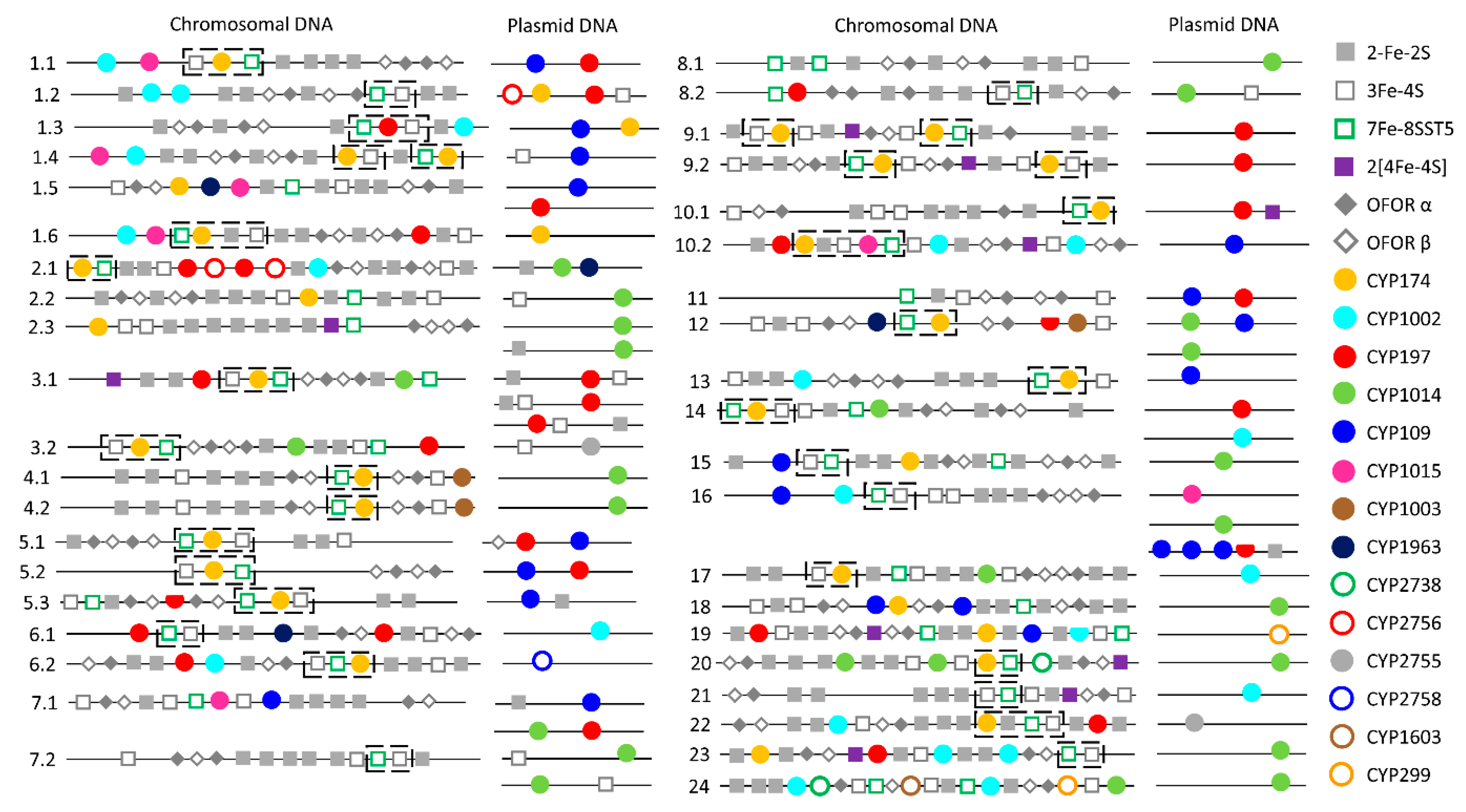
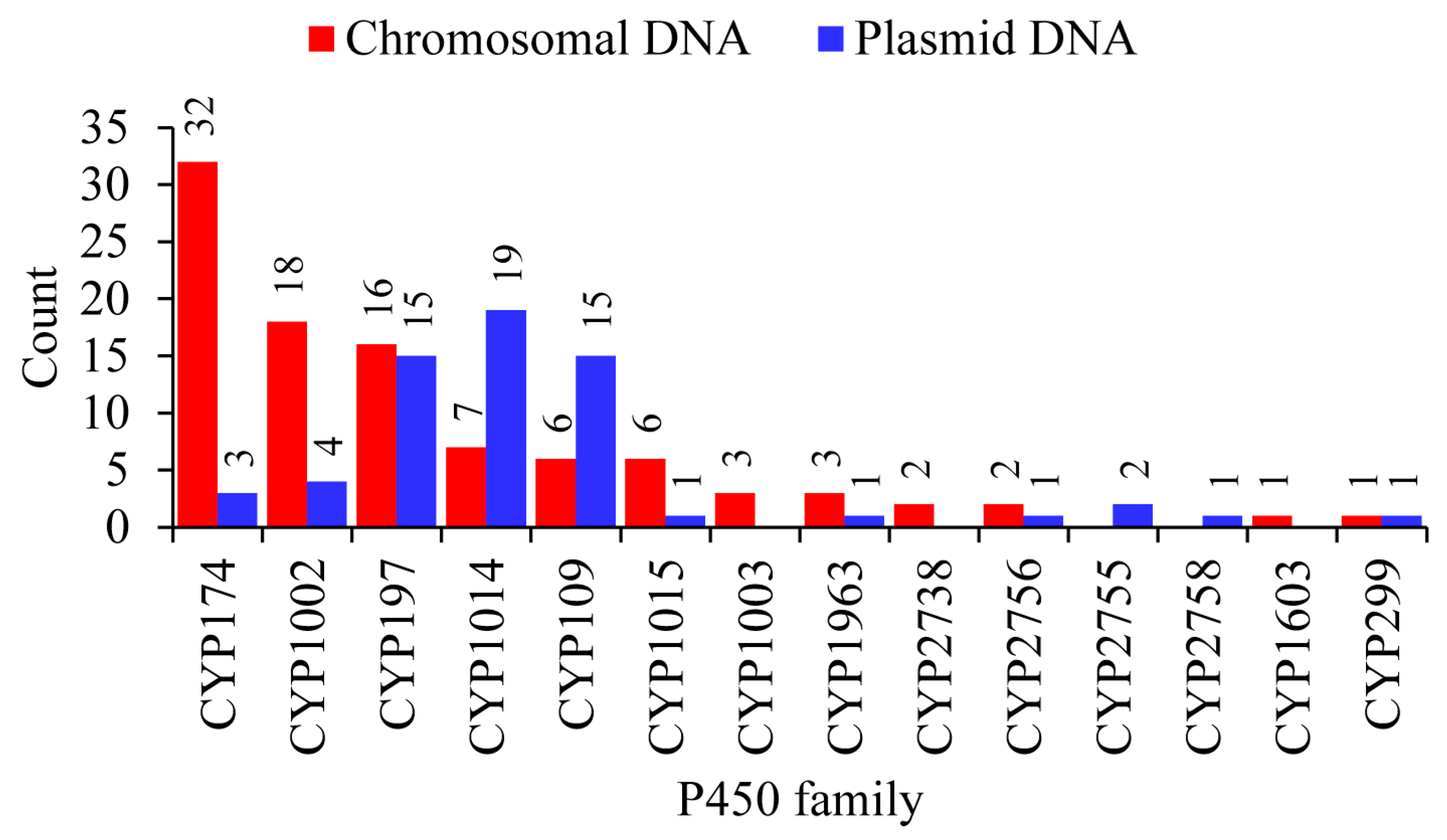
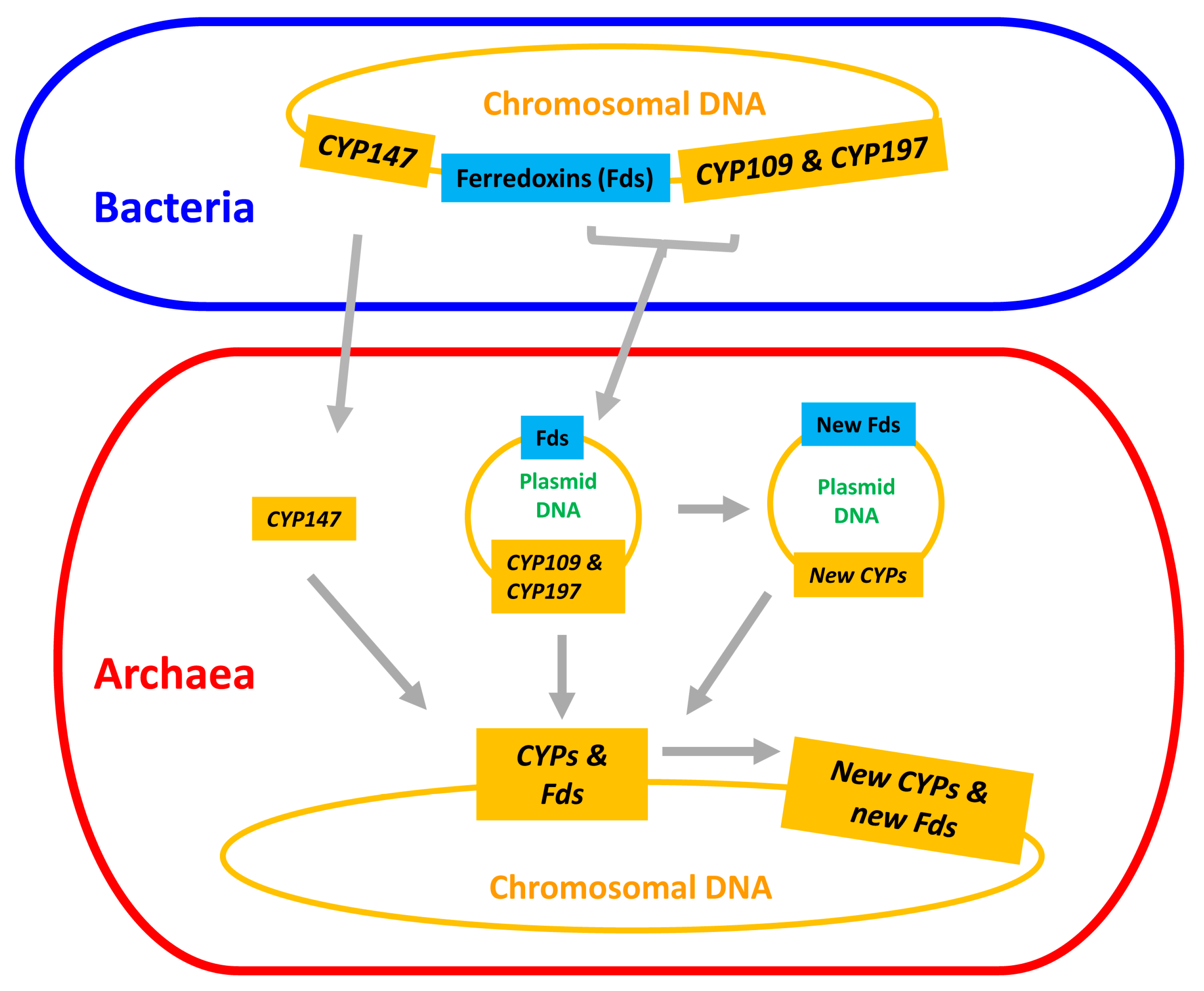
2.3. Plasmid-Mediated Lateral Transfer of P450s from Bacteria to Archaea
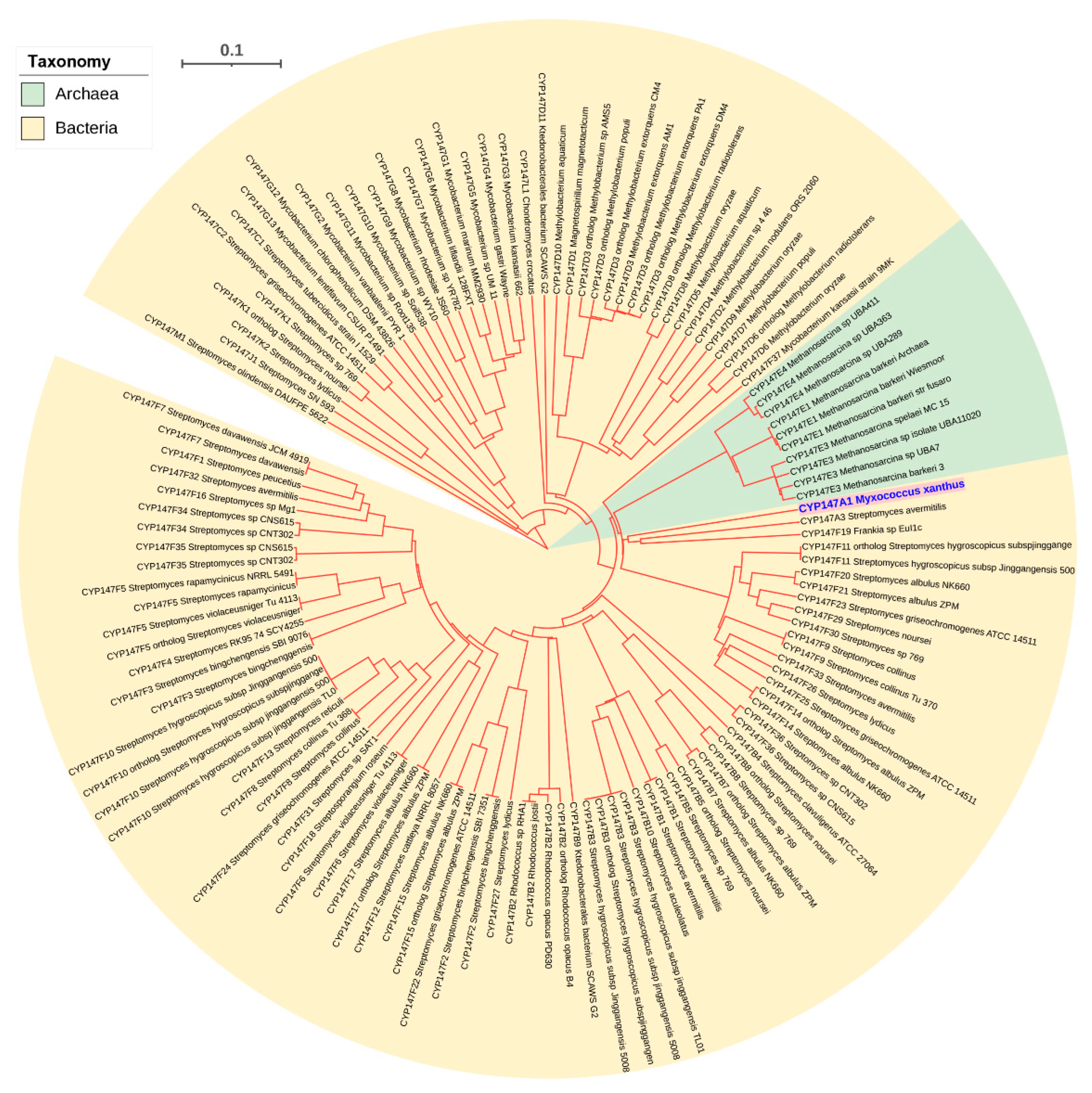
2.4. Archaeal P450s Are Bacterial in Origin, Not Vice Versa
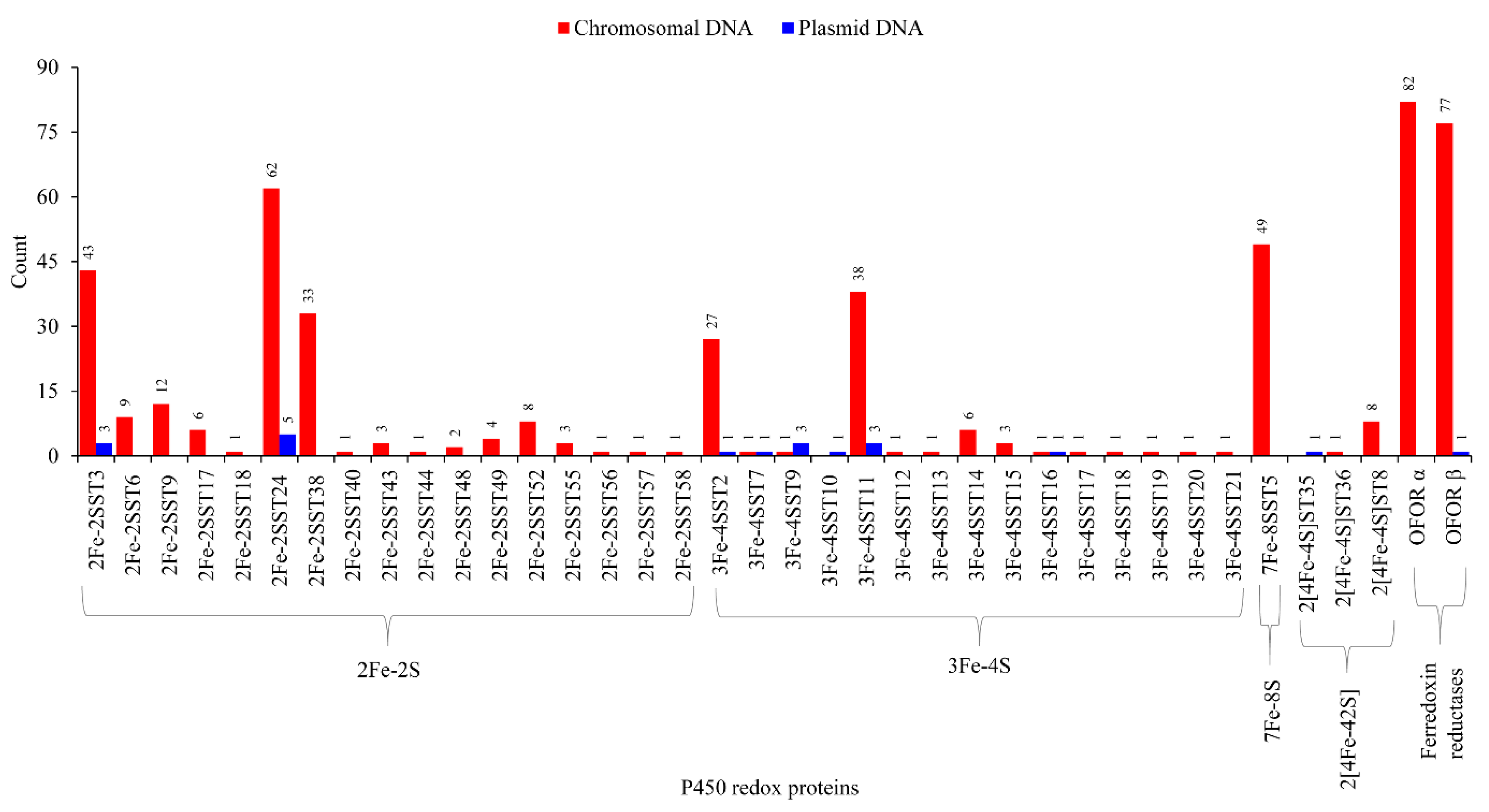
2.5. Lateral Transfer of Putative Redox Partners Is Independent of P450s
2.6. CYP109, CYP147 and CYP197 Gave Rise to Archaeal P450s
2.7. Most of the Archaeal P450s Are Orphans with No Known Function
3. Materials and Methods
3.1. Species and Their Genome Database Information
3.2. Genome Data Mining and Annotation of P450s
3.3. Analysis of P450s in Archaeal Plasmids
3.4. Phylogenetic Analysis of P450s
3.5. BLAST Analysis of Archaeal P450 Families for Affinity to CYP109 or CYP197
3.6. Comparative Analysis of P450s
3.7. Genome Data Mining and Annotation of Ferredoxins
3.8. Comparative Analysis of Ferredoxins
3.9. Genome Data Mining of Ferredoxin Reductases
3.10. Retrieving Protein Identification Numbers from NCBI
3.11. Operon Predictions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garfinkel, D. Studies on pig liver microsomes. I. Enzymic and pigment composition of different microsomal fractions. Arch. Biochem. Biophys. 1958, 77, 493–509. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, M. Pigments of rat liver microsomes. Arch. Biochem. Biophys. 1958, 75, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Omura, T. Recollection of the early years of the research on cytochrome P450. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 617–640. [Google Scholar] [CrossRef] [PubMed]
- Omura, T.; Sato, R. A new cytochrome in liver microsomes. J. Biol. Chem. 1962, 237, 1375–1376. [Google Scholar] [CrossRef]
- White, R.E.; Coon, M.J. Oxygen activation by cytochrome P-450. Annu. Rev. Biochem. 1980, 49, 315–356. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, R. Cytochromes P450 as versatile biocatalysts. J. Biotechnol. 2006, 124, 128–145. [Google Scholar] [CrossRef] [PubMed]
- Sono, M.; Roach, M.P.; Coulter, E.D.; Dawson, J.H. Heme-containing oxygenases. Chem. Rev. 1996, 96, 2841–2888. [Google Scholar] [CrossRef]
- Yan, Y.; Wu, J.; Hu, G.; Gao, C.; Guo, L.; Chen, X.; Liu, L.; Song, W. Current state and future perspectives of cytochrome P450 enzymes for C–H and C = C oxygenation. Synth. Syst. Biotechnol. 2022, 7, 887–899. [Google Scholar] [CrossRef]
- Kelly, S.L.; Kelly, D.E. Microbial cytochromes P450: Biodiversity and biotechnology. Where do cytochromes P450 come from, what do they do and what can they do for us? Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2013, 368, 20120476. [Google Scholar] [CrossRef]
- Girvan, H.M.; Munro, A.W. Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology. Curr. Opin. Chem. Biol. 2016, 31, 136–145. [Google Scholar] [CrossRef]
- Lepesheva, G.I.; Friggeri, L.; Waterman, M.R. CYP51 as drug targets for fungi and protozoan parasites: Past, present and future. Parasitology 2018, 145, 1820–1836. [Google Scholar] [CrossRef] [PubMed]
- Urlacher, V.B.; Girhard, M. Cytochrome P450 monooxygenases in biotechnology and synthetic biology. Trends Biotechnol. 2019, 37, 882–897. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, Y.; Guengerich, F.P.; Ma, L.; Li, S.; Zhang, W. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J. Biol. Chem. 2020, 295, 833–849. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 nomenclature. Methods Mol. Biol. 1998, 107, 15–24. [Google Scholar]
- Nelson, D.R. Cytochrome P450 nomenclature, 2004. Methods Mol. Biol. 2006, 320, 1–10. [Google Scholar]
- Nelson, D.R. The cytochrome p450 homepage. Hum. Genom. 2009, 4, 59–65. [Google Scholar] [CrossRef]
- Nelson, D.R.; Kamataki, T.; Waxman, D.J.; Guengerich, F.P.; Estabrook, R.W.; Feyereisen, R.; Gonzalez, F.J.; Coon, M.J.; Gunsalus, I.C.; Gotoh, O.; et al. The P450 superfamily: Update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993, 12, 1–51. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef]
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef]
- Lamb, D.C.; Lei, L.; Warrilow, A.G.; Lepesheva, G.I.; Mullins, J.G.; Waterman, M.R.; Kelly, S.L. The first virally encoded cytochrome p450. J. Virol. 2009, 83, 8266–8269. [Google Scholar] [CrossRef] [PubMed]
- Lamb, D.C.; Follmer, A.H.; Goldstone, J.V.; Nelson, D.R.; Warrilow, A.G.; Price, C.L.; True, M.Y.; Kelly, S.L.; Poulos, T.L.; Stegeman, J.J. On the occurrence of cytochrome P450 in viruses. Proc. Natl. Acad. Sci. USA 2019, 116, 12343–12352. [Google Scholar] [CrossRef] [PubMed]
- Lamb, D.C.; Hargrove, T.Y.; Zhao, B.; Wawrzak, Z.; Goldstone, J.V.; Nes, W.D.; Kelly, S.L.; Waterman, M.R.; Stegeman, J.J.; Lepesheva, G.I. Concerning P450 evolution: Structural analyses support bacterial origin of sterol 14α-demethylases. Mol. Biol. Evol. 2021, 38, 952–967. [Google Scholar] [CrossRef] [PubMed]
- McLean, M.A.; Maves, S.A.; Weiss, K.E.; Krepich, S.; Sligar, S.G. Characterization of a cytochrome P450 from the acidothermophilic Archaea Sulfolobus solfataricus. Biochem. Biophys. Res. Commun. 1998, 252, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Yano, J.K.; Koo, L.S.; Schuller, D.J.; Li, H.; de Montellano, P.R.O.; Poulos, T.L. Crystal structure of a thermophilic cytochrome P450 from the archaeon Sulfolobus solfataricus. J. Biol. Chem. 2000, 275, 31086–31092. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Yamane, K.; Adachi, S.-i.; Shiro, Y.; Weiss, K.E.; Maves, S.A.; Sligar, S.G. Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: High resolution structure and functional properties. J. Inorg. Biochem. 2002, 91, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Rabe, K.S.; Kiko, K.; Niemeyer, C.M. Characterization of the peroxidase activity of CYP119, a thermostable P450 from Sulfolobus acidocaldarius. Chem. Bio. Chem. 2008, 9, 420–425. [Google Scholar] [CrossRef]
- Basudhar, D.; Madrona, Y.; Kandel, S.; Lampe, J.N.; Nishida, C.R.; de Montellano, P.R.O. Analysis of cytochrome P450 CYP119 ligand-dependent conformational dynamics by two-dimensional NMR and X-ray crystallography. J. Biol. Chem. 2015, 290, 10000–10017. [Google Scholar] [CrossRef]
- Sakalli, T.; Surmeli, N.B. Functional characterization of a novel CYP119 variant to explore its biocatalytic potential. Biotechnol. Appl. Biochem. 2021, 69, 1741–1756. [Google Scholar] [CrossRef]
- Hannemann, F.; Bichet, A.; Ewen, K.M.; Bernhardt, R. Cytochrome P450 systems--biological variations of electron transport chains. Biochim. Et Biophys. Acta 2007, 1770, 330–344. [Google Scholar] [CrossRef]
- Puchkaev, A.V.; Wakagi, T.; Ortiz de Montellano, P.R. CYP119 Plus a Sulfolobus tokodaii Strain 7 ferredoxin and 2-oxoacid: Ferredoxin oxidoreductase constitute a high-temperature cytochrome P450 catalytic system. J. Am. Chem. Soc. 2002, 124, 12682–12683. [Google Scholar] [CrossRef]
- Zhang, Q.; Iwasaki, T.; Wakagi, T.; Oshima, T. 2-Oxoacid: Ferredoxin oxidoreductase from the thermoacidophilic archaeon, Sulfolobus sp. strain 7. J. Biochem. 1996, 120, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Venkateswara Rao, P.; Holm, R. Synthetic analogues of the active sites of iron−sulfur proteins. Chem. Rev. 2004, 104, 527–560. [Google Scholar] [CrossRef]
- Cammack, R. Evolution and diversity in the iron-sulphur proteins. Chem. Scr. 1983, 21, 87–95. [Google Scholar]
- Nzuza, N.; Padayachee, T.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. Diversification of ferredoxins across living organisms. Curr. Issues Mol. Biol. 2021, 43, 1374–1390. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.; Varghese, N.; Seshadri, R. The IMG/M data management and analysis system v. 6.0: New tools and advanced capabilities. Nucleic Acids Res. 2021, 49, D751–D763. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef]
- Baker, B.J.; De Anda, V.; Seitz, K.W.; Dombrowski, N.; Santoro, A.E.; Lloyd, K.G. Diversity, ecology and evolution of Archaea. Nat. Microbiol. 2020, 5, 887–900. [Google Scholar] [CrossRef]
- Rothman, D.H.; Fournier, G.P.; French, K.L.; Alm, E.J.; Boyle, E.A.; Cao, C.; Summons, R.E. Methanogenic burst in the end-Permian carbon cycle. Proc. Natl. Acad. Sci. USA 2014, 111, 5462–5467. [Google Scholar] [CrossRef]
- Nkosi, B.V.Z.; Padayachee, T.; Gront, D.; Nelson, D.R.; Syed, K. Contrasting health effects of Bacteroidetes and Firmicutes lies in their genomes: Analysis of P450s, ferredoxins, and secondary metabolite clusters. Int. J. Mol. Sci. 2022, 23, 5057. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Lepesheva, G.I.; Waterman, M.R. Sterol 14alpha-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim. Biophys. Acta 2007, 1770, 467–477. [Google Scholar] [CrossRef]
- Kelly, S.L.; Lamb, D.C.; Baldwin, B.C.; Corran, A.J.; Kelly, D.E. Characterization of Saccharomyces cerevisiae CYP61, sterol delta22-desaturase, and inhibition by azole antifungal agents. J. Biol. Chem. 1997, 272, 9986–9988. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, T.; Mizutani, M.; Aoki, N.; Watanabe, B.; Saga, H.; Saito, S.; Oikawa, A.; Suzuki, H.; Sakurai, N.; Shibata, D. Cytochrome P450 CYP710A encodes the sterol C-22 desaturase in Arabidopsis and tomato. Plant Cell. 2006, 18, 1008–1022. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 and the individuality of species. Arch. Biochem. Biophys 1999, 369, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Lee, M.-K.; Jefcoate, C.; Kim, S.-C.; Chen, F.; Yu, J.-H. Fungal cytochrome p450 monooxygenases: Their distribution, structure, functions, family expansion, and evolutionary origin. Genome. Biol. Evol. 2014, 6, 1620–1634. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.-R.; Eun, C.-Y.; Park, H.-G.; Han, S.-H.; Han, J.-S.; Cho, K.-S.; Chun, Y.-J.; Kim, D.-H. Regioselective oxidation of lauric acid by CYP119, an orphan cytochrome P450 from Sulfolobus acidocaldarius. J. Microbiol. Biotechnol. 2010, 20, 574–578. [Google Scholar]
- Koo, L.S.; Immoos, C.E.; Cohen, M.S.; Farmer, P.J.; Ortiz de Montellano, P.R. Enhanced electron transfer and lauric acid hydroxylation by site-directed mutagenesis of CYP119. J. Am. Chem. Soc. 2002, 124, 5684–5691. [Google Scholar] [CrossRef] [PubMed]
- Blair, E.; Greaves, J.; Farmer, P.J. High-temperature electrocatalysis using thermophilic P450 CYP119: Dehalogenation of CCl4 to CH4. J. Am. Chem. Soc. 2004, 126, 8632–8633. [Google Scholar] [CrossRef]
- Immoos, C.E.; Chou, J.; Bayachou, M.; Blair, E.; Greaves, J.; Farmer, P.J. Electrocatalytic reductions of nitrite, nitric oxide, and nitrous oxide by thermophilic cytochrome P450 CYP119 in film-modified electrodes and an analytical comparison of its catalytic activities with myoglobin. J. Am. Chem. Soc. 2004, 126, 4934–4942. [Google Scholar] [CrossRef]
- Başlar, M.S.; Sakallı, T.; Güralp, G.; Kestevur Doğru, E.; Haklı, E.; Surmeli, N.B. Development of an improved Amplex Red peroxidation activity assay for screening cytochrome P450 variants and identification of a novel mutant of the thermophilic CYP119. JBIC J. Biol. Inorg. Chem. 2020, 25, 949–962. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Hirakawa, H.; Nagamune, T. Electron donation to an archaeal cytochrome P450 is enhanced by PCNA-mediated selective complex formation with foreign redox proteins. Biotechnol. J. 2014, 9, 1573–1581. [Google Scholar] [CrossRef] [PubMed]
- Khatri, Y.; Hannemann, F.; Girhard, M.; Kappl, R.; Même, A.; Ringle, M.; Janocha, S.; Leize-Wagner, E.; Urlacher, V.B.; Bernhardt, R. Novel family members of CYP109 from Sorangium cellulosum So ce56 exhibit characteristic biochemical and biophysical properties. Biotechnol. Appl. Biochem. 2013, 60, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Girhard, M.; Klaus, T.; Khatri, Y.; Bernhardt, R.; Urlacher, V.B. Characterization of the versatile monooxygenase CYP109B1 from Bacillus subtilis. Appl. Microbiol. Biotechnol. 2010, 87, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Furuya, T.; Nishi, T.; Shibata, D.; Suzuki, H.; Ohta, D.; Kino, K. Characterization of orphan monooxygenases by rapid substrate screening using FT-ICR mass spectrometry. Chem. Biol. 2008, 15, 563–572. [Google Scholar] [CrossRef]
- Khatri, Y.; Girhard, M.; Romankiewicz, A.; Ringle, M.; Hannemann, F.; Urlacher, V.B.; Hutter, M.C.; Bernhardt, R. Regioselective hydroxylation of norisoprenoids by CYP109D1 from Sorangium cellulosum So ce56. Appl. Microbiol. Biotechnol. 2010, 88, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Putkaradze, N.; König, L.; Kattner, L.; Hutter, M.C.; Bernhardt, R. Highly regio-and stereoselective hydroxylation of vitamin D2 by CYP109E1. Biochem. Biophys. Res. Commun. 2020, 524, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Putkaradze, N.; Litzenburger, M.; Hutter, M.C.; Bernhardt, R. CYP109E1 from Bacillus megaterium acts as a 24-and 25-hydroxylase for cholesterol. Chem. Bio. Chem. 2019, 20, 655–658. [Google Scholar] [CrossRef] [PubMed]
- Child, S.A.; Rossi, V.P.; Bell, S.G. Selective ϖ-1 oxidation of fatty acids by CYP147G1 from Mycobacterium marinum. Biochim. Et. Biophys. Acta. Gen. Subj. 2019, 1863, 408–417. [Google Scholar] [CrossRef]
- Bhattarai, S.; Liou, K.; Oh, T.-J. Hydroxylation of long chain fatty acids by CYP147F1, a new cytochrome P450 subfamily protein from Streptomyces peucetius. Arch. Biochem. Biophys. 2013, 539, 63–69. [Google Scholar] [CrossRef]
- Padayachee, T.; Nzuza, N.; Chen, W.; Nelson, D.R.; Syed, K. impact of lifestyle on cytochrome P450 monooxygenase repertoire is clearly evident in the bacterial phylum Firmicutes. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Senate, L.M.; Tjatji, M.P.; Pillay, K.; Chen, W.; Zondo, N.M.; Syed, P.R.; Mnguni, F.C.; Chiliza, Z.E.; Bamal, H.D.; Karpoormath, R.; et al. Similarities, variations, and evolution of cytochrome P450s in Streptomyces vs. Mycobacterium. Sci. Rep. 2019, 9, 3962. [Google Scholar]
- Mnguni, F.C.; Padayachee, T.; Chen, W.; Gront, D.; Yu, J.-H.; Nelson, D.R.; Syed, K. More P450s are involved in secondary metabolite biosynthesis in Streptomyces compared to Bacillus, Cyanobacteria and Mycobacterium. Int. J. Mol. Sci. 2020, 21, 4814. [Google Scholar] [CrossRef] [PubMed]
- Zondo, N.M.; Padayachee, T.; Nelson, D.R.; Syed, K. Saprophytic to pathogenic mycobacteria: Loss of cytochrome P450s vis a vis their prominent involvement in natural metabolite biosynthesis. Int. J. Mol. Sci. 2022, 24, 149. [Google Scholar] [CrossRef] [PubMed]
- Malinga, N.A.; Nzuza, N.; Padayachee, T.; Syed, P.R.; Karpoormath, R.; Gront, D.; Nelson, D.R.; Syed, K. An unprecedented number of cytochrome P450s are involved in secondary metabolism in Salinispora Species. Microorganisms 2022, 10, 871. [Google Scholar] [CrossRef] [PubMed]
- Syed, K.; Mashele, S.S. Comparative analysis of P450 signature motifs EXXR and CXG in the large and diverse kingdom of fungi: Identification of evolutionarily conserved amino acid patterns characteristic of P450 family. PLoS ONE 2014, 9, e95616. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 1992, 267, 83–90. [Google Scholar] [CrossRef]
- Msomi, N.N.; Padayachee, T.; Nzuza, N.; Syed, P.R.; Kryś, J.D.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. In silico analysis of P450s and their role in secondary metabolism in the bacterial class Gammaproteobacteria. Molecules 2021, 26, 1538. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021, 49, D10. [Google Scholar] [CrossRef]
- wwPDB consortium. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Operon-mapper: A web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef] [PubMed]
| Species Category | Species Subcategory | Species Subcategory P450 Count | Species Category P450 Count | P450 Families |
|---|---|---|---|---|
| Animals | 37,149 | 1948 * | ||
| Mammals | 4558 | 18 | ||
| Other vertebrates | 3268 | 19 | ||
| Insects | 22,173 | 1031 | ||
| Non-insect invertebrates | 7150 | 880 | ||
| Plants | 42,102 | 819 | ||
| Fungi | 28,260 | 3204 | ||
| Protozoa | 5807 | 1374 | ||
| Bacteria | 17,236 | 1910 | ||
| Archaea | 1204 | 34 | ||
| Viruses | 37 | 13 | ||
| Total | 131,795 | 9302 |
| Species Names | Accession Number | Operon |
|---|---|---|
| Plasmid DNA | ||
| Halorussus sp. RC-68 | NZ_CP035120.1 | Epimerase/dehydratase→Thiamine pyrophosphate-binding protein→CYP109G24 |
| Halorussus sp. YCN54 | NZ_CP099994.1 | Cupin domain-containing→ProteinNAD(P)-dependent oxidoreductase→CYP109G32 |
| Halorussus sp. XZYJT49 | NZ_CP096661.1 | CYP174A39→PIN domain-containing protein→AbrB/MazE/SpoVT family DNA-binding domain-containing protein |
| Haloprofundus salinisoli strain SQT7-1 | NZ_CP083664.1 | Enoyl-CoA hydratase-related protein→CYP109G25 |
| Haloprofundus sp. MHR1 | NZ_MN918443.1 | CYP109G22→Enoyl-CoA hydratase-related protein |
| Haloprofundus halobius strain SEDH52 | NZ_CP083667.1 | MFS transporter→CYP109G26 |
| Saliphagus sp. WLHS1 | NZ_CP100356.1 | CYP1002B4→Putative protein |
| Natrinema sp. DC36 | NZ_CP084474.1 | CYP1014C9→Putative protein |
| Halorubrum lacusprofundi strain HLS1 | NZ_KX906370.1 | CYP109G27→SDR family oxidoreductase→Putative protein |
| Halocatena sp. AD-1 | NZ_CP096021.1 | ABC transporter permease subunit→CYP1014B9 |
| NZ_CP096022.1 | Hydroxypyruvate isomerase→dehydrogenase→CYP109AU2 | |
| Halobellus limi CGMCC 1.10331 | NZ_CP031313.1 | Amidohydrolase family protein→CYP1002B4 |
| Chromosomal DNA | ||
| Halorussus halophilus strain ZS-3 | NZ_CP044523.1 | SAM-dependent methyltransferases→CYP1002C17 |
| Halorussus sp. YCN54 | NZ_CP099993.1 | CYP1002C24→SAM-dependent methyltransferases |
| Halorussus sp. XZYJ18 | NZ_CP100400.1 | SAM-dependent methyltransferases→CYP1002C25 |
| Halorussus sp. XZYJT49 | NZ_CP096659.1 | Dehydrogenase→Flavoprotein→CYP174C3 |
| Haladaptatus sp. PSR5 | NZ_CP085335.1 | CYP174D1→Putative protein |
| Halomicroarcula sp. DT1 | NZ_CP100404.1 | Archaeal kinase→Mevalonate kinase→CYP174A38 |
| Halomicroarcula sp. YSSS71 | NZ_CP100407.1 | Archaeal kinase→Mevalonate kinase→CYP174A38 |
| Halomicrobium salinisoli strain TH30 | NZ_CP084466.1 | Putative protein→Putative protein→CYP174A35→Mevalonate kinase→Archaeal kinase |
| Halomicrobium salinisoli strain LT50 | NZ_CP084463.1 | Archaeal kinase→Mevalonate kinase→CYP174A36→Putative protein→Putative protein |
| Halosiccatus urmianus strain IBRC-M: 10911 | NZ_CP084090.1 | Archaeal kinase→Mevalonate kinase→CYP174A37→Putative protein→Tellurite resistance protein and related permeases→Ribosomal protein S2→Enolase→DNA-directed RNA polymerase, subunit K/omega→DNA-directed RNA polymerase, subunit N (RpoN/RPB10)→Ribosomal protein S9→Ribosomal protein L13→Ribosomal protein L18E→Putative protein |
| Halalkalicoccus jeotgali B3 | NC_014297.1 | Ketopantoate reductase→Zn finger protein HypA/HybF→CYP109G2 |
| Halocatena sp. AD-1 | NZ_CP096019.1 | CYP109F16→Putative protein |
| Halobellus limi CGMCC 1.10331 | NZ_CP031311.1 | CYP1014A7→Phosphoribosylaminoimidazole (AIR) synthetase |
| Halococcus dombrowskii H4 | NZ_CP095005.1 | ATP-dependent 26S proteasome regulatory subunit→CYP109G4 |
| Halosegnis sp. ZY10 | NZ_CP101161.1 | CYP2738A7→Putative protein |
| Natribaculum breve TRM20010 | NZ_CP095390.1 | Phytoene/squalene synthetase→CYP197C46 |
| Natronobiforma sp. CGA73 | NZ_CP101458.1 | Predicted metal-dependent membrane protease→Holliday junction resolvase-archaeal type→Putative protein→CYP174B67 |
| Natronomonas sp. ZY43 | NZ_CP101154.1 | CYP1603A5→Fibrillarin-like rRNA methylase→Protein implicated in ribosomal biogenesis, Nop56p |
| Putative protein→Putative protein→CYP1002C26 | ||
| CYP1014G24→Aspartate/tyrosine/aromatic aminotransferase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngcobo, P.E.; Nkosi, B.V.Z.; Chen, W.; Nelson, D.R.; Syed, K. Evolution of Cytochrome P450 Enzymes and Their Redox Partners in Archaea. Int. J. Mol. Sci. 2023, 24, 4161. https://doi.org/10.3390/ijms24044161
Ngcobo PE, Nkosi BVZ, Chen W, Nelson DR, Syed K. Evolution of Cytochrome P450 Enzymes and Their Redox Partners in Archaea. International Journal of Molecular Sciences. 2023; 24(4):4161. https://doi.org/10.3390/ijms24044161
Chicago/Turabian StyleNgcobo, Phelelani Erick, Bridget Valeria Zinhle Nkosi, Wanping Chen, David R. Nelson, and Khajamohiddin Syed. 2023. "Evolution of Cytochrome P450 Enzymes and Their Redox Partners in Archaea" International Journal of Molecular Sciences 24, no. 4: 4161. https://doi.org/10.3390/ijms24044161
APA StyleNgcobo, P. E., Nkosi, B. V. Z., Chen, W., Nelson, D. R., & Syed, K. (2023). Evolution of Cytochrome P450 Enzymes and Their Redox Partners in Archaea. International Journal of Molecular Sciences, 24(4), 4161. https://doi.org/10.3390/ijms24044161








