Addressing Critical Issues Related to Storage and Stability of the Vault Nanoparticle Expressed and Purified from Komagataella phaffi
Abstract
:1. Introduction
2. Results
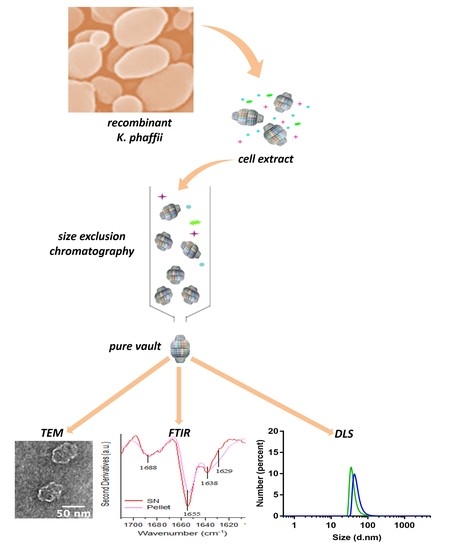
2.1. Production Procedure of Pure Vault Particle from K. phaffii
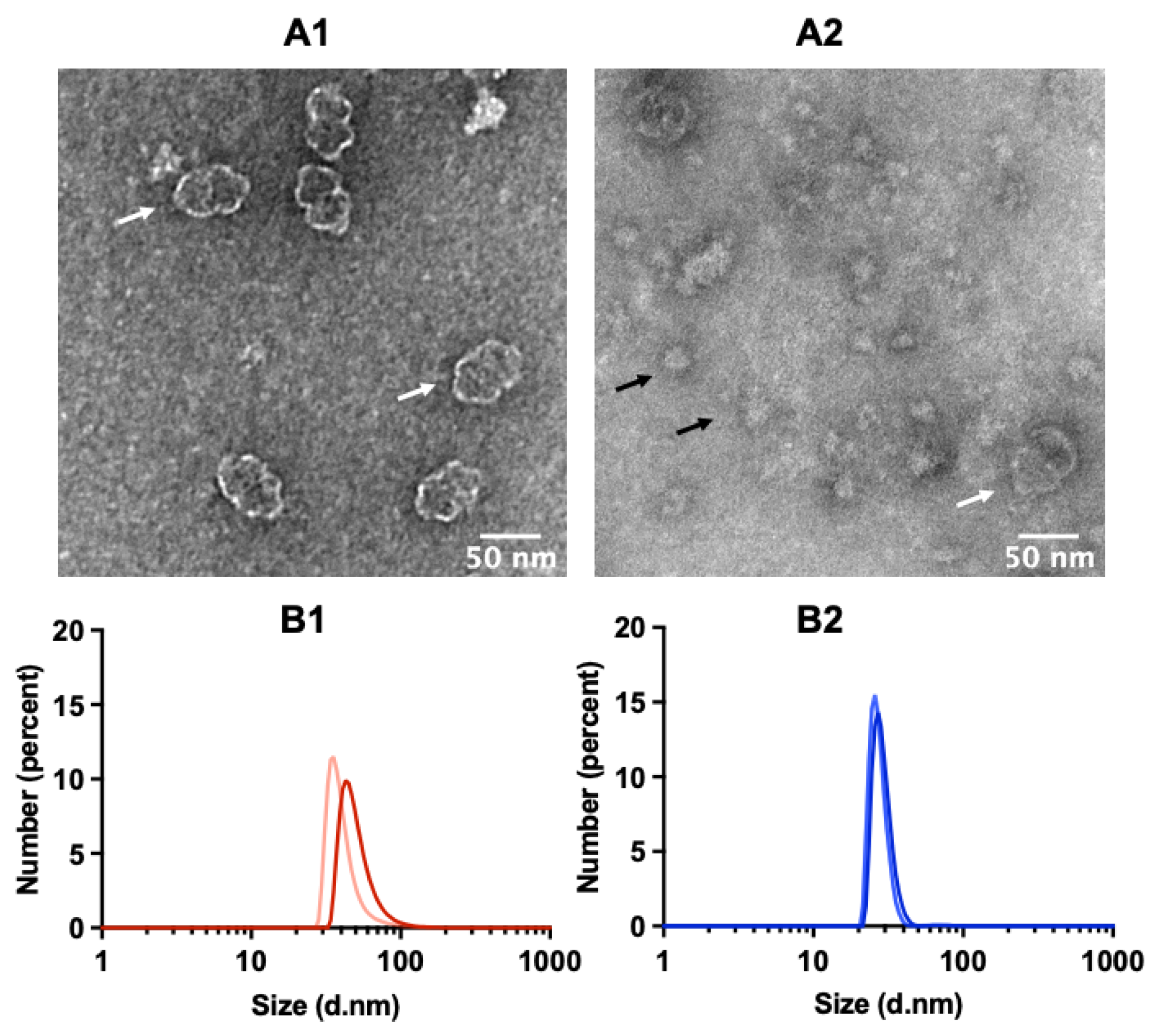
2.2. TEM Reveals Well-Assembled Vault Nanoparticles
2.3. Determining Optimal Conditions for Vault Storage and Stability using FTIR Spectroscopy and DLS
3. Discussion
4. Materials and Methods
4.1. Strain and Growth Media
4.2. MVP Gene Cloning
4.3. Transformation and Selection of Positive Clones
4.4. Cell Growth and Collection, and Cell-Free Extract Production
4.5. Vault Purification
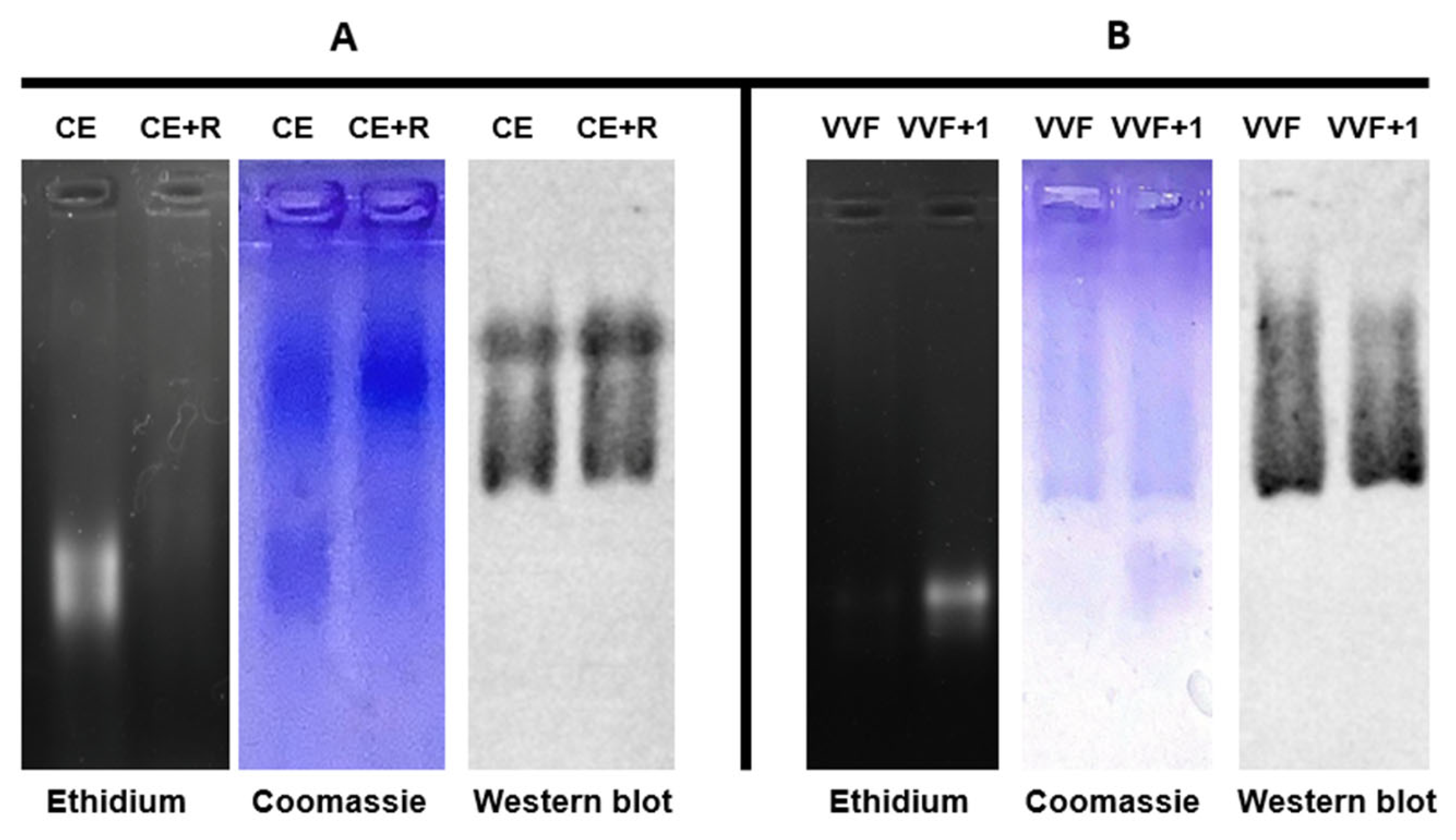
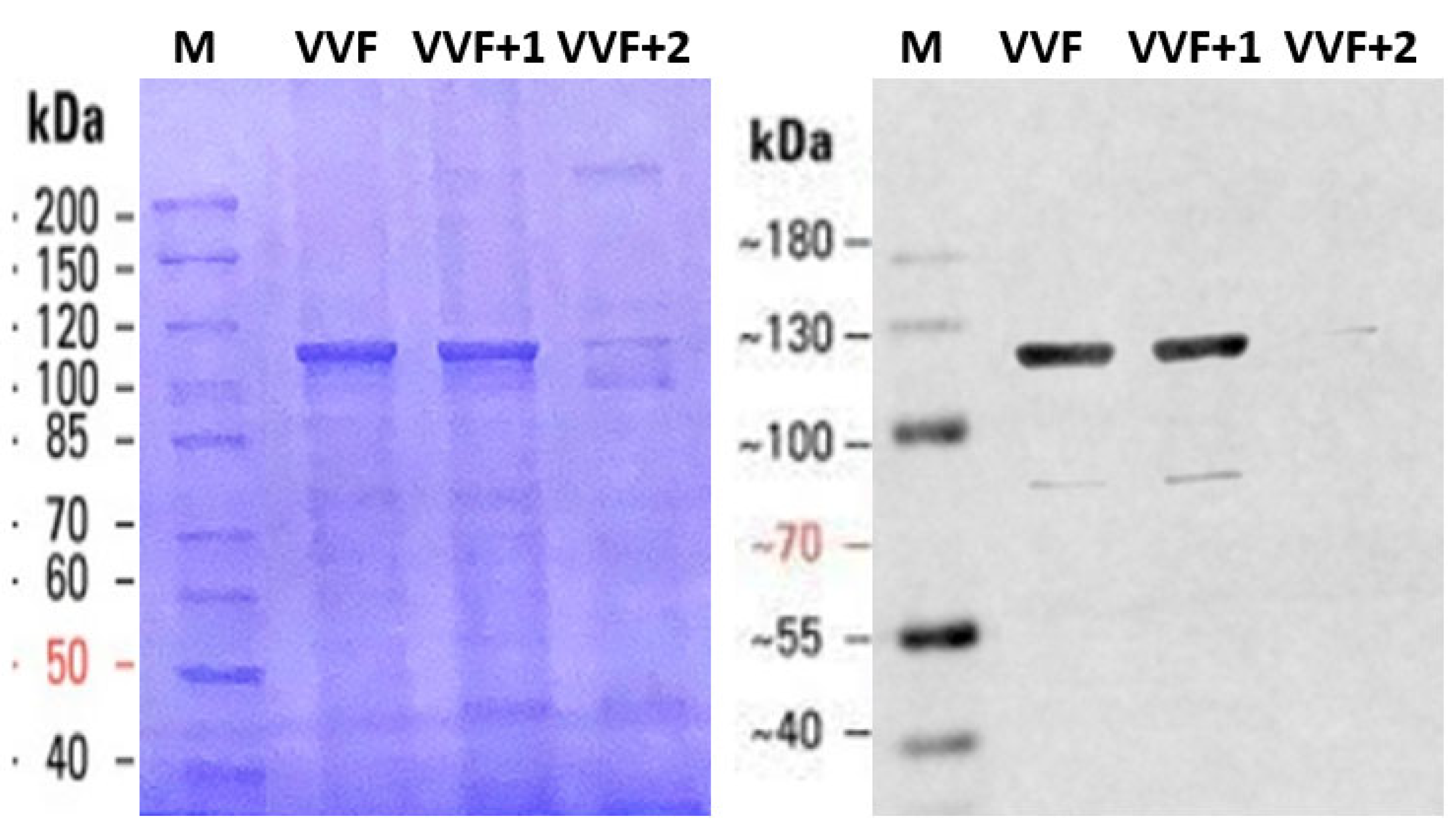
4.6. Electrophoretic Analyses
4.7. TEM
4.8. FTIR Spectroscopy
4.9. DLS
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kedersha, N.L.; Rome, L.H. Isolation and characterization of a novel ribonucleoprotein particle: Large structures contain a single species of small RNA. J. Cell Biol. 1986, 103, 699–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frascotti, G.; Galbiati, E.; Mazzucchelli, M.; Pozzi, M.; Salvioni, L.; Vertemara, J.; Tortora, P. The vault nanoparticle: A gigantic ribonucleoprotein assembly involved in diverse physiological and pathological phenomena and an ideal nanovector for drug delivery and therapy. Cancers 2021, 13, 707. [Google Scholar] [CrossRef]
- Kedersha, N.L.; Heuser, J.E.; Chugani, D.C.; Rome, L.H. Vaults. III. Vault ribonucleoprotein particles open into flower-like structures with octagonal symmetry. J. Cell Biol. 1991, 112, 225–235. [Google Scholar] [CrossRef] [Green Version]
- Kickhoefer, V.A.; Siva, A.C.; Kedersha, N.L.; Inman, E.M.; Ruland, C.; Streuli, M.; Rome, L.H. Vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 1999, 146, 917–928. [Google Scholar] [CrossRef] [Green Version]
- Kickhoefer, V.A.; Stephen, A.G.; Harrington, L.; Robinson, M.O.; Rome, L.H. Vaults and telomerase share a common subunit, TEP1. J. Biol. Chem. 1999, 274, 32712–33271. [Google Scholar] [CrossRef] [Green Version]
- Kickhoefer, V.A.; Searles, R.P.; Kedersha, N.L.; Garber, M.E.; Johnson, D.L.; Rome, L.H. Vault ribonucleoprotein particles from rat and bullfrog contain a related small RNA that is transcribed by RNA polymerase III. J. Biol. Chem. 1993, 268, 7868–7873. [Google Scholar] [CrossRef]
- Kickhoefer, V.A.; Rajavel, K.S.; Scheffer, G.L.; Dalton, W.S.; Scheper, R.J.; Rome, L.H. Vaults are up-regulated in multidrug-resistant cancer cell lines. J. Biol. Chem. 1998, 273, 8971–8974. [Google Scholar] [CrossRef] [Green Version]
- van Zon, A.; Mossink, M.H.; Schoester, M.; Scheffer, G.L.; Scheper, R.J.; Sonneveld, P.; Wiemer, E.A. Multiple human vault RNAs. Expression and association with the vault complex. J. Biol. Chem. 2001, 276, 37715–37721. [Google Scholar] [CrossRef] [Green Version]
- Stephen, A.G.; Raval-Fernandes, S.; Huynh, T.; Torres, M.; Kickhoefer, V.A.; Rome, L.H. Assembly of vault-like particles in insect cells expressing only the major vault protein. J. Biol. Chem. 2001, 276, 23217–23220. [Google Scholar] [CrossRef] [Green Version]
- Mrazek, J.; Toso, D.; Ryazantsev, S.; Zhang, X.; Zhou, Z.H.; Fernandez, B.C.; Kickhoefer, V.A.; Rome, L.H. Polyribosomes are molecular 3D nanoprinters that orchestrate the assembly of vault particles. ACS Nano 2014, 8, 11552–11559. [Google Scholar] [CrossRef] [Green Version]
- Berger, W.; Steiner, E.; Grusch, M.; Elbling, L.; Micksche, M. Vaults and the major vault protein: Novel roles in signal pathway regulation and immunity. Cell. Mol. Life Sci. 2009, 66, 43–61. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.H.; Kickhoefer, V.A.; Slevers, S.A.; Rome, L.H.; Eisenberg, D. Draft crystal structure of the vault shell at 9-Å resolution. PLoS Biol. 2007, 5, e318. [Google Scholar] [CrossRef] [PubMed]
- Ng, B.C.; Yu, M.; Gopal, A.; Rome, L.H.; Monbouquette, H.G.; Tolbert, S.H. Encapsulation of semiconducting polymers in vault protein cages. Nano Lett. 2008, 8, 3503–3509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rome, L.H.; Kickhoefer, V.A. Development of the vault particle as a platform technology. ACS Nano 2013, 7, 889–902. [Google Scholar] [CrossRef]
- Buehler, D.C.; Toso, D.B.; Kickhoefer, V.A.; Zhou, Z.H.; Rome, L.H. Vaults engineered for hydrophobic drug delivery. Small 2011, 7, 1432–1439. [Google Scholar] [CrossRef] [Green Version]
- Benner, N.L.; Zang, X.; Buehler, D.C.; Kickhoefer, V.A.; Rome, M.E.; Rome, L.H.; Wender, P.A. Vault nanoparticles: Chemical modifications for imaging and enhanced delivery. ACS Nano 2017, 11, 872–881. [Google Scholar] [CrossRef] [Green Version]
- Kickhoefer, V.A.; Han, M.; Raval-Fernandes, S.; Poderycki, M.J.; Moniz, R.J.; Vaccari, D.; Silvestry, M.; Stewart, P.L.; Kelly, K.A.; Rome, L.H. Targeting vault nanoparticles to specific cell surface receptors. ACS Nano 2009, 3, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Wiethoff, C.M.; Wodrich, H.; Gerace, L.; Nemerow, G.R. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 2005, 79, 1992–2000. [Google Scholar] [CrossRef] [Green Version]
- Martín, F.; Carreño, A.; Mendoza, R.; Caruana, P.; Rodriguez, F.; Bravo, M.; Benito, A.; Ferrer-Miralles, N.; Céspedes, M.V.; Corchero, J.L. All-in-one biofabrication and loading of recombinant vaults in human cells. Biofabrication 2022, 14, 025018. [Google Scholar] [CrossRef]
- Fernández, R.; Carreño, A.; Mendoza, R.; Benito, A.; Neus Ferrer-Miralles, N.; Céspedes, M.V.; Corchero, J.L. Escherichia coli as a new platform for the fast production of vault-like nanoparticles: An optimized protocol. Int. J. Mol. Sci. 2022, 23, 15543. [Google Scholar] [CrossRef]
- Wang, M.; Kickhoefer, V.A.; Rome, L.H.; Foellmer, O.K.; Mahendra, S. Synthesis and assembly of human vault particles in yeast. Biotechnol. Bioeng. 2018, 115, 2941–2950. [Google Scholar] [CrossRef] [PubMed]
- Galbiati, E.; Avvakumova, S.; La Rocca, A.; Pozzi, M.; Messali, S.; Magnaghi, P.; Colombo, M.; Prosperi, D.; Tortora, P. A fast and straightforward procedure for vault nanoparticle purification and the characterization of its endocytic uptake. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2254–2260. [Google Scholar] [CrossRef] [PubMed]
- Aarum, J.; Cabrera, C.P.; Jones, T.A.; Rajendran, S.; Adiutori, R.; Giovannoni, G.; Barnes, M.R.; Malaspina, A.; Sheer, D. Enzymatic degradation of RNA causes widespread protein aggregation in cell and tissue lysates. EMBO Rep. 2020, 21, e49585. [Google Scholar] [CrossRef] [PubMed]
- Nobbmann, U.; Morfesis, A. Light scattering and nanoparticles. Mater. Today 2009, 12, 52–54. [Google Scholar] [CrossRef]
- Kerwin, B.A.; Heller, M.C.; Levin, S.H.; Randolph, T.W. Effects of Tween 80 and sucrose on acute short-term stability and long-term storage at -20 degrees C of a recombinant hemoglobin. J. Pharm. Sci. 1998, 87, 1062–1068. [Google Scholar] [CrossRef]
- Chou, D.K.; Krishnamurthy, R.; Randolph, T.W.; Carpenter, J.F.; Manning, M.C. Effects of Tween 20 and Tween 80 on the stability of Albutropin during agitation. J. Pharm. Sci. 2005, 94, 1368–1381. [Google Scholar] [CrossRef]
- Ami, D.; Natalello, A. Characterization of the conformational properties of soluble and insoluble proteins by Fourier transform infrared spectroscopy. Methods Mol. Biol. 2022, 2406, 439–454. [Google Scholar]
- Ami, D.; Mereghetti, P.; Natalello, A. Contribution of infrared spectroscopy to the understanding of amyloid protein aggregation in complex systems. Front. Mol. Biosci. 2022, 9, 822852. [Google Scholar] [CrossRef]
- Ding, K.; Zhang, X.; Mrazek, J.; Kickhoefer, V.A.; Lai, M.; Ng, H.L.; Yang, O.O.; Rome, L.H.; Zhou, Z.H. Solution structures of engineered vault particles. Structure 2018, 26, 619–626. [Google Scholar] [CrossRef]
- Goormaghtigh, E.; Raussens, V.; Ruysschaert, J.M. Attenuated total reflection infrared spectroscopy of proteins and lipids in biological membranes. Biochim. Biophys. Acta 1999, 1422, 105–185. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- van Zon, A.; Mossink, M.H.; Schoester, M.; Houtsmuller, A.B.; Scheffer, G.L.; Scheper, R.J.; Sonneveld, P.; Wiemer, E.A. The formation of vault-tubes: A dynamic interaction between vaults and vault PARP. J. Cell Sci. 2003, 116, 4391–4400. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Kickhoefer, V.A.; Ng, B.C.; Gopal, A.; Bentolila, L.A.; John, S.; Tolbert, S.H.; Rome, L.H. Vaults are dynamically unconstrained cytoplasmic nanoparticles capable of half vault exchange. ACS Nano 2010, 4, 7229–7240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerra, P.; González-Alamos, M.; Llauró, A.; Casañas, A.; Querol-Audí, J.; de Pablo, P.J.; Verdaguer, N. Symmetry disruption commits vault particles to disassembly. Sci. Adv. 2022, 8, eabj7795. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, N.M.; Buchman, G.W.; Rome, L.H.; Maynard, H.D. Dual pH- and temperature-responsive protein nanoparticles. Eur. Polym. J. 2015, 69, 532–539. [Google Scholar] [CrossRef] [Green Version]
- Aivaliotis, M.; Samolis, P.; Neofotistou, E.; Remigy, H.; Rizos, A.K.; Tsiotis, G. Molecular size determination of a membrane protein in surfactants by light scattering. Biochim. Biophys. Acta. 2003, 1615, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Juan, A.; Carreño, A.; Mendoza, R.; Corchero, I.L. Latest advances in the development of eukaryotic vaults as targeted drug delivery systems. Pharmaceutics 2019, 11, 300. [Google Scholar] [CrossRef] [Green Version]
- Linke, D. Detergents: An overview. Methods Enzymol. 2009, 463, 603–661. [Google Scholar]
- Gleeson, M.A.G.; White, C.E.; Meininger, D.P.; Komives, E.A. Generation of protease-deficient strains and their use in heterologous protein expression. In Pichia Protocols; Higgins, D.R., Cregg, J.M., Eds.; Humana Press: Totowa, NJ, USA, 1998; Volume 103, pp. 81–94. [Google Scholar]
- Weidner, M.; Taupp, M.; Hallam, S.J. Expression of recombinant proteins in the methylotrophic yeast Pichia pastoris. JoVE 2010, 36, e1862. [Google Scholar]
- Wu, D.; Chu, J.; Hao, Y.Y.; Wang, Y.H.; Zhuang, Y.P.; Zhang, S.L. Incomplete protein disulphide bond conformation and decreased protein expression result from high cell growth during heterologous protein expression in Pichia pastoris. J. Biotechnol. 2012, 157, 107–112. [Google Scholar] [CrossRef]
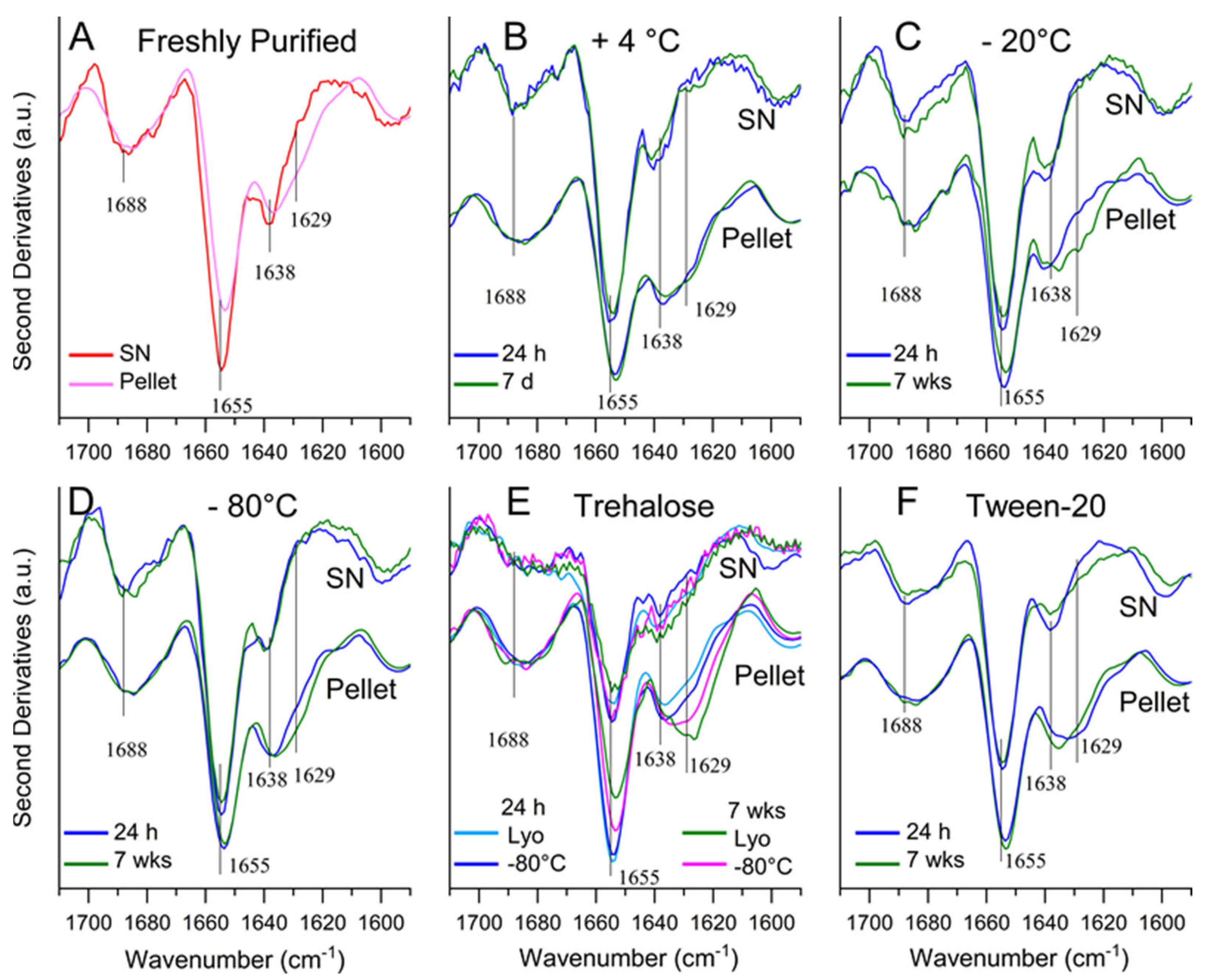
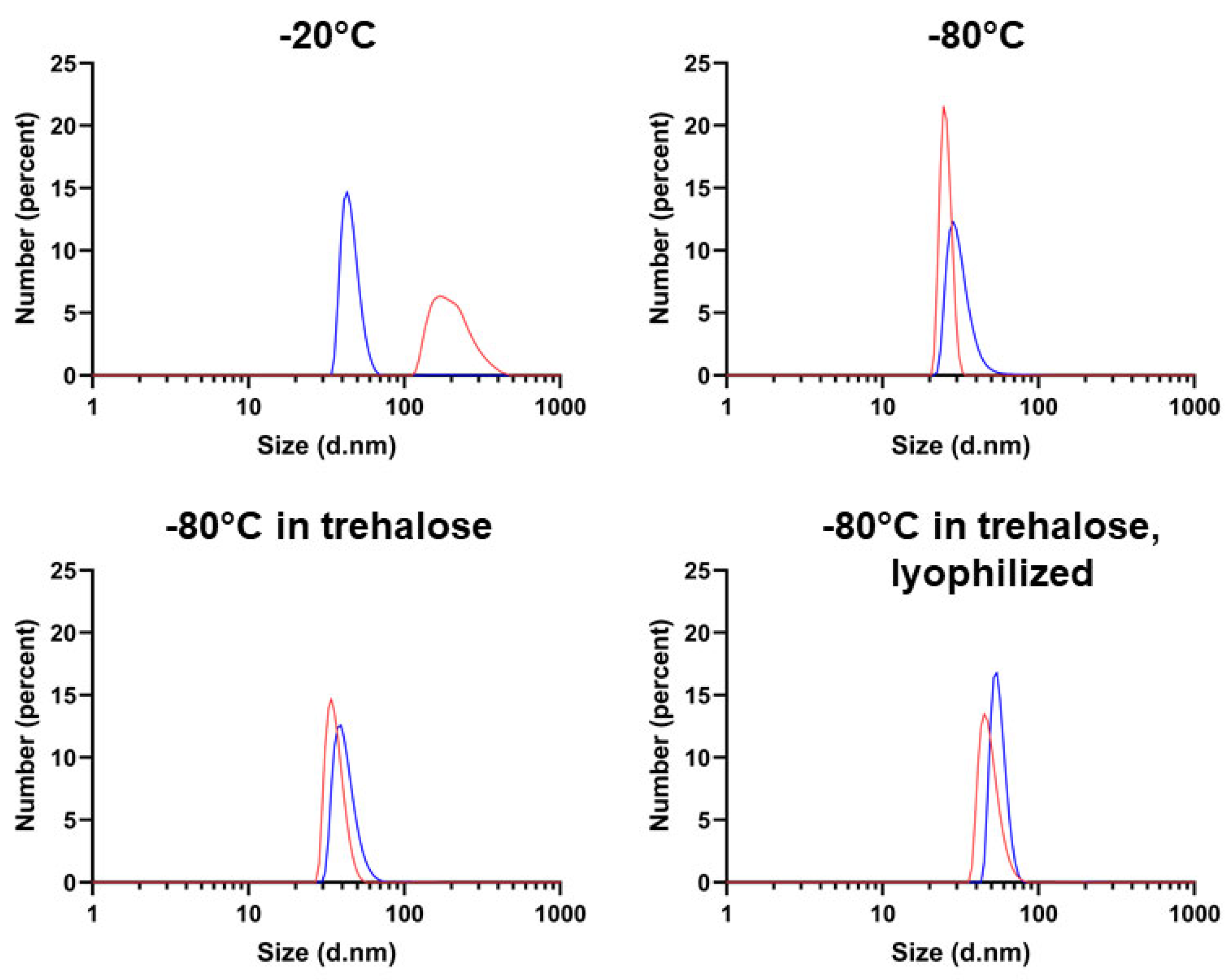
| Storage Conditions 1 | 1 Day | 1 Week | 7 Weeks |
|---|---|---|---|
| +4 °C | 51 ± 4 | 35 ± 2 | |
| −20 °C | 44 ± 2 | 31 ± 7 | |
| −80 °C | 52 ± 5 | 43 ± 6 | |
| −80 °C in 10 mg/mL trehalose | 56 ± 6 | 50 ± 3 | |
| −80 °C in 10 mg/mL trehalose, lyophilized and reconstituted | 50 ± 8 | 45 ± 5 | |
| −80 °C in 0.05% Tween 20 | 64 ± 4 | 59 ± 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomaino, G.; Pantaleoni, C.; Ami, D.; Pellecchia, F.; Dutriaux, A.; Barbieri, L.; Garbujo, S.; Natalello, A.; Tortora, P.; Frascotti, G. Addressing Critical Issues Related to Storage and Stability of the Vault Nanoparticle Expressed and Purified from Komagataella phaffi. Int. J. Mol. Sci. 2023, 24, 4214. https://doi.org/10.3390/ijms24044214
Tomaino G, Pantaleoni C, Ami D, Pellecchia F, Dutriaux A, Barbieri L, Garbujo S, Natalello A, Tortora P, Frascotti G. Addressing Critical Issues Related to Storage and Stability of the Vault Nanoparticle Expressed and Purified from Komagataella phaffi. International Journal of Molecular Sciences. 2023; 24(4):4214. https://doi.org/10.3390/ijms24044214
Chicago/Turabian StyleTomaino, Giulia, Camilla Pantaleoni, Diletta Ami, Filomena Pellecchia, Annie Dutriaux, Linda Barbieri, Stefania Garbujo, Antonino Natalello, Paolo Tortora, and Gianni Frascotti. 2023. "Addressing Critical Issues Related to Storage and Stability of the Vault Nanoparticle Expressed and Purified from Komagataella phaffi" International Journal of Molecular Sciences 24, no. 4: 4214. https://doi.org/10.3390/ijms24044214
APA StyleTomaino, G., Pantaleoni, C., Ami, D., Pellecchia, F., Dutriaux, A., Barbieri, L., Garbujo, S., Natalello, A., Tortora, P., & Frascotti, G. (2023). Addressing Critical Issues Related to Storage and Stability of the Vault Nanoparticle Expressed and Purified from Komagataella phaffi. International Journal of Molecular Sciences, 24(4), 4214. https://doi.org/10.3390/ijms24044214