Fatty Acid Sensing in the Gastrointestinal Tract of Rainbow Trout: Different to Mammalian Model?
Abstract
:1. Introduction
2. Results
2.1. Fatty Acid Receptors and Transporters mRNAs Are Differentially Expressed along the Rainbow Trout Gastrointestinal Tract
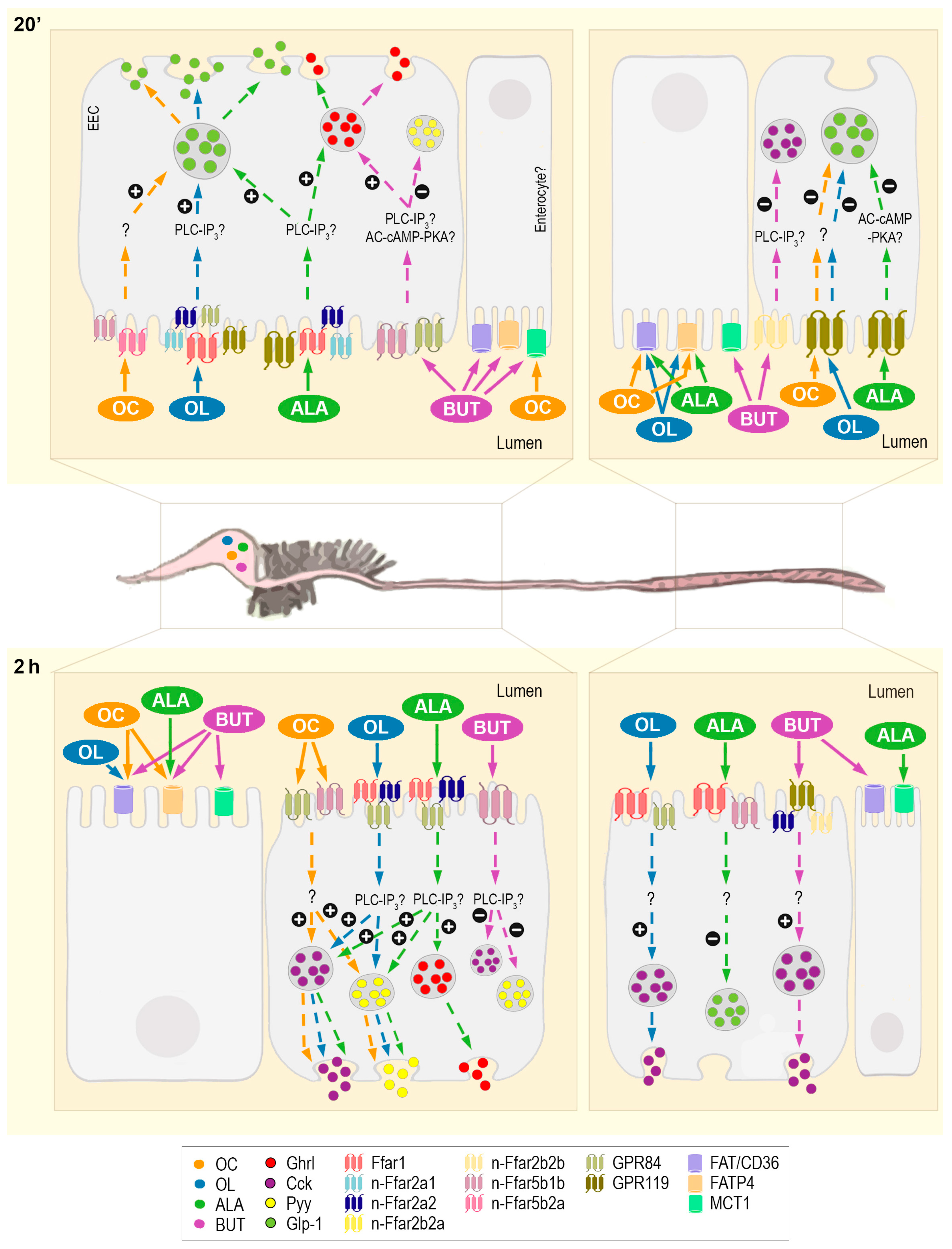
2.2. Luminal Fatty Acids Modulate Fatty Acid Receptors and Transporters mRNA Expression in the Gastrointestinal Tract
2.3. Gastrointestinal mRNA Expression of Intracellular Signaling Molecules Is Altered by the Luminal Presence of Fatty Acids
2.4. Abundance of Gastrointestinal Hormones Responds to Luminal Fatty Acids
3. Discussion
3.1. Fatty Acid Transporters and Carriers in Rainbow Trout GIT and Their Involvement in Sensing Different Types of FAs
3.2. Intracellular Mechanisms Triggered and Hormone Release as a Consequence of Gastrointestinal Fatty Acid Receptor Activation
4. Materials and Methods
4.1. Animals
4.2. Expression and Distribution of Fatty Acid Receptors and Transporters mRNAs along the Rainbow Trout Gastrointestinal Tract
4.3. Characterization of the Response of Gastrointestinal Fatty Acid Sensing Mechanisms to the Luminal Presence of Fatty Acids
4.4. Assessment of Plasma Metabolite Levels
4.5. Quantification of mRNA Abundance by Reverse Transcription—Quantitative Polymerase Chain Reaction (RT-qPCR)
4.6. Analysis of Protein Levels by Western Blot
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moura-Assis, A.; Friedman, J.M.; Velloso, L.A. Gut-to-brain signals in feeding control. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E326–E332. [Google Scholar] [CrossRef]
- Li, Y.; Kokrashvili, Z.; Mosinger, B.; Margolskee, R.F. Gustducin Couples Fatty Acid Receptors to GLP-1 Release in Colon. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E651–E660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, I.; Ichimura, A.; Ohue-Kitano, R.; Igarashi, M. Free Fatty Acid Receptors in Health and Disease. Physiol. Rev. 2020, 100, 171–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauffer, L.M.; Iakoubov, R.; Brubaker, P.L. GPR119 Is Essential for Oleoylethanolamide-Induced Glucagon-like Peptide-1 Secretion from the Intestinal Enteroendocrine L-Cell. Diabetes 2009, 58, 1058–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicol, L.S.C.; Dawes, J.M.; La Russa, F.; Didangelos, A.; Clark, A.K.; Gentry, C.; Grist, J.; Davies, J.B.; Malcangio, M.; McMahon, S.B. The Role of G-Protein Receptor 84 in Experimental Neuropathic Pain. J. Neurosci. 2015, 35, 8959–8969. [Google Scholar] [CrossRef] [Green Version]
- Peters, A.; Rabe, P.; Krumbholz, P.; Kalwa, H.; Kraft, R.; Schöneberg, T.; Stäubert, C. Natural Biased Signaling of Hydroxycarboxylic Acid Receptor 3 and G Protein-Coupled Receptor 84. Cell Commun. Signal. 2020, 18, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, S.; Depoortere, I. Nutrient Sensing in the Gut: New Roads to Therapeutics? Trends Endocrinol. Metab. 2013, 24, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Raka, F.; Farr, S.; Kelly, J.; Stoianov, A.; Adeli, K. Metabolic Control via Nutrient-Sensing Mechanisms: Role of Taste Receptors and the Gut-Brain Neuroendocrine Axis. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E559–E572. [Google Scholar] [CrossRef]
- Rasoamanana, R.; Darcel, N.; Fromentin, G.; Tomé, D. Nutrient Sensing and Signalling by the Gut. Proc. Nutr. Soc. 2012, 71, 446–455. [Google Scholar] [CrossRef] [Green Version]
- Tacon, A.G.J. The Essential Nutrients. In The Nutrition and Feeding of Farmed Fish and Shrimp—A Training Manual; FAO: Brasilia, Brazil, 1987; p. GCP/RLA/075/ITA. [Google Scholar]
- Bell, J.; Koppe, W. Lipids in Aquafeeds. In Fish Oil Replacement and Alternative Lipid Sources in Aquaculture Feeds; Turchini, G., Ng, W.-K., Tocher, D., Eds.; CRC Press: Boca Raton, FL, USA, 2010; pp. 21–59. ISBN 978-1-4398-0862-7. [Google Scholar]
- Kuah, M.-K.; Jaya-Ram, A.; Shu-Chien, A.C. The Capacity for Long-Chain Polyunsaturated Fatty Acid Synthesis in a Carnivorous Vertebrate: Functional Characterisation and Nutritional Regulation of a Fads2 Fatty Acyl Desaturase with Δ4 Activity and an Elovl5 Elongase in Striped Snakehead (Channa Striata). Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2015, 1851, 248–260. [Google Scholar] [CrossRef]
- Korkus, E.; Dąbrowskib, G.; Szustaka, M.; Czaplicki, S.; Madaj, R.; Chworośc, A.; Koziołkiewicz, M.; Konopka, I.; Gendaszewska-Darmacha, E. Evaluation of the anti-diabetic activity of sea buckthorn pulp oils prepared with different extraction methods in human islet EndoC-betaH1 cells. NFS J. 2022, 27, 54–66. [Google Scholar] [CrossRef]
- Tseng, Y.-C.; Liu, S.-T.; Hu, M.Y.; Chen, R.-D.; Lee, J.-R.; Hwang, P.-P. Brain Functioning under Acute Hypothermic Stress Supported by Dynamic Monocarboxylate Utilization and Transport in Ectothermic Fish. Front. Zool. 2014, 11, 53. [Google Scholar] [CrossRef] [Green Version]
- Hao, T.; Li, J.; Liu, Q.; Cui, K.; Chen, Q.; Xu, D.; Liu, Y.; Zhou, Y.; Mai, K.; Ai, Q. Fatty Acid Translocase (FAT/CD36) in Large Yellow Croaker (Larimichthys Crocea): Molecular Cloning, Characterization and the Response to Dietary Fatty Acids. Aquaculture 2020, 528, 735557. [Google Scholar] [CrossRef]
- Yu, H.-X.; Li, Y.; Ezeorba, T.; Mo, H.-L.; Zhang, Z.-H.; Yang, Q.-Y.; Wang, L.-X. Molecular Characterization and Functional Exploration of GPR84 in Chinese Giant Salamander (Andrias Davidianus). Dev. Comp. Immunol. 2022, 137, 104526. [Google Scholar] [CrossRef]
- Luscombe, V.B.; Lucy, D.; Bataille, C.J.R.; Russell, A.J.; Greaves, D.R. 20 Years an Orphan: Is GPR84 a Plausible Medium-Chain Fatty Acid-Sensing Receptor? DNA Cell Biol. 2020, 39, 1926–1937. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; van de Wetering, M.; Clevers, H. The Intestinal Stem Cell. Genes Dev. 2008, 22, 1856–1864. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.J.; Goldsworthy, S.M.; Barnes, A.A.; Eilert, M.M.; Tcheang, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The Orphan G Protein-Coupled Receptors GPR41 and GPR43 Are Activated by Propionate and Other Short Chain Carboxylic Acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichimura, A.; Hirasawa, A.; Hara, T.; Tsujimoto, G. Free Fatty Acid Receptors Act as Nutrient Sensors to Regulate Energy Homeostasis. Prostaglandins Other Lipid Mediat. 2009, 89, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Omlin, T.; Weber, J.-M. Exhausting Exercise and Tissue-Specific Expression of Monocarboxylate Transporters in Rainbow Trout. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 304, R1036–R1043. [Google Scholar] [CrossRef] [Green Version]
- Iwanaga, T.; Takebe, K.; Kato, I.; Karaki, S.-I.; Kuwahara, A. Cellular Expression of Monocarboxylate Transporters (MCT) in the Digestive Tract of the Mouse, Rat, and Humans, with Special Reference to Slc5a8. Biomed. Res. 2006, 27, 243–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirat, D.; Masuoka, J.; Hayashi, H.; Iwano, H.; Yokota, H.; Taniyama, H.; Kato, S. Monocarboxylate Transporter 1 (MCT1) Plays a Direct Role in Short-Chain Fatty Acids Absorption in Caprine Rumen. J. Physiol. 2006, 576, 635–647. [Google Scholar] [CrossRef]
- Bergman, E.N. Energy Contributions of Volatile Fatty Acids from the Gastrointestinal Tract in Various Species. Physiol. Rev. 1990, 70, 567–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parshukov, A.N.; Kashinskaya, E.N.; Simonov, E.P.; Hlunov, O.V.; Izvekova, G.I.; Andree, K.B.; Solovyev, M.M. Variations of the Intestinal Gut Microbiota of Farmed Rainbow Trout, Oncorhynchus Mykiss (Walbaum), Depending on the Infection Status of the Fish. J. Appl. Microbiol. 2019, 127, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Sivaprakasam, S.; Bhutia, Y.D.; Yang, S.; Ganapathy, V. Short-Chain Fatty Acid Transporters: Role in Colonic Homeostasis. Compr. Physiol. 2017, 8, 299–314. [Google Scholar] [CrossRef]
- Calo, J.; Blanco, A.M.; Comesaña, S.; Conde-Sieira, M.; Morais, S.; Soengas, J.L. First Evidence for the Presence of Amino Acid Sensing Mechanisms in the Fish Gastrointestinal Tract. Sci. Rep. 2021, 11, 4933. [Google Scholar] [CrossRef]
- Ohmoto, M.; Okada, S.; Nakamura, S.; Abe, K.; Matsumoto, I. Mutually Exclusive Expression of Gαia and Gα14 Reveals Diversification of Taste Receptor Cells in Zebrafish. J. Comp. Neurol. 2011, 519, 1616–1629. [Google Scholar] [CrossRef] [Green Version]
- Recio, C.; Lucy, D.; Purvis, G.S.D.; Iveson, P.; Zeboudj, L.; Iqbal, A.J.; Lin, D.; O’Callaghan, C.; Davison, L.; Griesbach, E.; et al. Activation of the Immune-Metabolic Receptor GPR84 Enhances Inflammation and Phagocytosis in Macrophages. Front. Immunol. 2018, 9, 1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgoyne, R.D.; Morgan, A. Secretory Granule Exocytosis. Physiological. Rev. 2003, 83, 581–632. [Google Scholar] [CrossRef] [PubMed]
- Coll, A.P.; Farooqi, I.S.; O’Rahilly, S. The Hormonal Control of Food Intake. Cell 2007, 129, 251–262. [Google Scholar] [CrossRef] [Green Version]
- Volkoff, H. The Neuroendocrine Regulation of Food Intake in Fish: A Review of Current Knowledge. Front. Neurosci. 2016, 10, 540. [Google Scholar] [CrossRef] [Green Version]
- Beglinger, S.; Drewe, J.; Schirra, J.; Göke, B.; D’Amato, M.; Beglinger, C. Role of Fat Hydrolysis in Regulating Glucagon-like Peptide-1 Secretion. J. Clin. Endocrinol. Metab. 2010, 95, 879–886. [Google Scholar] [CrossRef] [Green Version]
- Feinle-Bisset, C.; Patterson, M.; Ghatei, M.A.; Bloom, S.R.; Horowitz, M. Fat Digestion Is Required for Suppression of Ghrelin and Stimulation of Peptide YY and Pancreatic Polypeptide Secretion by Intraduodenal Lipid. Am. J. Physiol. Endocrinol. Metab. 2005, 289, E948–E953. [Google Scholar] [CrossRef]
- McLaughlin, J.; Grazia Lucà, M.; Jones, M.N.; D’Amato, M.; Dockray, G.J.; Thompson, D.G. Fatty Acid Chain Length Determines Cholecystokinin Secretion and Effect on Human Gastric Motility. Gastroenterology 1999, 116, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Brooks, L.; Viardot, A.; Tsakmaki, A.; Stolarczyk, E.; Howard, J.K.; Cani, P.D.; Everard, A.; Sleeth, M.L.; Psichas, A.; Anastasovskaj, J.; et al. Fermentable Carbohydrate Stimulates FFAR2-Dependent Colonic PYY Cell Expansion to Increase Satiety. Mol. Metab. 2017, 6, 48–60. [Google Scholar] [CrossRef] [Green Version]
- Samuel, B.S.; Shaito, A.; Motoike, T.; Rey, F.E.; Backhed, F.; Manchester, J.K.; Hammer, R.E.; Williams, S.C.; Crowley, J.; Yanagisawa, M.; et al. Effects of the Gut Microbiota on Host Adiposity Are Modulated by the Short-Chain Fatty-Acid Binding G Protein-Coupled Receptor, Gpr41. Proc. Natl. Acad. Sci. USA 2008, 105, 16767–16772. [Google Scholar] [CrossRef] [Green Version]
- Tolhurst, G.; Heffron, H.; Lam, Y.S.; Parker, H.E.; Habib, A.M.; Diakogiannaki, E.; Cameron, J.; Grosse, J.; Reimann, F.; Gribble, F.M. Short-Chain Fatty Acids Stimulate Glucagon-like Peptide-1 Secretion via the G-Protein-Coupled Receptor FFAR2. Diabetes 2012, 61, 364–371. [Google Scholar] [CrossRef] [Green Version]
- Christiansen, C.B.; Gabe, M.B.N.; Svendsen, B.; Dragsted, L.O.; Rosenkilde, M.M.; Holst, J.J. The Impact of Short-Chain Fatty Acids on GLP-1 and PYY Secretion from the Isolated Perfused Rat Colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 315, G53–G65. [Google Scholar] [CrossRef] [Green Version]
- Blanco, A.M.; Calo, J.; Soengas, J.L. The gut-brain axis in vertebrates: Implications in food intake regulation. J. Exp. Biol. 2021, 224, jeb231571. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Pinilla, F.; Ying, Z. Differential Effects of Exercise and Dietary Docosahexaenoic Acid on Molecular Systems Associated with Control of Allostasis in the Hypothalamus and Hippocampus. Neuroscience 2010, 168, 130–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greco, J.A.; Oosterman, J.E.; Belsham, D.D. Differential Effects of Omega-3 Fatty Acid Docosahexaenoic Acid and Palmitate on the Circadian Transcriptional Profile of Clock Genes in Immortalized Hypothalamic Neurons. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 307, R1049–R1060. [Google Scholar] [CrossRef] [Green Version]
- Ross, R.A.; Rossetti, L.; Lam, T.K.T.; Schwartz, G.J. Differential Effects of Hypothalamic Long-Chain Fatty Acid Infusions on Suppression of Hepatic Glucose Production. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E633–E639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Librán-Pérez, M.; Polakof, S.; López-Patiño, M.A.; Míguez, J.M.; Soengas, J.L. Evidence of a Metabolic Fatty Acid-Sensing System in the Hypothalamus and Brockmann Bodies of Rainbow Trout: Implications in Food Intake Regulation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, R1340–R1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velasco, C.; Bonacic, K.; Soengas, J.L.; Morais, S. Orally Administered Fatty Acids Enhance Anorectic Potential but Do Not Activate Central Fatty Acid Sensing in Senegalese Sole Post-Larvae. J. Exp. Biol. 2017, 220, 677–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Librán-Pérez, M.; Otero-Rodiño, C.; López-Patiño, M.A.; Míguez, J.M.; Soengas, J.L. Central Administration of Oleate or Octanoate Activates Hypothalamic Fatty Acid Sensing and Inhibits Food Intake in Rainbow Trout. Physiol. Behav. 2014, 129, 272–279. [Google Scholar] [CrossRef]
- Librán-Pérez, M.; Velasco, C.; Otero-Rodiño, C.; López-Patiño, M.A.; Míguez, J.M.; Soengas, J.L. Effects of Insulin Treatment on the Response to Oleate and Octanoate of Food Intake and Fatty Acid-Sensing Systems in Rainbow Trout. Domest. Anim. Endocrinol. 2015, 53, 124–135. [Google Scholar] [CrossRef]
- Librán-Pérez, M.; Geurden, I.; Dias, K.; Corraze, G.; Panserat, S.; Soengas, J.L. Feeding Rainbow Trout with a Lipid-Enriched Diet: Effects on Fatty Acid Sensing, Regulation of Food Intake and Cellular Signaling Pathways. J. Exp. Biol. 2015, 218, 2610–2619. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]





| Gene | GenBank Accession Number | Sequence (5′ to 3′) | Reverse Primer (5′ to 3′) | Amplico-n Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|---|---|
| ac | MF670431.1 | CACCAGAAGTGTGCCAGCTA | GAGCAAACTCGGGTGGATCT | 129 | 60 |
| actb | NM 001124235.1 | GATGGGCCAGAAAGACAGCTA | TCGTCCCAGTTGGTGACGAT | 105 | 59 |
| ccka | NM_001124345.1 | GGGTCCCAGCCACAAGATAA | TGGATTTAGTGGTGGTGCGT | 120 | 60 |
| cd36 | AY606034.1 | CAAGTCAGCGACAAACCAGA | ACTTCTGAGCCTCCACAGGA | 106 | 60 |
| ef1a | AF498320 | TCCTCTTGGTCGTTTCGCTG | ACCCGAGGGACATCCTGTG | 159 | 59 |
| fatp4 | XM_014138749.1 | GTAGCCTGGGAAACTTCGACA | TTCTTGCTGTTGGCTCCTTCG | 244 | 60 |
| ffar1 | XM_036951038.1 | CTGTGGTCATGCTGATGCTCT | CTTGGAAATGTTTGCTCCTGTC | 188 | 60 |
| ffar2b1.1 | XM_021571760.2 | CTTCCTCAGCGTGGCGTATC | CAGGTAGTGTTGTCGGCATCT | 153 | 62 |
| ffar2b1.2 | XM_021571759.2 | AGGCTGTTGATGACATGCACT | ATCTGATAGGGAAGGCCACA | 147 | 60 |
| ffar2b2a | XM_021561043.1 | CACCTGAGCATTGTCGTCATC | TAATGAGCACGTTGGAGACGTTG | 115 | 60 |
| ffar2b2b | XM_021595167.2 | ATGCCCTACTACAACCCACC | ACGTCACTAAGAGGCGCAATG | 101 | 60 |
| ffar2a1b | XM_021584265.2 | CCTACCGCCAACTCAGCAAAC | AGTTCTCGTAGCAGACGGAG | 147 | 62 |
| ffar2a2 | XM_021560940.2 | CCCTTGTACGGAGTGGTGAG | CCAGCAGTGGCACGATGTAT | 196 | 60 |
| gcg | NM_001124698.1 | AGGAGTGGTGCTCCATCCAAA | TCCTGATTTGAGCCAGGAAACA | 111 | 59 |
| ghrl | AB096919.1 | GGTCCCCTTCACCAGGAAGAC | GGTGATGCCCATCTCAAAAGG | 63 | 60 |
| gnai1 | CU073912 | GCAAGACGTGCTGAGGACCA | ATGGCGGTGACTCCCTCAAA | 150 | 60 |
| gpr84 | XM_021609929.1 | GTTTTCGTGGGCTGTTTTGTC | CTGTTGAGCCAGGTGAGGTT | 109 | 60 |
| gpr119 | NC_035086.1 | TGAGATTGGCACCCGACTCT | CACAGAAGGAGTGGATGTTGGT | 143 | 60 |
| itpr1 | XM_021569164.1 | AGAAGAACGCCATGAGAGTGA | ACCACTTTGTCCCCTATCACC | 121 | 60 |
| itpr3 | XM_021616029.1 | GCAGGGGACCTGGACTATCCT | TCATGGGGCACACTTTGAAGA | 64 | 59 |
| plcb1 | XM_036985415.1 | GGAGTTGAAGCAGCAGAAGG | GGTGGTGTTTCCTGACCAAC | 83 | 60 |
| plcb3 | XM_021577635.1 | ATAGTGGACGGCATCGTAGC | TGTGTCAGCAGGAAGTCCAA | 120 | 60 |
| plcb4 | XM_021600840.1 | ACCTCTCTGCCATGGTCAAC | CGACATGTTGTGGTGGATGT | 89 | 60 |
| pyy | XM_021557532.1 | GGCTCCCGAAGAGCTGGCCAAATA | CCTCCTGGGTGGACCTCTTTCCA | 95 | 60 |
| slc16a1a | XM_036947863.1 | TGTTCGCCCGTCCTTCTATG | ACACAGGTAGGTCCACTGGT | 347 | 60 |
| slc16a1b | KF032405.1 | CCACAGCCTGCAGTGAAAAGT | GCCAGAACAGACAGCAGGAAG | 233 | 60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calo, J.; Comesaña, S.; Alonso-Gómez, Á.L.; Soengas, J.L.; Blanco, A.M. Fatty Acid Sensing in the Gastrointestinal Tract of Rainbow Trout: Different to Mammalian Model? Int. J. Mol. Sci. 2023, 24, 4275. https://doi.org/10.3390/ijms24054275
Calo J, Comesaña S, Alonso-Gómez ÁL, Soengas JL, Blanco AM. Fatty Acid Sensing in the Gastrointestinal Tract of Rainbow Trout: Different to Mammalian Model? International Journal of Molecular Sciences. 2023; 24(5):4275. https://doi.org/10.3390/ijms24054275
Chicago/Turabian StyleCalo, Jessica, Sara Comesaña, Ángel L. Alonso-Gómez, José L. Soengas, and Ayelén M. Blanco. 2023. "Fatty Acid Sensing in the Gastrointestinal Tract of Rainbow Trout: Different to Mammalian Model?" International Journal of Molecular Sciences 24, no. 5: 4275. https://doi.org/10.3390/ijms24054275
APA StyleCalo, J., Comesaña, S., Alonso-Gómez, Á. L., Soengas, J. L., & Blanco, A. M. (2023). Fatty Acid Sensing in the Gastrointestinal Tract of Rainbow Trout: Different to Mammalian Model? International Journal of Molecular Sciences, 24(5), 4275. https://doi.org/10.3390/ijms24054275






