Abstract
Since its outbreak in December 2019, the COVID-19 pandemic has caused the death of more than 6.5 million people around the world. The high transmissibility of its causative agent, the SARS-CoV-2 virus, coupled with its potentially lethal outcome, provoked a profound global economic and social crisis. The urgency of finding suitable pharmacological tools to tame the pandemic shed light on the ever-increasing importance of computer simulations in rationalizing and speeding up the design of new drugs, further stressing the need for developing quick and reliable methods to identify novel active molecules and characterize their mechanism of action. In the present work, we aim at providing the reader with a general overview of the COVID-19 pandemic, discussing the hallmarks in its management, from the initial attempts at drug repurposing to the commercialization of Paxlovid, the first orally available COVID-19 drug. Furthermore, we analyze and discuss the role of computer-aided drug discovery (CADD) techniques, especially those that fall in the structure-based drug design (SBDD) category, in facing present and future pandemics, by showcasing several successful examples of drug discovery campaigns where commonly used methods such as docking and molecular dynamics have been employed in the rational design of effective therapeutic entities against COVID-19.
1. The COVID-19 Pandemic
In December 2019, a cluster of pneumonia cases of unknown etiology emerged in the Chinese city of Wuhan [1]. Soon after, analyses of patients’ lung fluid, blood, and throat swabs reconducted this outbreak to a newly identified virus, tentatively named 2019-new coronavirus (2019-nCoV) [2].
Phylogenetic analyses performed on viral genomes isolated from patients’ samples revealed a close relationship between the new virus with several bat coronaviruses isolated in China (>90%). A lower degree of similarity was also found with SARS-CoV (80%) and MERS-CoV (50%), the causative agents of two recent coronavirus-related epidemics [3]. Based on phylogeny, taxonomy, and established practice, the virus was renamed SARS-CoV-2 [4], while the associated illness was defined as COVID-19 by the World Health Organization (WHO) [5].
The striking similarity between the SARS-CoV-2 genome and several bat coronaviruses led to the hypothesis that bats could be the animal reservoir for SARS-CoV-2, with pangolins or other mammals acting as the intermediate host before human transmission [6]. The assumption that bats could be the animal reservoir of SARS-CoV-2 was further reaffirmed at a later stage by the work of Temmam et al., which identified in the caverns of North Laos a series of bat coronaviruses that share a high level of sequence similarity (96%) with the SARS-CoV-2 genome [7].
From a clinical perspective, the spectrum of COVID-19 manifestation is broad, ranging from asymptomatic infections to severe viral pneumonia with respiratory failure and even death [8]. The most common symptoms, similar to influenza, are related to mild upper respiratory tract affection, such as fever, cough, myalgia, and headache [9]. Less common but still relevant ones include gastrointestinal manifestations, such as diarrhea, and more severe respiratory illnesses, such as dyspnea, and multiorgan failure [10].
The long incubation time compared to similar infections [11], the capability of asymptomatic [12] or paucisymptomatic [13] patients to transmit the virus even before the eventual symptoms’ manifestation, and the aerial transmission modality [14,15] all concurred to determine a higher transmissibility index (estimated between 2.5 and 3.0) for the SARS-CoV-2 virus, compared to similar viral infections [16]. These factors contributed to the rapid spread of SARS-CoV-2 worldwide, resulting in more than 650 million cases and more than 6.5 million deaths globally [17].
In the first stages of the COVID-19 pandemic, extraordinary sanitary measures, such as physical and social distancing, wearing face masks, and eye protection devices [18,19] were adopted to prevent the collapse of the public healthcare system [20], due to the imbalance between the high demand and the low availability of critical supplies [21,22]. Although this short-term plan has proven helpful in gaining time [23,24], more sustainable and long-term oriented strategies were needed to better cope with the socio-economic [25] and psychological [26] consequences of the pandemic, other than ensuring fair and efficient resource management [27].
1.1. Drug Repurposing
Considering that bringing a brand-new drug on the market is usually a very long and expensive process [28], the so-called “drug repurposing” was the first approach to finding suitable therapeutic options for COVID-19 patients [29,30]. This strategy extends the applicability domain of already marketed drugs for treating diseases other than the one it was conceived for [31]. This approach is appealing because it involves using derisked compounds, with potentially lower overall development costs and shorter development timelines [32]. Unfortunately, despite all the promising premises [33], this approach was largely unsuccessful [34]. Indeed, several investigated drugs showed little to no efficacy in randomized clinical trials [34]. The few successful cases were primarily symptomatic treatments, mostly limited to hospital usage for the most severe cases due to the therapy’s high costs or route of administration [35].
Failure of the drug repurposing strategy against COVID-19 can be mostly traced to the very first stages of the pandemic, where few clinical pieces of evidence were available for the rational elaboration of therapy plans. For example, the combination of HIV protease inhibitors Lopinavir and Ritonavir was examined [36], despite a suboptimal predicted recognition pattern towards the SARS-CoV-2 main protease (Mpro) compared to other compounds of the same class [37]. Another example is the combined use of an antimalaria drug (hydroxychloroquine) and an antibiotic (azithromycin) despite no clear indication of the possible mechanism of action [38,39].
With more and more clinical observations becoming available, more fine-tuned treatments, especially symptomatologic ones, were adopted. This is the case, for example, of corticosteroids such as dexamethasone [40], employed to tame the inflammatory response associated with severe COVID-19 cases, and low molecular weight heparins [41], used to prevent or treat thrombo-embolic events associated caused by interference with the cardiocirculatory system.
A group of antiarthritis drugs represents another successful example of drug repurposing to their ability to modulate the immune response [42] and cytokine storm [43] caused by severe SARS-CoV-2 infection. This family includes the monoclonal antibodies Tocilizumab [44] and Sarilumab [45], which both inhibit Interleukin-6 (IL-6) signaling; Anakinra [46], which interferes instead with IL-1 signaling; and the Janus Kinase (JAK) inhibitor Baricitinib [47], alone or in conjunction with Remdesivir [48], with the latest representing maybe the most successful example of drug repurposing against COVID-19 being the first approved drug against this illness [49].
Originally designed against Ebola virus, Remdesivir is a nucleotide analog prodrug that acts as a viral polymerase inhibitor [50] and is efficient in shortening the recovery time in hospitalized adult patients affected by COVID-19 [51]. Unfortunately, as previously mentioned, Remdesivir and the other repurposed drugs need parenteral administration, thereby limiting their massive-scale adoption as pharmacological treatments against COVID-19 [35].
1.2. Convalescent Plasma and Monoclonal Antibodies
With the first round of spontaneously healed patients, doctors started flanking standard treatment with the use of convalescent plasma (CP), i.e., the plasma derived from recently recovered donors with a sufficiently high neutralizing antibody titer [52]. A similar protocol was previously adopted to face Ebola [53] and MERS [54] outbreaks, justifying its emergency use in the first stages of the COVID-19 pandemic. Unfortunately, despite promising observational data from the first studies performed on small-size patient cohorts [55], more thorough investigations from more extensive clinical trials demonstrated the inefficacy of this treatment [56,57], leading to its dismission from routine clinical practices. Despite this failure, CP inspired the design of safer and more targeted immunological treatments in the form of monoclonal antibodies (mAbs) [58,59]. Since the beginning of the pandemic, several mAbs directed against COVID-19 have been developed, with some obtaining approval from regulatory agencies [60]. Multiple of these mAbs are often used in conjunction to combine their neutralizing power and boost their therapeutic efficiency, exploiting their ability to bind at different epitopes [61].
The list of approved ones contains the therapeutic combinations of casirivimab and imdevimab (Regeneron/Roche), redanvimab (Celltrion Healthcare), sotrovimab (GSK), and the combination of tixagevimab and cilgavimab [62,63]. Furthermore, the association of bamlanivimab and etesevimab is nearly approved, despite the previous failure of trials investigating bamlanivimab on its own [63].
1.3. Vaccines
As seen in the case of CP and mAbs, a targeted immune response against SARS-CoV-2 can be a beneficial treatment for patients [64]. While immunoglobulins are limited to treating ongoing infections in hospital settings due to the high costs and the parenteral administration route, a more economical and scalable approach would be instructing the human body to produce this type of response without needing external intervention [65]. Based on this assumption and parallel to the drug repurposing approach, the industry and academia spent a consistent joint effort on developing preventive tools to avoid the infection in the first place or at least mitigate the most detrimental effects of the illness. This endeavor resulted in the quick approval by regulatory agencies of several vaccines [66].
Three different classes of these therapeutic entities can be recognized [67]. The first one, related to inactivated virus vaccines, comprises the Chinese CoronaVac (Sinovac) and the Russian CoviVac. The second group is formed by adenovirus vector vaccines such as Vaxzevria/ChAdOx1-S (AstraZeneca), Sputnik V/Gam-COVID-Vac, and Jcovden/Ad26.COV2.S (Janssen). Finally, the third one is composed of mRNA-based vaccines, including Comirnaty/BNT162b2 (Pfizer-BioNTech) and Spikevax/mRNA-1273 (Moderna) [68,69].
Despite the poor performances of the first class of vaccines [70,71], several independent studies have asserted worldwide the efficacy of vaccination campaigns based on the other two types of vaccines, particularly in the case of mRNA-based ones [72,73].
1.4. Spike Protein
The ability of the SARS-CoV-2 virus to infect human cells heavily depends on a surface glycoprotein known as the S/spike protein [74], named after its peculiar shape [75]. For this reason, both mRNA vaccines and mAbs are designed to target this protein and prevent the virus’s entry into the cell, thereby limiting its replication [76].
Concerning these, although different pathways for SARS-CoV-2 cell entry are possible [77,78], the principal and better-characterized one involves binding to the human ACE2 receptor (hACE2) [79], a membrane-anchored metallopeptidase that is abundantly present in various districts of the human body, from the vascular endothelium to the epithelia of lungs and small intestine [80]. On its own, host cell receptor binding is not sufficient to ensure entrance within host cells. Priming and activating the S protein by host proteases is required to enhance its cell–cell and virus–cell fusion processes and increase viral shielding from neutralizing antibodies [79,81]. The list of priming proteases includes, but is not limited to, TMPRSS2, a transmembrane serine protease that is often co-expressed with ACE2 in SARS-CoV-2 target cells; Furin; and cathepsin B/L [79,82,83]. The priming process entails the exposure of a lipophilic fusion peptide (FP), which penetrates the host cell membrane triggering the viral fusion [84] thanks to its strong membrane-perturbing capacities [85]
From a structural perspective, the spike is a trimeric transmembrane glycoprotein composed of 1273 amino acids organized in two main subunits, S1 and S2, and several functional domains [86].
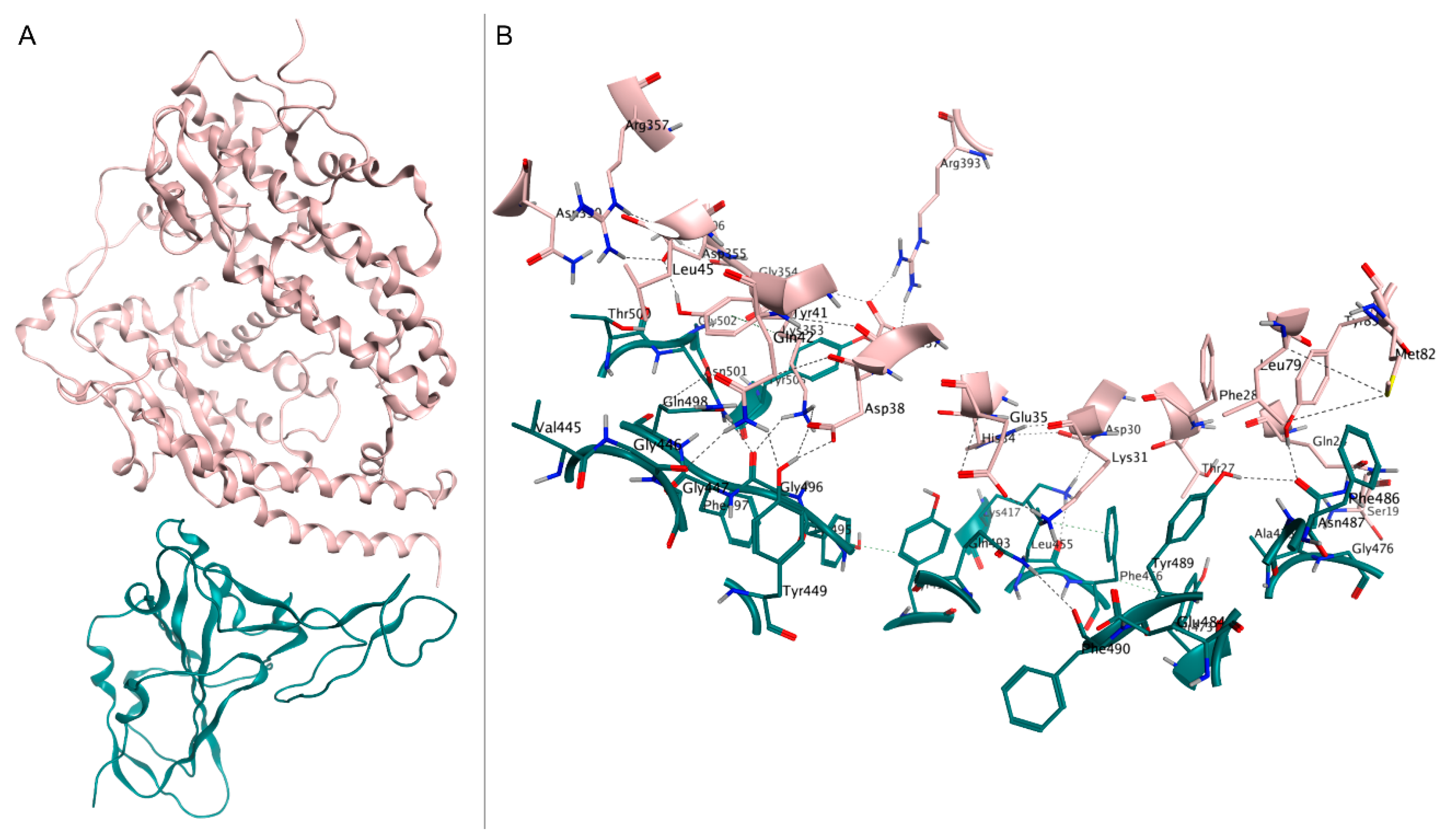
The S1 subunit comprises two main domains, specifically the N-terminal and C-terminal domains (NTD and CTD, respectively), which are both involved in the binding to host cell receptors [86]. The CTD contains the receptor-binding domain (RBD, residues 319–541), consisting of two motifs. Firstly, a core structure is formed by a twisted five-stranded antiparallel β sheet (β1, β2, β3, β4, and β7), with three short helices (α1, α2, and α3). Secondly, an extended loop (receptor binding motif, RBM) is formed by a two-stranded β sheet (β5 and β6), lying at one edge of the core and containing most of the residues involved in binding to hACE2 [87] (Figure 1).
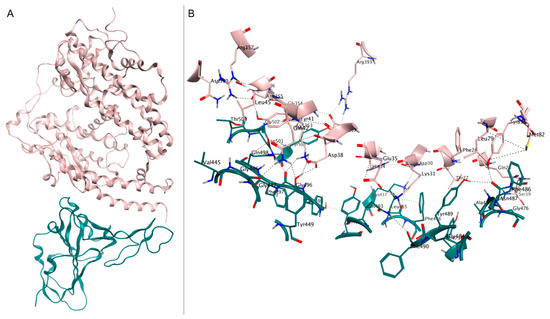
Figure 1.
(A) Crystal structure of spike RBD (teal) in complex with hACE2 (pink), deposited in the Protein Data Bank with accession code 6M0J. (B) Close-up view of interface contacts between the spike RBD and hACE2: hydrogen bonds are represented as black dashed lines.
The S2 subdomain has significant roles in spike protein trimerization and in mediating the virion entry into the host cell once the molecular contacts have been established [88]. It is formed by relevant subdomains such as the transmembrane domain (TD) (residues 1296–1317), which exerts both the spike anchoring to the outer side of the viral membrane and the maintenance of the trimeric quaternary structure [89,90], and a cytoplasm domain (CD) (residues 1318–1353), which mediates viral assembly and cell–cell fusion [91]. Furthermore, the previously mentioned fusion peptide, a cleavage S2′ site (residues 815/816), and two heptad-repeat domains (HR1/HR2) (residues 984–1104/1246–1295) are also part of S2 [92].
1.5. Viral Variants
Due to its exposition on the external surface of the SARS-CoV-2 membrane and its pivotal role in the virus’s ability to infect host cells, the spike protein is often subjected to mutations that alter the virus’s infectivity and antigenicity [93,94]. Therefore, since the spreading of the original viral strain (Wuhan-Hu-1) began, several viral variants appeared on the scene [95], particularly in third-world nations where collective sanitary practices such as social and physical distancing [96] or wearing face masks in public places [18] were hardly implementable [97].
The insurgence of novel viral strains with different susceptibility to the protective effect of vaccines [98] demands periodical updates of their original formulations coupled with multiple booster shots to maintain their efficacy [99], thus hampering the management of the pandemic based on massive vaccination of the world population [100,101].
Among the large pool of SARS-CoV-2 mutations [102], some gathered the scientific community’s attention due to their increased fitness, gaining the “variant of concern” (VOC) status [103].
The first ever SARS-CoV-2 VOC was the B.1.1.7 variant, more commonly referred to as the “Alpha” or “English” variant due to being first identified in November 2020 in the Kent region of the United Kingdom [104,105]. Despite worries about the higher transmissibility compared to other circulating variants at the time [106,107], clinical studies demonstrated how mAbs, CP, and especially vaccines, were still able to confer protection against B.1.1.7 [108,109,110], containing its impact on the sanitary system [111].
Unfortunately, soon after the emergence of the Alpha variant, a more threatening VOC arose. The B.1.617.2 variant, commonly known as the “Delta” or “Indian” variant, due to being first identified in India in late 2020, quickly overthrew B.1.1.7 thanks to its strikingly increased transmissibility [105]. The advent of the Delta variant was associated with the first signs of reduced protection provided by mAbs, CP, and most importantly, vaccines [112,113,114], thanks to its increased immune system evasion capability [115], posing a heavier workload on the sanitary system [116].
The latest hallmark in the history of SARS-CoV-2 variants is represented by the B.1.1.529 variant, first detected in South Africa and more often called the Omicron variant [117]. The combination of increased transmissibility [118] and immune system evasion [119] conferred this variant a net selective advantage in bypassing the protection provided by the complete primary vaccination cycle and a variety of clinically utilized mAbs [120,121,122] compared to other circulating strains. The ground-breaking impact the Omicron variant had on the worldwide spread of SARS-CoV-2 even led to the introduction of the “booster dose” to compensate for the reduced coverage of the primary vaccine cycle [98,123].
Lately, several subvariants germinated from the original Omicron strain (also labeled as BA.1), namely BA.2, BA.3, BA.4, and BA.5 [124,125,126]. Although different studies indicated how the first identified Omicron subvariants (BA.2 and BA.3) were similarly susceptible to existing treatments despite their increased transmissibility [127,128,129], it also emerged how the most recently identified ones (BA.4 and BA.5) are significantly more efficient in evading the immune response [130,131,132].
These findings indicate that SARS-CoV-2 continued to evolve by increasing its immune-evasion capability rather than counting on sheer higher transmissibility, sustaining the virus spread even in populations with high vaccination frequency and recovery rates [130,131,132].
1.6. Main Protease (3CLpro)
Considering the uncertainty about the efficacy of existing treatments [133] and booster vaccinations [134] against present and future Omicron subvariants, the need to find more reliable and variant-agnostic therapeutic tools against COVID-19 is emerging. The previously mentioned issues with the continuously mutating spike protein, which affects most present gold-standard COVID-19 treatments, indicate that different viral targets should be explored for developing novel antiviral drugs [135]. Generally speaking, an ideal target would have to play a pivotal role in the virus replication cycle and be highly conserved across different viral strains [136]. Within SARS-CoV-2, this role is portrayed by its main protease [137] (Mpro, or 3C-like protease / 3CLpro due to similarities with the picornavirus 3C protease [138]), thanks to its conserved fold across different coronaviruses [138,139,140,141] (including SARS-CoV [142]) and essentiality for the replication of this virus’s subfamily [143].
SARS-CoV-2 Mpro, also called nsp5, is a cysteine protease composed of 306 residues [144] that steers the maturation of two partially overlapping polyproteins (pp1a and pp1ab) into individual mature nonstructural proteins (including Mpro itself) through their proteolytic cleavage [145].
Functionally speaking, Mpro exists in equilibrium between a monomeric and a homodimer form [146,147,148]. This dimerization directly influences the shape of the catalytic site [147], thus altering the enzymatic activity [138] and playing an indirect regulatory role during the virus replication cycle [149,150].
Within the Mpro functional dimer, each protomer is composed of three structural domains. The chymotrypsin-like fold, including β-barrel domain I (residues 1–99) and II (residues 100–182), hosts the active site and thus has direct control over the catalytic event [138,147], while the α-helical domain III (residues 198–306) is mainly involved in the direct regulation of dimerization, exerting only a secondary and indirect role on regulating Mpro’s enzymatic activity [151]. Between the second and third domains lies a flexible 16-residue loop (residues 183–197) [152].
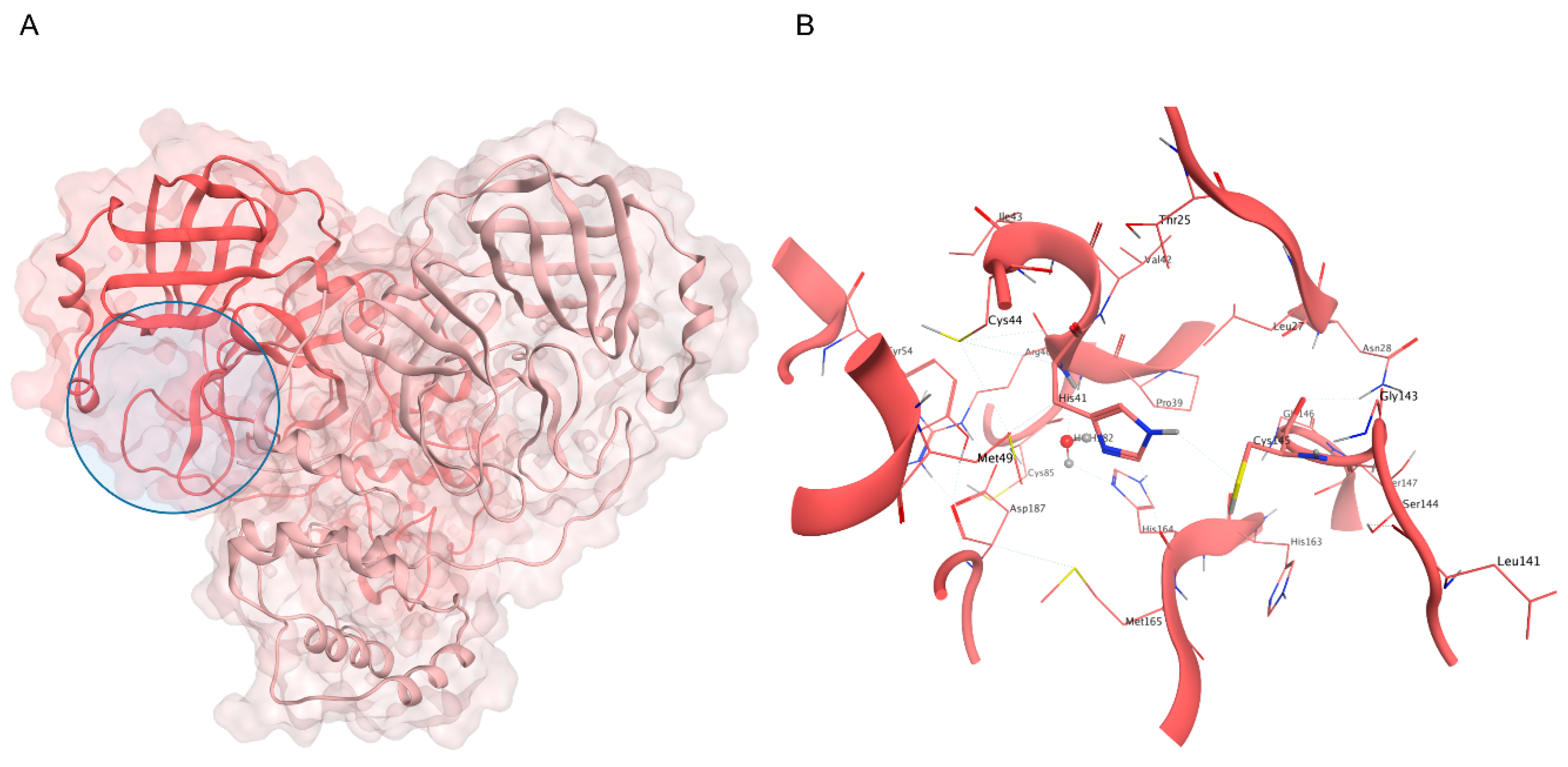
As anticipated, the catalytic site is located between domains I and II, bordered by the N-terminal domain I of the second protomer in the dimer (Figure 2). Notably, the N-finger (residues 1–7) interacts with the binding site through a salt bridge between the positively charged end of Ser1 and the negatively charged end of Glu166 [153]. The latter is also involved in forming a hydrogen bond with His172, an essential interaction for the enzyme’s proteolytic activity [154]. These interactions are so crucial in stabilizing the catalytic site [155] that N-finger deletion impairs dimerization and abolishes the protease’s enzymatic activity [156].
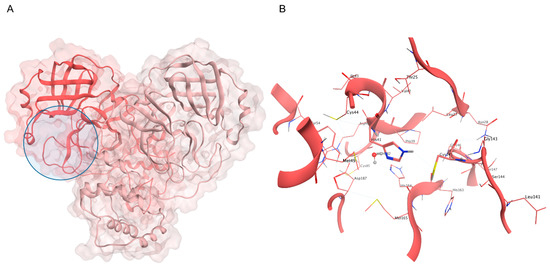
Figure 2.
(A) Crystal structure of SARS-CoV-2 Mpro (PDB ID: 6Y2E): the first protomer is colored in salmon, while the second protomer is colored in pink, and the active site position is highlighted with a blue circle. (B) Close-up view of the catalytic site: the H41-C145 dyad is highlighted, alongside the conserved water molecule that substitutes the third member of the canonical catalytic triad diffused in several cysteine proteases.
Mpro’s shallow, plastic, and solvent-exposed active site [152,157] comprises several subpockets (ranging from S6 to S3′), hosting the corresponding substrate residues (which vary from P6 to P3′) [139]. Speaking of substrates, the SARS-CoV-2 Mpro cleaves peptide bonds at the C-terminus end of a glutamine residue (P1) [137], which is conserved across different SARS-CoV-2, SARS-CoV, and even MERS-CoV substrate sequences [152].
SARS-CoV-2 Mpro recognizes sequences as long as ten residues (P6–P5–P4–P3–P2–P1↓P1′–P2′–P3′ P4′, where ↓ indicates the scissile bond [139]), but only shows remarkable selectivity at four subsites: S4, S2, S1, and S1′ [158]. On the contrary, prime recognition subsites located at the C-terminus of the conserved P2 (Leu/Val/Phe), P1 (Gln) ↓-P1′ (Ser/Ala) sequence are not conserved and show remarkable plasticity [152,159]. Furthermore, the main structural alterations of the binding site derive from flexibility at residues that line the S1 subpocket and segments incorporating methionine 49 and glutamine 189 [152,160].
Different from many other chymotrypsin-like proteases, Mpro exerts its enzymatic functions through a catalytic dyad instead of the usual triad, where His41 and Cys145 are flanked by a conserved water molecule that substitutes the sidechain of the third component (usually an aspartate or an asparagine) [138,161].
Aside from the catalytic dyad, another vital component of the catalytic machinery is represented by a set of conserved residues contouring the S1 subpocket known as the oxyanion loop (138–145) [152,162]. Notably, the correct conformation [87,163,164] of the oxyanion hole (Gly143-Ser144-Cys145) is required for stabilizing the tetrahedral transition state through a coordinated series of hydrogen bonds involving the backbone amides [138,155,165]. Accordingly, alternative oxyanion loop conformations are associated with catalytically incompetent/inactive proteases [140,152,154,166,167].
1.7. Rational Design of COVID-19 Drugs
Several characteristics of the viral proteases family, including SARS-CoV-2 Mpro, make them an attractive target for the rational development of tailored drugs against COVID-19. First, the low sequence identity with human proteases coupled with distinct cleavage-site specificities reduces the possibility of off-target/side effects associated with the therapy [168]. Second, the striking conservation of protein fold and structural organization of the active site among different members of the same family leads to the possibility of developing pan-coronaviral drugs [169]. Third, the abundance of structural data about the SARS-CoV-2 main protease (659 structures have been deposited in the Protein Data Bank [170] to date) makes it possible to exploit the state-of-the-art structure-based approaches in drug design [171]. Furthermore, a similar strategy has already proved successful in finding efficient treatments against the hepatitis C virus [172,173] and human immunodeficiency virus (HIV) [174,175]. Finally, the experience acquired studying the original SARS-CoV protease [176], in conjunction with the rapid release to the scientific community of the SARS-CoV-2 protease [164], certainly played a major role in determining its prominent place within most COVID-19 drug discovery campaigns. A detailed report on structural features of the 3CLpro protease that can guide the design of novel inhibitors can be found in the work of Xiong et al. [177].
The first attempts at finding SARS-CoV-2 Mpro inhibitors involved the repurposing of existing protease inhibitors. Particularly, the hepatitis C protease inhibitor Boceprevir [178,179] and the feline coronavirus 3CLpro inhibitor GC373 (derived from its prodrug GC376) [180] were found to be active in the low µM potency range against Mpro [181], with the latter being particularly interesting due its promiscuous anticoronaviral activity [182]. Both candidate drugs share a similar peptidomimetic scaffold, which entails the most prominent interaction features of the first identified ones [164].
Although these primary hit compounds present a good binding pattern, their evolution towards clinical candidates and drugs is prevented by two main factors: first, covalent inhibitors are usually associated with selectivity problems, due to their ability to react promiscuously with a plethora of nucleophile moieties [183]; second, the peptidomimetic scaffold is usually associated with suboptimal pharmacokinetic properties that affect the preferred route of administration [184].
In this regard, a step forward was obtained when the first SARS-CoV-2 Mpro inhibitors were able to reach clinical stage experimentation, namely PF-07304814 (lately renamed as Lufotrelvir), a prodrug for the active principle PF-00835231, and PF-07321332 (Nirmatrelvir).
Lufotrelvir was originally developed by Pfizer in 2002–2003 for the SARS-CoV virus and later repurposed against the SARS-CoV-2 due to the high similarities between the two proteases [185]. Due to its efficacy against several viral strains in preclinical studies [186,187], it was advanced to the clinical stages of experimentation, albeit quickly overcome by Nirmatrelvir thanks to its more favorable pharmacokinetic profile [188].
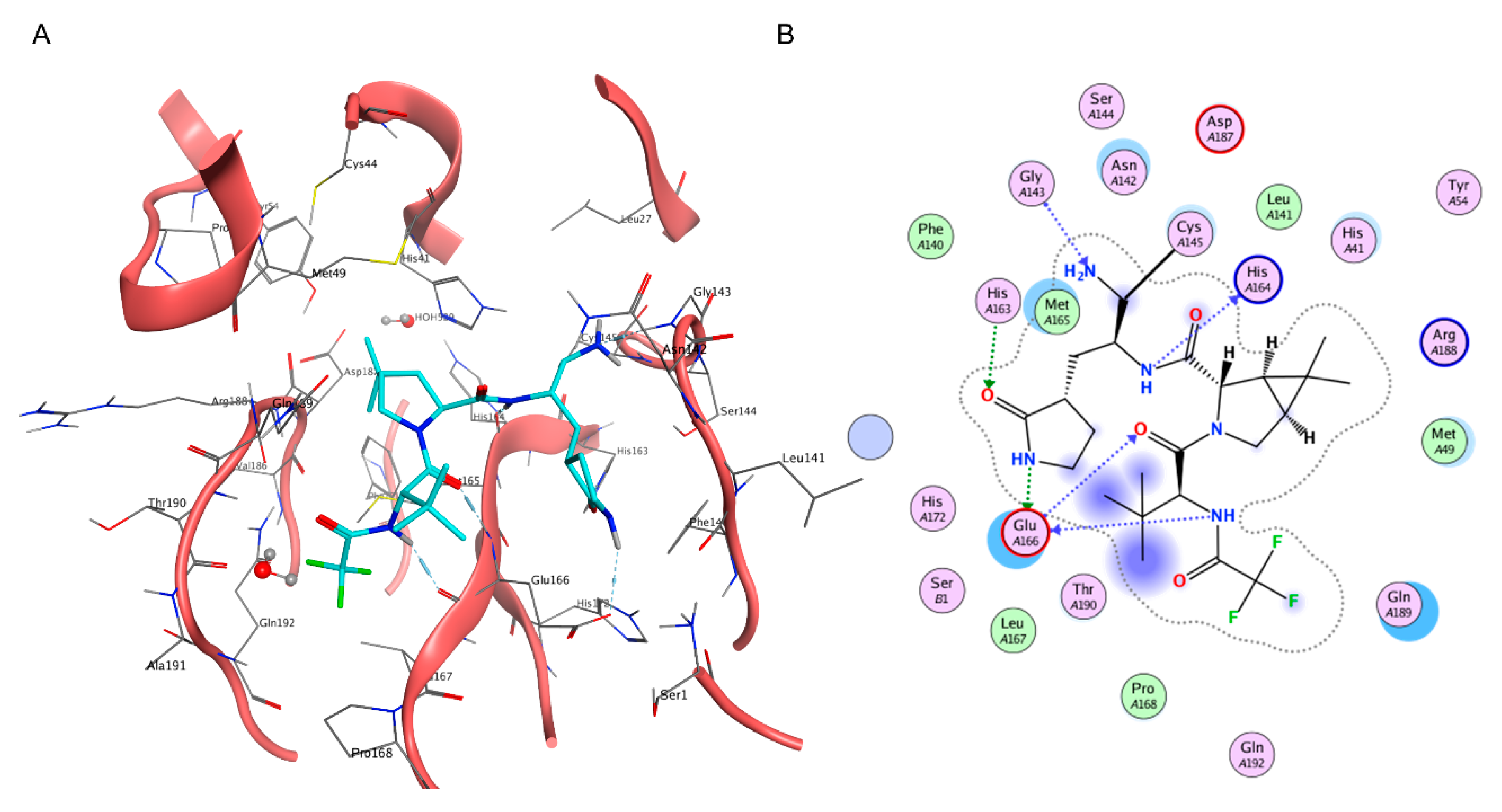
Contrary to Lufotrelvir, which, similar to Remdesivir, requires parenteral administration, Nirmatrelvir can be administered orally [189], a must-have characteristic for the widespread adoption of drugs [190,191]. Designed by Pfizer amid the pandemic through the rational modification of Lufotrelvir [192], the structure of Nirmatrelvir was officially presented to the general audience on 6 April at the American Chemical Society Spring 2021 meeting [193], only one year after the official start of its development process [192] (Figure 3).
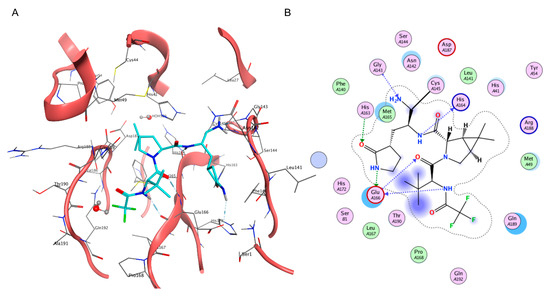
Figure 3.
(A) Three-dimensional depiction of Nirmatrelvir orientation within the catalytic site of SARS-CoV-2 Mpro (PDB ID: 7RFW). (B) Bidimensional representation of intermolecular interactions of Nirmatrelvir–SARS-CoV-2 Mpro 7RFW complex.
This peptidomimetic inhibitor, which is administered in association with the pharmacokinetic enhancer Ritonavir and sold under the commercial name of Paxlovid, represents a hallmark in the history of both the COVID-19 pandemic and structure-based drug discovery, due to the groundbreaking speed of its discovery campaign [194]. Although clinical studies highlighted the remarkable therapeutic efficacy of Paxlovid in preventing the most severe COVID-19 cases [195], its effectiveness on more mild infections remains unclear [196]. Furthermore, the impact of viral mutations on present and future protease inhibitors has yet to be disclosed [197,198], thus justifying the current effort to find novel and diverse drugs that can enlarge the pool of pharmacological tools available against COVID-19.
An important step in this direction is represented by the development of Ensitrelvir (formerly known as S-217622), the first noncovalent, nonpeptidomimetic, orally available Mpro inhibitor to reach clinical stage experimentation [199]. This compound has successfully reached the third and final stage of clinical experimentation, thanks to its proven efficacy against mild-to-moderate or even asymptomatic infections [200,201]. Possible approval of this active principle by regulatory agencies would provide an additional and orthogonal therapeutic tool to Nirmatrelvir in the treatment of COVID-19 cases, thus reducing the impact of resistance mechanisms associated with the emergence of mutated viral strains [197,198].
1.8. Potential Targets of Interest
Although targeting the SARS-CoV-2 main protease was successful in individuating several clinical candidate drugs, even leading to the first approval of a COVID-19 specifically designed drug, other drug discovery campaigns aimed at different viral targets are needed for therapy diversification, potentially combined and synergic treatment, and resistance prevention [202,203,204].
Altogether, the SARS-CoV-2 genome encodes four major structural proteins, including nucleocapsid (N), membrane (M), envelope (E), and the spike as mentioned earlier (S), plus 16 nonstructural proteins, encompassing the previously mentioned main protease [205].
Although Mpro plays a pivotal role in processing the SARS-CoV-2 viral polyproteins, it is not the only component of the functional replicase complex that is required for the viral spread process [206]. Alongside this, a secondary but still relevant enzyme operates, namely the papain-like protease (PLpro, the catalytic domain of protein nsp3) [207]. Despite being a cysteine protease similar to Mpro, PLpro exerts its enzymatic functions through a catalytic triad composed of Cys111, His272, and Asp286 [208]. Further, PLpro processes peptide bonds located at the C-terminal end of LXGG motifs [209]. Functionally speaking, this 343-residue segment, which is part of the multidomain nsp3 protein, is responsible for cleaving the SARS-CoV-2 polyproteins at three different sites, resulting in the liberation of nsp1, nsp2, and nsp3 proteins [210]. Moreover, PLpro is also responsible for cleaving post-translational modifications on known regulators of host innate immune response [211].
As demonstrated by the approval of Remdesivir by regulatory agencies, another valuable target for the development of COVID-19 drugs is represented by the RNA-dependent RNA polymerase (RdRp) [49]. This complex machinery comprises four subunits, including one nsp12, responsible for the catalytic activity of the assembly; one nsp7; and two nsp8, with the latest two acting as cofactors [212]. The assembled holoenzyme presides RNA replication, a process that results in the formation of nine subgenomic RNAs [213]. The active site of nsp12 resides in its C-terminal RdRp domain and includes residues spanning from Thr611 to Met626, which are involved in binding one turn of double-stranded RNA, while residues D760 and D761 are required for recognition of the 3′ end and are essential for RNA synthesis [214,215]. Remdesivir binds within the active site, forming direct contact with residues K545, R553, D623, S682, T687, N691, S759, D760, and D761 and blocking the catalytic machinery by delaying the chain termination process [216,217].
During the RNA synthesis process, the RdRp also interoperates with nsp13 (helicase) [218], an enzyme involved in unwinding the RNA secondary structure of the 5′ untranslated section of the viral genome [219] to increase the efficiency of the copy process [220,221]. From a structural perspective, the nsp13 is a 596 residue, triangular pyramid-shaped helicase, which exploits its function thanks to the energy provided by its NTPase domain composed of six conserved residues (K288, S289, D374, E375, Q404, and R567) [222]. Adding to its helicase activity, the nsp13 active site also exerts RNA 5′ triphosphatase activity, further highlighting its importance in the maturation process of the viral mRNA [223].
The 5′ end of the newly synthetized mRNA is then subjected to post-translational modifications to boost both its stability (preventing cleavage from exonucleases), protein translation, and viral immune escape [224]. This activity is sequentially carried out by two S-adenosyl-L-methionine-dependent methyltransferases, namely nsp14 and nsp16 [225].
Specifically, the 527 residues’ nsp14 encompass both a proofreading exoribonuclease (ExoN) and an N7-methyltransferase enzymatic activity [226]. Furthermore, it has recently been suggested that it could encompass also a third, essential function for the viral replication cycle, based on the fact that SARS-CoV-2 ExoN knockout mutants are nonviable despite the 95% sequence identity with SARS-CoV [227] and the conservation of important active site amino acids including both the cap-binding residues (N306, C309, R310, W385, N386, N422, and F426) and the S-adenosyl methionine (SAM) binding residues (D352, Q354, F367, Y368, and W385) [228,229].
After its cleavage by the Mpro, evidence suggests that it forms a binary complex with nsp10, which cooperatively exerts the proofreading activity on fresh RNAs produced by the RdRp machinery [230,231]. Although the binary complex theory is the most prominent one, an alternative hypothesis based on the formation of a ternary nsp10-nsp14-nsp16 has been proposed due to the flexibility of the lid subdomain of nsp14 and the fact that nsp10 also forms a heterocomplex with nsp16 [231].
Particularly, the nsp16-nsp10 heterodimer is responsible for the 2′ O-methyltransferase activity that is required to complete the cap-0 ➔ cap-1 conversion of mRNA that is initiated by nsp14 [225]. While the catalytic activity entirely resides on nsp16, nsp10 provides a support role, aiding the recruitment of both the m7GppA-RNA substrate (which happens at a binding site defined by residues K24, C25, L27, Y30, K46, Y132, K137, K170, T172, E173, H174, S201, and S202) and the SAM cofactor (which binds in a pocket defined by N43, G71, G73, G81, D99, D114, C115, D130, and M131), thus enhancing nsp16′s catalytic activity [232,233,234].
Lastly, another essential target for coronavirus biology is represented by nsp15, a uridine-specific endoribonuclease (NendoU) [235]. The active form of this enzyme is a dimer of trimers, with each monomer composed of 345 residues organized in three different domains: N-terminal, middle, and C-terminal NendoU, where the catalytic activity resides [236].
The active site contains six conserved residues: His250, His250, and Lys290, which compose the catalytic triad, and Thr341, Tyr343, and Ser294, with the latest associated with selectivity in substrate recognition [237]. Due to their localization within the hexamer, cooperativity or anticooperativity between different binding sites is possible [238]. Nsp15 enzymatic activity involves the cleavage of both single- and double-stranded RNA at uridine sites producing 2′,3′-cyclic phosphodiester, and 5′-hydroxyl termini [239].
Functionally speaking, Nsp15 seems to directly participate in viral replication through interference with the innate immune response [237]. Indeed, to evade host pattern recognition receptor MDA5 responsible for activating the host defenses, the Nsp15 cleaves the 5′-polyuridine tracts in (-) sense viral RNAs [240], though it has also been suggested that Nsp15 degrades viral RNA to hide it from the host defenses [238]. More detailed structural information about potentially druggable SARS-CoV-2 protein targets can be found in the works of Littler et al. [241] and Wu et al. [242].
2. Computer Simulations for Rational Drug Design
For most of its existence, the human genre has exploited natural products such as leaves, seeds, roots, bark, and flowers as medicines, based on empirical observations purely based on symptom relief [243,244].
Nevertheless, throughout the latest two centuries, the process of drug discovery has evolved rapidly from the serendipitous discovery of novel active principles derived from or inspired by natural compounds [245,246] to the rational design of brand-new chemical entities [247].
The major turning point in the history of modern drug discovery can be traced back to the 1980s when experimentally solved macromolecular structures become routinely available [248]. The enhanced accessibility of structural data about biological targets is reflected in a rapid interest in the development of computational methods that could valorize this information and aid medicinal chemists’ work [249].
Today, computer simulations are a staple point of drug discovery campaigns, thanks to their ability to streamline and reduce their attrition rate [250]. From a functional perspective, computer-aided drug discovery (CADD) techniques are employed in the earliest stages of the pipeline for hit identification, hit-to-lead optimization, and pharmacokinetic evaluations [251].
CADD methodologies can either fall into one of two subgroups, based on the rationale behind them: the first group is represented by ligand-based (LBDD) approaches, while the second one includes structure-based (SBDD) methods [252]. The main difference between these two orthogonal and complementary approaches is that the first one does not exploit any information about the target macromolecule structure (e.g., a protein or a nucleic acid), while the second one does [253].
Nowadays, with the advent of cryo-electron microscopy (cryo-EM) [254] and groundbreaking tools for de novo prediction of protein structures such as AlphaFold [255], the second approach has become the gold standard [171].
2.1. CADD Strategies against COVID-19
The starting point of every SBDD campaign is the identification of a target macromolecule (a protein or a nucleic acid) that is involved in the etiology and or pathogenesis of a disease of interest, whose function can be opportunely modulated through a specifically designed ligand, usually a small organic molecule [171].
Once the target has been identified, its structure must be retrieved, either through experimental methods such as X-Ray crystallography (XRC, the gold standard) [256], nuclear magnetic resonance (NMR) [257], and cryo-EM [258] or hypothesized through homology modeling or de novo prediction [259].
Homology modeling involves the use of a homologous protein with a high primary sequence identity with the target as a template for the construction of its three-dimensional model [260,261]. De novo prediction, instead, does not rely on any information about other proteins’ structures and outputs a structural hypothesis that is solely based on the primary sequence of the target of interest [262].
While the second approach has gained a lot of momentum during the last two years, thanks to its unprecedentedly high accuracy [263,264], the first one is still relevant in those cases where important structural rearrangements occur between different states of the target functional cycle, other than predicting ligand-bound conformations [265,266].
In the context of the COVID-19 pandemic, where the extraordinary effort promoted by the scientific community quickly made several experimentally determined structures available, the relevance of structural modeling was highlighted by the ability to keep up with the high mutation rate of the virus [135,207], other than providing a useful starting point for drug discovery campaigns for a target whose structure had yet to be elucidated [267,268]. For example, several studies were conducted to investigate the impact of mutations found in both the spike protein [135,269,270,271,272,273] and the main protease [135,198,274,275] of emerging strains on viral fitness and resistance to existing therapies. These studies showed that relatively inexpensive approaches such as homology modeling and positional scanning can be reliable tools to rationalize the origin of the virus [274,276,277,278], quickly track the evolution of the original strain [135,279,280], predict the impact of future possible mutations [270,272] and adjust existing therapeutics tools accordingly [198,281].
The huge amount of structural information available on several SARS-CoV-2 druggable targets was fertile terrain for various COVID-19 SBDD campaigns [282,283], both in academia and in industry, with the most effort aimed at hitting well-characterized and pivotal viral targets such as Mpro or spike [284,285].
A remarkable example is represented by the COVID Moonshot Consortium, a drug discovery campaign driven by a collaborative effort among different research groups across the world aimed at targeting the SARS-CoV-2 main protease. This project led to the advancement of novel noncovalent orally available nanomolar Mpro inhibitors to clinical stage experimentation [286].
2.2. The Swiss Knife of SBDD: Molecular Docking
Within every SBDD campaign, available information about the target structure is exploited to fetch molecules able to recognize it selectively and potently [287]. Usually, this involves the identification of molecules that have good steric and electrostatic complementarity with the active site [288]. Depending on the steric and volumetric features of the binding site, the ligand type can be chosen accordingly, with small organic molecules being a better solution for buried cavities [289] and peptides, aptamers, or antibodies a better one for larger, flatter, and solvent-exposed interaction surfaces [290].
To narrow down the list of potentially active molecules to experimentally test to a feasible number, and to avoid wasting resources on compounds that do not possess the appropriate features to interact with the target, most SBDD campaigns start with a virtual screening process (SBVS) [291]. The most widely and successfully adopted method for SBVS is molecular docking, a computational protocol developed in the 1980s by Kuntz et al. [292] for predicting the preferred orientation of a certain ligand within the active site of a receptor [293].
Each docking program has two major components, which cooperate to find the solution to the protein–ligand docking problem [294]. The first part is the search algorithm (SA), which explores the ligand degrees of freedom within a user-defined search space centered around the active site of the protein [295]. The SA generates several ligand conformations (poses) that are fed to the second element of the program, i.e., the scoring function (SF), which qualitatively evaluates subsisting protein–ligand interaction features [296].
In the context of the COVID-19 pandemic, docking was also the king of computational methods used for drug discovery, thanks to the combination of its accuracy [297] and rapidity, which allows it to virtually screen billions of compounds in just a few days [298,299,300].
For example, Corona et al. reported the discovery of four low micromolar nsp13 inhibitors through a virtual screening carried out with the LiGen [301] docking program on an in-house natural compounds library [302].
Kolarič et al. identified two micromolar SARS-CoV-2 cell-entry inhibitors that act by binding human neuropilin-1 (nrp-1) and preventing its interaction with the spike protein, by performing a virtual screening with the GOLD [303] program on a library of commercially available compounds [304].
Vatansever et al. performed a virtual screening based on the Autodock [305] program on a library of drugs approved by the Food and Drug Administration and by the European Medical Agency (EMA) to discover six micromolar Mpro inhibitors [306].
Kao et al. reported the discovery of three sub-micromolar, synergistic nsp1 inhibitors identified through two independently executed virtual screenings with ICM [307,308] and Vina [309] software on a library of FDA-approved drugs [310].
Zhang et al. identified 11 natural compound Mpro inhibitors active in the low micromolar range through a virtual screening purely based on the commercial software Glide [311], developed by Schrödinger [312]. Another strategic use of docking-based virtual screening based on the Glide program is portrayed by the work of Huff et al., which designed six mixed covalent and noncovalent nanomolar Mpro inhibitors [313]. Another Glide-based virtual screening performed by Liu et al. led to the repurposing of histone deacetylase (HDAC) inhibitors as SARS-CoV-2 cell entry inhibitors through allosteric modulation of ACE2 and alteration of its ability to recognize the spike protein [314].
Wang et al. used LibDock [315] to perform a virtual screening on a library composed of FDA-approved peptides, which led to the identification of a nanomolar SARS-CoV-2 cell entry inhibitor that exerts its effect by binding the human ACE2 receptor [316].
A remarkable result was obtained by Luttens et al., which identified eight Mpro inhibitors (including a nanomolar compound with pan coronaviral activity) by combining fragment-based drug design with ultralarge virtual screening based on the DOCK [292] program [317].
Welker et al. exploited the molecular docking pipeline of the LeadIT [318] program to repurpose previously identified SARS-CoV PLpro inhibitors towards its SARS-CoV-2 homolog, demonstrating their activity on viral replication in cell-based assays [319].
Otava et al. utilized docking calculations with the GOLD [303] software to rationalize the structure–activity relationship of a series of rationally designed S-adenosyl-L-homocysteine derivatives, some of which showed inhibitory activity towards SARS-CoV-2 nsp14 in the low nanomolar potency range [320].
Similarly, Wang et al. exploited docking with Vina to rationalize the SAR of a series of rationally designed phenanthridine nucleocapsid protein (NPro) inhibitors, including two compounds showing low micromolar inhibitory activity [321].
2.3. Complementary Strategies to Address Docking Limitations
Although a very efficient and useful tool, molecular docking is rarely used on its own within SBDD campaigns and, indeed, is most often coupled with other methods to compensate for its weak points, such as neglecting receptor flexibility or the role of solvents [322], thus increasing the virtual screening success rate [323]. Another major limitation is represented by the poor ranking capabilities of classical scoring functions [324], which is the main cause of the high false positive rate of docking-based virtual screenings [325]. Indeed, in order to be universally applicable across different biological targets and computationally efficient enough to evaluate a large number of compounds, scoring functions have some limitations in the physical description of the binding event, which prevent any correlation between docking scores and experimentally determined affinity values [296]. Furthermore, little to no difference in score exists between top-ranking compounds derived from large virtual screening campaigns, making it practically impossible to distinguish active from inactive compounds solely based on the docking score [326]. For these reasons, each docking-based virtual screening cannot be blindly executed and fully automatized, and a careful setup of the experiment must be executed based on the available literature data and the knowledge of the target [326,327]. For COVID-19, the importance of this common-sense medicinal chemistry practice has been highlighted by the retrospective literature analysis provided by Llanos et al., which showcased the poor performances of structure-based virtual screenings solely based on ranking provided by docking scoring functions [323].
A possible solution to the limited physical description of the protein–ligand binding event of docking is to couple it with molecular dynamics (MD) simulations [294,328]. Molecular dynamics is a computational technique that allows investigating the time-dependent evolution of biological systems following the rules of molecular mechanics, i.e., determining the atomic trajectories by numerically solving Newton’s equation of motion, where forces between the particles and their potential energies are calculated according to molecular mechanical force fields [329]. Due to the heavy computational workload required to run these types of simulations, MD is rarely used for screening purposes, while it is more frequently exploited for the refinement of docking results, i.e., evaluating the pose stability or optimizing the protein–ligand complex geometry for a more accurate estimation of the free binding energy [330,331].
Regarding the pitfalls of the scoring component of docking programs, one possible strategy is to apply some form of knowledge-based filter upon docking results, in a similar fashion to what would happen if each pose were visually inspected [332]. For example, experimental information about critical protein–ligand interactions required for binding can be encoded within a pharmacophore filter or an interaction fingerprint, both of which can be used as constraints in the pose selection process [333]. In the case of pharmacophore filters, poses are filtered based on their ability to place a given functional group within a defined volume [334,335], while in the case of protein–ligand interaction fingerprint, the selection is usually based on the similarity between the reference and the query vector, representing the interaction features of the reference compound (a true active) and the investigated molecule respectively [336,337].
For instance, Wang et al. used a combination of structure-based pharmacophore screening, docking (both performed with the appropriate tools of the Molecular Operating Environment suite), and postdocking molecular dynamics refinement to identify a set of four sub-micromolar Mpro inhibitors among a database of in-house compounds [338].
The same protocol was successfully exploited by Tian et al. to identify four sub-micromolar PLpro inhibitors in the same in-house library [339].
Furthermore, a slight variation of the protocol was also employed by Yin et al. to discover a noncovalent cyclic peptide that simultaneously inhibits both SARS-CoV-2 Mpro and nrp-1 with an activity in the low nanomolar range [340]. Within this scientific work, pharmacophore constraints were used for scoring peptide poses on Mpro, while traditional docking scores were used for the nrp-1 screening.
A remarkable joint computational work by Gossen et al. led to the molecular dynamics-driven design of a structure-based pharmacophore filter, which was then exploited to identify two nanomolar Mpro inhibitors among a library of publicly available compounds [341].
A similar approach was exploited by Hu et al., which exploited the combination between MD-based pharmacophore filtering, docking-based virtual screening within the Molecular Operating Environment suite, and MD-based postdocking refinement to identify micromolar SARS-CoV-2 cell entry inhibitors targeting the FP of the spike protein [342].
Jang et al. used protein–ligand interaction fingerprint similarity as a postdocking filter for their double virtual screening on both Mpro and RdRp with the Vina program to identify seven compounds inhibiting SARS-CoV-2 replication in cell-based assays among a library of approved drugs [343].
Due to the static nature of molecular docking, which does not consider receptor flexibility, the choice of the input structure is vital for the success rate of a virtual screening [344]. Although molecular dynamics can be a useful posterior refinement of poses, a wrong input conformation of the target macromolecule could prevent the sampling of native-like poses for active compounds, leading to a reduced hit-finding rate [345]. For this reason, multiple conformations of the same receptor derived from MD simulations or experimentally solved in different conditions can be used in parallel in a process defined as ensemble docking (ED) [346]. When this approach is used, docking calculations are independently run on each structure, with virtual hit compounds being identified either through consensus scoring or a consensus ranking approach [347,348]. In the case of consensus scoring, the docking score of the same molecule is averaged across the different virtual screenings, with the final ranking based on the consensus score [349]. Differently, consensus ranking involves the selection of top-ranking hit compounds across different virtual screenings, regardless of congruence between scores [350]. A consensus approach can also be utilized to rank molecules based on virtual screening executed on the same receptor structures with different docking protocols [351].
For example, Gimeno et al. applied a consensus scoring approach to three independently executed virtual screenings through Glide, FRED [352], and Vina software to identify two Mpro micromolar inhibitors within the Drugbank database, a library that includes all drugs approved by the Food and Drug Administration (FDA) [353].
Yang et al., instead, employed an ensemble docking approach with the Glide docking software to identify six Mpro inhibitors among a library of commercially available peptidomimetic compounds, two of which demonstrated sub-micromolar potency [354].
Rubio-Martinez et al. used a combination of ensemble docking based on QVina2 [355] and postdocking molecular dynamics refinement to identify five Mpro micromolar inhibitors within a library of commercially available natural compounds [356].
A mixture of the previous two approaches was exploited by Clyde et al. for their High-Throughput Virtual Screening (HTVS), based on both ensemble docking and consensus scoring between the FRED and Vina docking programs, that led to the discovery seven micromolar Mpro inhibitors among a set of commercially available compounds [357].
Further, a combination of consensus ranking among Autodock, Hybrid, and FlexX and postdocking molecular dynamics refinement was utilized by Glaab et al. to virtually screen a library of commercially available compounds and identify two micromolar Mpro inhibitors [358].
Similarly, Ghahremanpour et al. applied both consensus ranking among three independent virtual screenings performed with the Glide, Autodock, and Vina software and postdocking molecular dynamics refinement to identify 14 micromolar Mpro inhibitors within the Drugbank database [359].
Another possible solution to cope with inaccuracy in free binding energy determination by traditional scoring functions is to rescore docking poses using more computationally intensive and accurate methods such as Free Energy Perturbation (FEP) [360] or MMGBSA/MMPBSA [361]. The first approach relies on performing a series of alchemical transformations across a set of ligands that need to be evaluated. This conversion cycle allows calculating relative differences in the free binding energy that can be used for a more accurate ranking of hit compounds derived from a virtual screening [362]. The second approach relies instead on correcting the gas phase interaction energy calculated according to the molecular mechanics force field with a term accounting for the desolvation-free energy, where the polar component is estimated either by numerically solving the Poisson–Boltzmann equation (MMPBSA) or through the Generalized Born method (MMGBSA) [363].
Intriguingly, one of the hit compounds identified in the work of Ghahremanpour et al. was then used by Zhang et al. for the FEP-driven design of multiple nanomolar Mpro inhibitors [364].
A similar combination of Glide docking and FEP to determine the absolute binding free energy was also employed by Li et al. to identify 15 micromolar Mpro inhibitors within the Drugbank database [365]. The efficacy of FEP in estimating the binding energy of potential Mpro inhibitors was also highlighted by a retrospective study by Ngo et al. [366].
A multistep virtual screening involving semiflexible docking with Glide, Schrödinger induced-fit docking [367], MD-based postdocking refinement, and binding free energy estimation with the MMGBSA [368] protocol was exploited by Ibrahim et al. to identify one low micromolar nsp15 inhibitor [369].
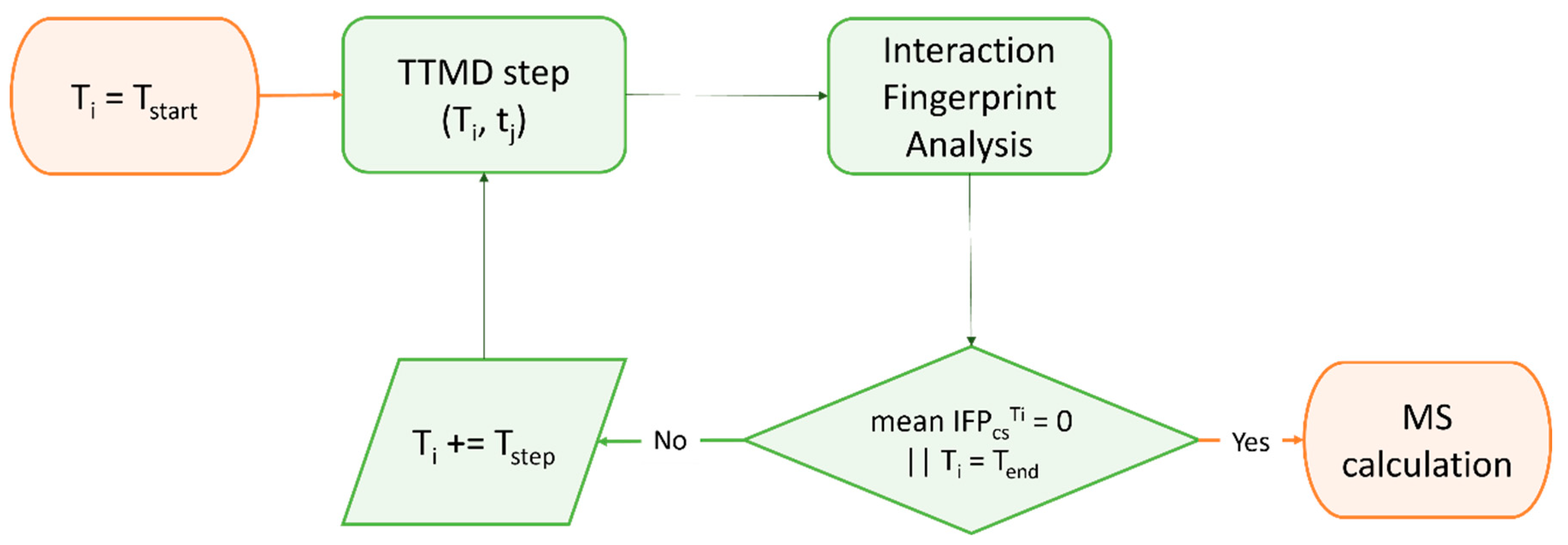
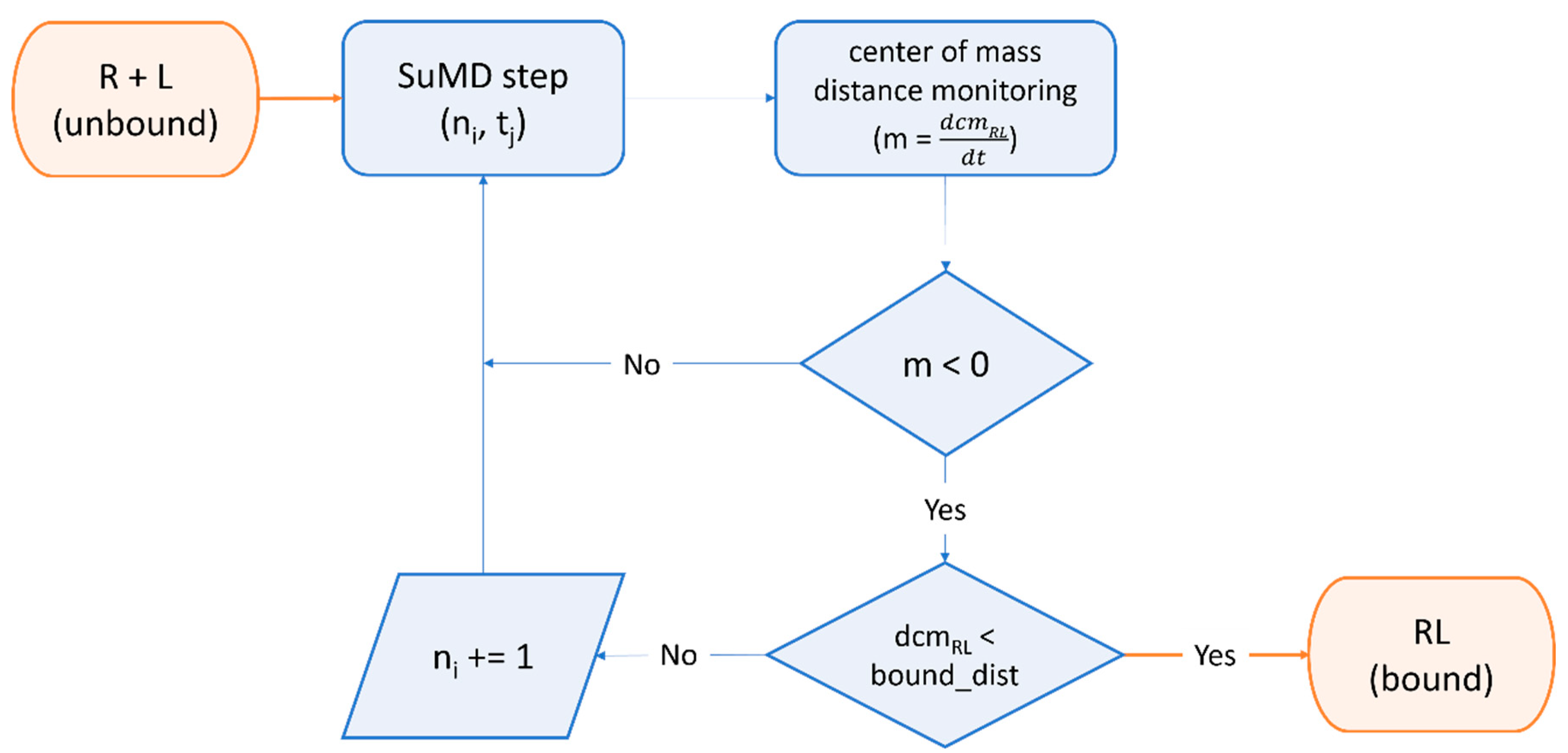
Although the estimation of thermodynamic properties such as the free binding energy has been a staple point of drug discovery campaigns, both from a computational and an experimental perspective, lately there has been a major interest shift towards the determination of kinetic parameters since they better correlate with in vivo efficacy [370]. Specifically, several MD-based methods have been developed throughout the years to rank compounds based on their predicted residence time, i.e., the time that the ligand spends in the receptor-bound state [371]. Among those, Pavan et al. developed Thermal Titration Molecular Dynamics (TTMD), a new method for qualitative estimation of protein–ligand complex stability (Figure 4), which was successfully applied for correctly discriminating tight, low nanomolar binders from weak, micromolar SARS-CoV-2 Mpro inhibitors [372].
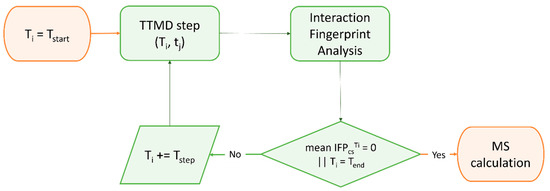
Figure 4.
Workflow of a Thermal Titration Molecular Dynamics (TTMD) simulation. The time-dependent conservation of the native binding mode within a protein–ligand complex of interest is monitored with a scoring function based on interaction fingerprint through a series of short molecular dynamics simulations performed at progressively increasing temperatures. The simulation is carried out until the target temperature is reached or the dissociation process is completed. A coefficient called MS is then calculated and used to rank ligands based on the persistence of their native binding mode.
2.4. Beyond Protein–Ligand Docking: Alternative Strategies for Rational Drug Development
Despite the indisputable relevance of molecular docking within most SARS-CoV-2 drug discovery campaigns, other approaches were successfully implemented, especially for projects which deviate from the design of a standard small molecule noncovalent binder.
For example, Zaidman et al. developed Covalentizer, an automated pipeline for the conversion of noncovalent binders to irreversible ones, which was successfully applied to the conversion of a SARS-CoV Mpro reversible inhibitor to a sub-micromolar SARS-CoV-2 Mpro irreversible one [373].
Valiente et al. reported the discovery of D-peptides that bind the spike RBD with low nanomolar affinity, hence blocking SARS-CoV-2 infection in cell-based assays. These ACE2-mimicking peptides were selected within the starting library through a combination of structural alignment, MD-based post docking refinement, and binding free energy estimation [374].
Similarly, a series of peptides mimicking the HR2 domain of the spike protein able to prevent SARS-CoV-2 infection in cell-based assays with low micromolar potency were designed through the combination between structural alignment, mutational scanning with the BeAtMuSiC [375] tool, and MD-based postdocking refinement [376].
Jeong et al. used Rosetta [377] to rationally design a mAb that recognizes a conserved surface on the spike RBD of various coronaviruses with picomolar binding affinities, thereby strongly inhibiting SARS-CoV-2 replication in cell-based assay [378].
A similar strategy was exploited by Miao et al., which employed Rosetta docking and MD-based postdocking refinement to design an RNA aptamer that binds with picomolar affinity to the spike RBD and inhibits SARS-CoV-2 replication with sub-micromolar potency in cell-based assay [379].
Further, Cao et al. utilized a combination of modeling with Rosetta and docking with RifDock [380] to design ten mini proteins which bind with picomolar affinity to the spike RBD thus inhibiting SARS-CoV-2 infection within cell-based assays [381].
Moreover, Glasgow et al. combined modeling with Rosetta and computational alanine scanning with Robetta [382,383] to rationally design “ACE2 receptor traps”, i.e., engineered proteins that bind the spike RBD with high affinity and neutralize SARS-CoV-2 infection as effectively as clinically used mAbs [384].
As thoroughly discussed in previous paragraphs, many SARS-CoV-2 drug discovery campaigns favored static, time-independent approaches such as docking or structural alignment, over time-dependent methods such as molecular dynamics. This can be attributed to the long calculation times, the reduced conformational sampling capabilities, and the lower accessibility of MD simulations to the general medicinal chemistry audience [331,385]. Despite these issues, several works demonstrated the potential of using full-fledged MD-based drug discovery pipelines, especially when smart enhanced-sampling strategies are employed [385].
For example, Bissaro et al. showed how high-throughput supervised molecular dynamics (HT-SuMD) [386], a virtual screening platform based on an enhanced sampling MD protocol, could be successfully exploited for docking fragments to the active site of SARS-CoV-2 Mpro, overcoming accuracy limitations of most docking protocols [387] in identifying the native-like binding mode for frag-like compounds [388].
Furthermore, the SuMD [389,390] algorithm (Figure 5) was successfully exploited by Pavan et al. to decipher details about the recognition mechanism of Nirmatrelvir upon the SARS-CoV-2 Mpro catalytic site before any structural detail was revealed by the drug developer, with successive structural [189] and molecular medicine [198] studies confirming the prediction validity [391].
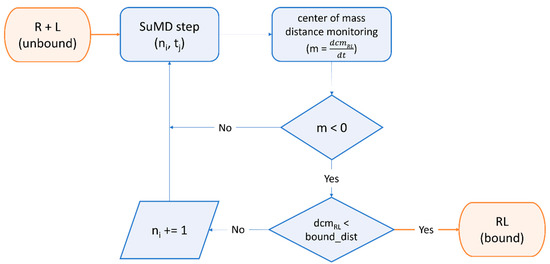
Figure 5.
Workflow of a Supervised Molecular Dynamics (SuMD) simulation. The ligand is dynamically docked within a user-defined binding site through a series of short, unbiased molecular dynamics simulations. At the end of each step, the distance of mass between the ligand and the receptor binding site is computed for each trajectory frame and is fed to a tabu-like algorithm. If the slope of the straight line that interpolates the data is negative, indicating the ligand is approaching the binding site, the step is retained, and the simulation continues with the next “SuMD-step”. If not, the step is discarded and repeated, randomly reassigning particles’ velocities through the Langevin thermostat. This cycle is repeated until a threshold distance is reached or other user-defined termination criteria are met.
Moreover, an evolved version of the SuMD protocol was developed by Pavan et al. and successfully applied to the study of the recognition mechanism between RNA aptamers and proteins, including an RNA-aptamer that binds to the spike RBD with picomolar affinity thus preventing the viral infection of host cells [392].
3. Conclusions and Future Perspectives
Despite an unprecedented vaccination effort, which brought at least one vaccine shot to 70% of the world’s population [101,393], the battle against COVID-19 is far from won. Indeed, there is still a huge disparity between vaccination rates across first-world and low-income countries [101,393]. Furthermore, aside from the vaccines’ availability, several cultural and sociological factors contribute to the worldwide asymmetric vaccination coverage [394,395]. Finally, even in countries with the highest vaccination rates, the continuous emergence of novel viral variants [396] with enhanced immune escape capability sustains the viral spread even among the vaccinated population [397], so that to date a hundred thousand new COVID-19 cases are reported each day, leading, on an average, to a daily toll of hundreds of deaths globally [398]. Although the task of predicting the insurgence of novel variants of concern is not trivial [399], and the debate on the mechanism behind the genesis of these viral variants is still heated [400], it is reasonable to assume, based on the history of COVID-19 so far and other virus-related illnesses such as flu [401,402], that this phenomenon will continue to occur at least into the near future, forcing the scientific community to adapt existing treatments to emerging viral strains, other than developing novel therapeutics complementary to the existing ones [403]. Moreover, even if massive vaccination sensibly lowered the harmful effect on patients’ health caused by acute infection, long-term consequences of COVID-19 infections can still manifest at later stages [404], further reaffirming the need for tools that can effectively treat the disease other than preventing it.
In conclusion, the take-home message from the present pandemic situation is that, among the strategies for identifying new therapeutic classes, with timescales compatible with those marked as a health emergency caused by a shapeshifting pathogen, the integration of structural biology information and new computational approaches probably represents the most promising one. The abundant amount of information provided by structural biologists coupled with the good predictive power of established computational workflows provides a quick platform for finding temporary solutions in the form of drug repurposing, allowing necessary time to develop more specific and tailored therapeutic entities. Although this strategy is not always successful in promoting hit compounds for clinical use [405], it can serve as a rational hypothesis generator for clinical studies, identify molecules to use as pharmacological tools to expand the knowledge on the etiopathogenesis of an emerging illness, and set the basis for the development of derivatives that can overcome the limitations of first-generation hits.
Despite all the scientific advancements in the field of computer-aided drug discovery, indeed, the time required for the release to the market of a new drug has not been sensibly reduced. Indeed, as highlighted in the work of Gupta et al. [406], many active compounds identified through structure-based drug design and computational techniques possess comparable activity to compounds in clinical trials. Many of these compounds, however, despite showing good antiviral activity and having a well-defined mechanism of action, fail to survive clinical stages of experimentation, due to the lack of good pharmacokinetic properties, which are essential for ensuring both good therapeutic efficacy and lack of intolerable side effects.
This fact further stresses the necessity for developing novel and complementary tools to the existing ones, especially in the evaluation of pharmacokinetic properties and off-target effects, which are usually the main causes of failure for candidate drugs in the clinical stages of experimentation. Accordingly, because the presented in silico approaches serve to provide candidates for preliminary selection, to extract the most value from these tools, predictions generated from computational approaches must be verified with biological confirmation, with both in vitro and in vivo models. Furthermore, with the increasing amount of curated experimental datasets becoming available to the scientific community, physics-based methods will be flanked more and more by artificial intelligence methods, both for evaluating the pharmacodynamic and pharmacokinetic properties of investigated compounds [407,408].
Finally, as estimated by a recent study [409], the likelihood of a highly infectious disease epidemic could double in the coming decades, indicating that the successful computational strategies applied in the biology domain that have been adopted against COVID-19 will most likely come in handy soon, providing us with robust and efficient solutions in tackling challenging diseases including new pandemics.
Author Contributions
Conceptualization, M.P.; data curation, M.P.; writing—original draft preparation, M.P.; writing—review and editing, S.M.; supervision, S.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
MMS Lab is very grateful to Chemical Computing Group, OpenEye, and Acellera for their scientific and technical partnership.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lu, H.; Stratton, C.W.; Tang, Y.W. Outbreak of Pneumonia of Unknown Etiology in Wuhan, China: The Mystery and the Miracle. J. Med. Virol. 2020, 92, 401. [Google Scholar] [CrossRef] [PubMed]
- Guarner, J. Three Emerging Coronaviruses in Two Decades: The Story of SARS, MERS, and Now COVID-19. Am. J. Clin. Pathol. 2020, 153, 420–421. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- WHO Director-General’s Remarks at the Media Briefing on 2019-NCoV on 11 February 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020 (accessed on 5 December 2022).
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The Proximal Origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Temmam, S.; Vongphayloth, K.; Salazar, E.B.; Munier, S.; Bonomi, M.; Regnault, B.; Douangboubpha, B.; Karami, Y.; Chrétien, D.; Sanamxay, D.; et al. Bat Coronaviruses Related to SARS-CoV-2 and Infectious for Human Cells. Nature 2022, 604, 330–336. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical Course and Risk Factors for Mortality of Adult Inpatients with COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Lauer, S.A.; Grantz, K.H.; Bi, Q.; Jones, F.K.; Zheng, Q.; Meredith, H.R.; Azman, A.S.; Reich, N.G.; Lessler, J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) from Publicly Reported Confirmed Cases: Estimation and Application. Ann. Intern. Med. 2020, 172, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Lau, E.H.Y.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; et al. Temporal Dynamics in Viral Shedding and Transmissibility of COVID-19. Nat. Med. 2020, 26, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Rothan, H.A.; Byrareddy, S.N. The Epidemiology and Pathogenesis of Coronavirus Disease (COVID-19) Outbreak. J. Autoimmun. 2020, 109, 102433. [Google Scholar] [CrossRef] [PubMed]
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 324, 782–793. [Google Scholar] [CrossRef]
- Petersen, E.; Koopmans, M.; Go, U.; Hamer, D.H.; Petrosillo, N.; Castelli, F.; Storgaard, M.; Al Khalili, S.; Simonsen, L. Comparing SARS-CoV-2 with SARS-CoV and Influenza Pandemics. Lancet Infect. Dis.. 2020, 20, e238–e244. [Google Scholar] [CrossRef]
- COVID Live—Coronavirus Statistics—Worldometer. Available online: https://www.worldometers.info/coronavirus/ (accessed on 5 December 2022).
- Chu, D.K.; Akl, E.A.; Duda, S.; Solo, K.; Yaacoub, S.; Schünemann, H.J.; El-harakeh, A.; Bognanni, A.; Lotfi, T.; Loeb, M.; et al. Physical Distancing, Face Masks, and Eye Protection to Prevent Person-to-Person Transmission of SARS-CoV-2 and COVID-19: A Systematic Review and Meta-Analysis. Lancet 2020, 395, 1973–1987. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Freedman, D.O. Isolation, Quarantine, Social Distancing and Community Containment: Pivotal Role for Old-Style Public Health Measures in the Novel Coronavirus (2019-NCoV) Outbreak. J. Travel Med. 2020, 27, 1–4. [Google Scholar] [CrossRef]
- Remuzzi, A.; Remuzzi, G. COVID-19 and Italy: What Next? Lancet 2020, 395, 1225–1228. [Google Scholar] [CrossRef]
- Ranney, M.L.; Griffeth, V.; Jha, A.K. Critical Supply Shortages—The Need for Ventilators and Personal Protective Equipment during the COVID-19 Pandemic. N. Engl. J. Med. 2020, 382, e41. [Google Scholar] [CrossRef]
- Grasselli, G.; Pesenti, A.; Cecconi, M. Critical Care Utilization for the COVID-19 Outbreak in Lombardy, Italy: Early Experience and Forecast During an Emergency Response. JAMA 2020, 323, 1545–1546. [Google Scholar] [CrossRef]
- Pan, A.; Liu, L.; Wang, C.; Guo, H.; Hao, X.; Wang, Q.; Huang, J.; He, N.; Yu, H.; Lin, X.; et al. Association of Public Health Interventions With the Epidemiology of the COVID-19 Outbreak in Wuhan, China. JAMA 2020, 323, 1915–1923. [Google Scholar] [CrossRef] [PubMed]
- Giordano, G.; Blanchini, F.; Bruno, R.; Colaneri, P.; di Filippo, A.; di Matteo, A.; Colaneri, M. Modelling the COVID-19 Epidemic and Implementation of Population-Wide Interventions in Italy. Nat. Med. 2020, 26, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Nicola, M.; Alsafi, Z.; Sohrabi, C.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, M.; Agha, R. The Socio-Economic Implications of the Coronavirus Pandemic (COVID-19): A Review. Int. J. Surg. 2020, 78, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, R.P. COVID-19 and Mental Health: A Review of the Existing Literature. Asian J. Psychiatry 2020, 52, 102066. [Google Scholar] [CrossRef]
- Emanuel, E.J.; Persad, G.; Upshur, R.; Thome, B.; Parker, M.; Glickman, A.; Zhang, C.; Boyle, C.; Smith, M.; Phillips, J.P. Fair Allocation of Scarce Medical Resources in the Time of COVID-19. N. Engl. J. Med. 2020, 382, 2049–2055. [Google Scholar] [CrossRef]
- Schlander, M.; Hernandez-Villafuerte, K.; Cheng, C.Y.; Mestre-Ferrandiz, J.; Baumann, M. How Much Does It Cost to Research and Develop a New Drug? A Systematic Review and Assessment. Pharmacoeconomics 2021, 39, 1243–1269. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, Q.; Li, Y.; Garner, L.V.; Watkins, S.P.; Carter, L.J.; Smoot, J.; Gregg, A.C.; Daniels, A.D.; Jervey, S.; et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent. Sci. 2020, 6, 315–331. [Google Scholar] [CrossRef]
- Tu, Y.F.; Chien, C.S.; Yarmishyn, A.A.; Lin, Y.Y.; Luo, Y.H.; Lin, Y.T.; Lai, W.Y.; Yang, D.M.; Chou, S.J.; Yang, Y.P.; et al. A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Int. J. Mol. Sci. 2020, 21, 2657. [Google Scholar] [CrossRef]
- Ashburn, T.T.; Thor, K.B. Drug Repositioning: Identifying and Developing New Uses for Existing Drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discov. 2018, 18, 41–58. [Google Scholar] [CrossRef]
- Mani, D.; Wadhwani, A.; Krishnamurthy, P.T. Drug Repurposing in Antiviral Research: A Current Scenario. J. Young Pharm. 2019, 11, 117–121. [Google Scholar] [CrossRef]
- Viveiros Rosa, S.G.; Santos, W.C. Clinical Trials on Drug Repositioning for COVID-19 Treatment. Pan Am. J. Public Health 2020, 44, e40. [Google Scholar] [CrossRef]
- Treatments to Be Used in COVID-19 Patients in a Hospital Setting. Available online: https://www.aifa.gov.it/documents/20142/1307084/Informativa_hcp.pdf (accessed on 5 December 2022).
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir–Ritonavir in Adults Hospitalized with Severe COVID-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Bolcato, G.; Bissaro, M.; Pavan, M.; Sturlese, M.; Moro, S. Targeting the Coronavirus SARS-CoV-2: Computational Insights into the Mechanism of Action of the Protease Inhibitors Lopinavir, Ritonavir and Nelfinavir. Sci. Rep. 2020, 10, 20927. [Google Scholar] [CrossRef] [PubMed]
- Gautret, P.; Lagier, J.C.; Parola, P.; Hoang, V.T.; Meddeb, L.; Mailhe, M.; Doudier, B.; Courjon, J.; Giordanengo, V.; Vieira, V.E.; et al. Hydroxychloroquine and Azithromycin as a Treatment of COVID-19: Results of an Open-Label Non-Randomized Clinical Trial. Int. J. Antimicrob. Agents 2020, 56, 105949. [Google Scholar] [CrossRef]
- Arshad, S.; Kilgore, P.; Chaudhry, Z.S.; Jacobsen, G.; Wang, D.D.; Huitsing, K.; Brar, I.; Alangaden, G.J.; Ramesh, M.S.; McKinnon, J.E.; et al. Treatment with Hydroxychloroquine, Azithromycin, and Combination in Patients Hospitalized with COVID-19. Int. J. Infect. Dis. 2020, 97, 396–403. [Google Scholar] [CrossRef]
- Recovery Collaborative Group. Dexamethasone in Hospitalized Patients with COVID-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar] [CrossRef]
- Tang, N.; Bai, H.; Chen, X.; Gong, J.; Li, D.; Sun, Z. Anticoagulant Treatment Is Associated with Decreased Mortality in Severe Coronavirus Disease 2019 Patients with Coagulopathy. J. Thromb. Haemost. 2020, 18, 1094–1099. [Google Scholar] [CrossRef]
- Giamarellos-Bourboulis, E.J.; Netea, M.G.; Rovina, N.; Akinosoglou, K.; Antoniadou, A.; Antonakos, N.; Damoraki, G.; Gkavogianni, T.; Adami, M.E.; Katsaounou, P.; et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe 2020, 27, 992–1000.e3. [Google Scholar] [CrossRef]
- Moore, J.B.; June, C.H. Cytokine Release Syndrome in Severe COVID-19. Science 2020, 368, 473–474. [Google Scholar] [CrossRef]
- Xu, X.; Han, M.; Li, T.; Sun, W.; Wang, D.; Fu, B.; Zhou, Y.; Zheng, X.; Yang, Y.; Li, X.; et al. Effective Treatment of Severe COVID-19 Patients with Tocilizumab. Proc. Natl. Acad. Sci. USA 2020, 117, 10970–10975. [Google Scholar] [CrossRef]
- Remap-Cap Investigators. Interleukin-6 Receptor Antagonists in Critically Ill Patients with COVID-19. N. Engl. J. Med. 2021, 384, 1491–1502. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; de Luca, G.; Campochiaro, C.; Della-Torre, E.; Ripa, M.; Canetti, D.; Oltolini, C.; Castiglioni, B.; Tassan Din, C.; Boffini, N.; et al. Interleukin-1 Blockade with High-Dose Anakinra in Patients with COVID-19, Acute Respiratory Distress Syndrome, and Hyperinflammation: A Retrospective Cohort Study. Lancet Rheumatol. 2020, 2, e325–e331. [Google Scholar] [CrossRef] [PubMed]
- Marconi, V.C.; Ramanan, A.V.; de Bono, S.; Kartman, C.E.; Krishnan, V.; Liao, R.; Piruzeli, M.L.B.; Goldman, J.D.; Alatorre-Alexander, J.; de Cassia Pellegrini, R.; et al. Efficacy and Safety of Baricitinib for the Treatment of Hospitalised Adults with COVID-19 (COV-BARRIER): A Randomised, Double-Blind, Parallel-Group, Placebo-Controlled Phase 3 Trial. Lancet Respir. Med. 2021, 9, 1407–1418. [Google Scholar] [CrossRef]
- Kalil, A.C.; Patterson, T.F.; Mehta, A.K.; Tomashek, K.M.; Wolfe, C.R.; Ghazaryan, V.; Marconi, V.C.; Ruiz-Palacios, G.M.; Hsieh, L.; Kline, S.; et al. Baricitinib plus Remdesivir for Hospitalized Adults with COVID-19. N. Engl. J. Med. 2021, 384, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Rubin, D.; Chan-Tack, K.; Farley, J.; Sherwat, A. FDA Approval of Remdesivir—A Step in the Right Direction. N. Engl. J. Med. 2020, 383, 2598–2600. [Google Scholar] [CrossRef] [PubMed]
- Malin, J.J.; Suárez, I.; Priesner, V.; Fätkenheuer, G.; Rybniker, J. Remdesivir against COVID-19 and Other Viral Diseases. Clin. Microbiol. Rev. 2021, 34, e00162-20. [Google Scholar] [CrossRef]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of COVID-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef]
- Chen, L.; Xiong, J.; Bao, L.; Shi, Y. Convalescent Plasma as a Potential Therapy for COVID-19. Lancet Infect. Dis. 2020, 20, 398–400. [Google Scholar] [CrossRef]
- Use of Convalescent Whole Blood or Plasma Collected from Patients Recovered from Ebola Virus Disease for Transfusion, as an Empirical Treatment during Outbreaks: Interim Guidance for National Health Authorities and Blood Transfusion Services. Available online: https://apps.who.int/iris/handle/10665/135591 (accessed on 6 December 2022).
- Arabi, Y.; Balkhy, H.; Hajeer, A.H.; Bouchama, A.; Hayden, F.G.; Al-Omari, A.; Al-Hameed, F.M.; Taha, Y.; Shindo, N.; Whitehead, J.; et al. Feasibility, Safety, Clinical, and Laboratory Effects of Convalescent Plasma Therapy for Patients with Middle East Respiratory Syndrome Coronavirus Infection: A Study Protocol. Springerplus 2015, 4, 709. [Google Scholar] [CrossRef]
- Duan, K.; Liu, B.; Li, C.; Zhang, H.; Yu, T.; Qu, J.; Zhou, M.; Chen, L.; Meng, S.; Hu, Y.; et al. Effectiveness of Convalescent Plasma Therapy in Severe COVID-19 Patients. Proc. Natl. Acad. Sci. USA 2020, 117, 9490–9496. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, W.; Hu, Y.; Tong, X.; Zheng, S.; Yang, J.; Kong, Y.; Ren, L.; Wei, Q.; Mei, H.; et al. Effect of Convalescent Plasma Therapy on Time to Clinical Improvement in Patients with Severe and Life-Threatening COVID-19: A Randomized Clinical Trial. JAMA 2020, 324, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Simonovich, V.A.; Burgos Pratx, L.D.; Scibona, P.; Beruto, M.V.; Vallone, M.G.; Vázquez, C.; Savoy, N.; Giunta, D.H.; Pérez, L.G.; Sánchez, M.d.L.; et al. A Randomized Trial of Convalescent Plasma in COVID-19 Severe Pneumonia. N. Engl. J. Med. 2021, 384, 619–629. [Google Scholar] [CrossRef]
- Pinto, D.; Park, Y.J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; de Marco, A.; et al. Cross-Neutralization of SARS-CoV-2 by a Human Monoclonal SARS-CoV Antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent Binding of 2019 Novel Coronavirus Spike Protein by a SARS Coronavirus-Specific Human Monoclonal Antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Use of Monoclonal Antibodies for COVID-19 | Italian Medicines Agency. Available online: https://www.aifa.gov.it/en/uso-degli-anticorpi-monoclonali (accessed on 6 December 2022).
- Corti, D.; Purcell, L.A.; Snell, G.; Veesler, D. Tackling COVID-19 with Neutralizing Monoclonal Antibodies. Cell 2021, 184, 3086–3108. [Google Scholar] [CrossRef] [PubMed]
- Kaplon, H.; Reichert, J.M. Antibodies to Watch in 2021. MAbs 2021, 13, 1860476. [Google Scholar] [CrossRef]
- Kaplon, H.; Chenoweth, A.; Crescioli, S.; Reichert, J.M. Antibodies to Watch in 2022. MAbs 2022, 14, 2014296. [Google Scholar] [CrossRef] [PubMed]
- Zost, S.J.; Gilchuk, P.; Case, J.B.; Binshtein, E.; Chen, R.E.; Nkolola, J.P.; Schäfer, A.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; et al. Potently Neutralizing and Protective Human Antibodies against SARS-CoV-2. Nature 2020, 584, 443–449. [Google Scholar] [CrossRef]
- Thanh Le, T.; Andreadakis, Z.; Kumar, A.; Gómez Román, R.; Tollefsen, S.; Saville, M.; Mayhew, S. The COVID-19 Vaccine Development Landscape. Nat. Rev. Drug Discov. 2020, 19, 305–306. [Google Scholar] [CrossRef]
- Kashte, S.; Gulbake, A.; El-Amin, S.F.; Gupta, A. COVID-19 Vaccines: Rapid Development, Implications, Challenges and Future Prospects. Hum. Cell 2021, 34, 711–733. [Google Scholar] [CrossRef] [PubMed]
- Pollard, A.J.; Bijker, E.M. A Guide to Vaccinology: From Basic Principles to New Developments. Nat. Rev. Immunol. 2021, 21, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Mascellino, M.T.; di Timoteo, F.; de Angelis, M.; Oliva, A. Overview of the Main Anti-SARS-CoV-2 Vaccines: Mechanism of Action, Efficacy and Safety. Infect. Drug Resist. 2021, 14, 3459–3476. [Google Scholar] [CrossRef] [PubMed]
- Doroftei, B.; Ciobica, A.; Ilie, O.D.; Maftei, R.; Ilea, C. Mini-Review Discussing the Reliability and Efficiency of COVID-19 Vaccines. Diagnostics 2021, 11, 579. [Google Scholar] [CrossRef]
- Jara, A.; Undurraga, E.A.; González, C.; Paredes, F.; Fontecilla, T.; Jara, G.; Pizarro, A.; Acevedo, J.; Leo, K.; Leon, F.; et al. Effectiveness of an Inactivated SARS-CoV-2 Vaccine in Chile. N. Engl. J. Med. 2021, 385, 875–884. [Google Scholar] [CrossRef]
- Tanriover, M.D.; Doğanay, H.L.; Akova, M.; Güner, H.R.; Azap, A.; Akhan, S.; Köse, Ş.; Erdinç, F.Ş.; Akalın, E.H.; Tabak, Ö.F.; et al. Efficacy and Safety of an Inactivated Whole-Virion SARS-CoV-2 Vaccine (CoronaVac): Interim Results of a Double-Blind, Randomised, Placebo-Controlled, Phase 3 Trial in Turkey. Lancet 2021, 398, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Baden, L.R.; el Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the MRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Lwoff, A.; Tournier, P. The Classification of Viruses. Annu. Rev. Microbiol. 2003, 20, 45–74. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Gadanec, L.K.; McSweeney, K.R.; Qaradakhi, T.; Ali, B.; Zulli, A.; Apostolopoulos, V. Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells? Int. J. Mol. Sci. 2021, 22, 992. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Zhang, J.; Fang, Y.; Lu, S.; Wu, J.; Zheng, X.; Deng, F. SARS-CoV-2 Interacts with Platelets and Megakaryocytes via ACE2-Independent Mechanism. J. Hematol. Oncol. 2021, 14, 72. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Hamming, I.; Timens, W.; Bulthuis, M.; Lely, A.; Navis, G.; van Goor, H. Tissue Distribution of ACE2 Protein, the Functional Receptor for SARS Coronavirus. A First Step in Understanding SARS Pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Glowacka, I.; Bertram, S.; Müller, M.A.; Allen, P.; Soilleux, E.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence That TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J. Virol. 2011, 85, 4122–4134. [Google Scholar] [CrossRef]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Guillén, J.; Pérez-Berná, A.J.; Moreno, M.R.; Villalaín, J. Identification of the Membrane-Active Regions of the Severe Acute Respiratory Syndrome Coronavirus Spike Membrane Glycoprotein Using a 16/18-Mer Peptide Scan: Implications for the Viral Fusion Mechanism. J. Virol. 2005, 79, 1743–1752. [Google Scholar] [CrossRef]
- Zhu, C.; He, G.; Yin, Q.; Zeng, L.; Ye, X.; Shi, Y.; Xu, W. Molecular Biology of the SARS-CoV-2 Spike Protein: A Review of Current Knowledge. J. Med. Virol. 2021, 93, 5729–5741. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and Functional Properties of SARS-CoV-2 Spike Protein: Potential Antivirus Drug Development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Schroth-Diez, B.; Ludwig, K.; Baljinnyam, B.; Kozerski, C.; Huang, Q.; Herrmann, A. The Role of the Transmembrane and of the Intraviral Domain of Glycoproteins in Membrane Fusion of Enveloped Viruses. Biosci. Rep. 2000, 20, 571–595. [Google Scholar] [CrossRef] [PubMed]
- Reuven, E.M.; Dadon, Y.; Viard, M.; Manukovsky, N.; Blumenthal, R.; Shai, Y. HIV-1 Gp41 Transmembrane Domain Interacts with the Fusion Peptide: Implication in Lipid Mixing and Inhibition of Virus-Cell Fusion. Biochemistry 2012, 51, 2867–2878. [Google Scholar] [CrossRef]
- Petit, C.M.; Melancon, J.M.; Chouljenko, V.N.; Colgrove, R.; Farzan, M.; Knipe, D.M.; Kousoulas, K.G. Genetic Analysis of the SARS-Coronavirus Spike Glycoprotein Functional Domains Involved in Cell-Surface Expression and Cell-to-Cell Fusion. Virology 2005, 341, 215–230. [Google Scholar] [CrossRef]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Forster, P.; Forster, L.; Renfrew, C.; Forster, M. Phylogenetic Network Analysis of SARS-CoV-2 Genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 9241–9243. [Google Scholar] [CrossRef]
- Oran, D.P.; Topol, E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection. Ann. Intern. Med. 2020, 173, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Hellewell, J.; Abbott, S.; Gimma, A.; Bosse, N.I.; Jarvis, C.I.; Russell, T.W.; Munday, J.D.; Kucharski, A.J.; Edmunds, W.J.; Sun, F.; et al. Feasibility of Controlling COVID-19 Outbreaks by Isolation of Cases and Contacts. Lancet Glob. Health 2020, 8, e488–e496. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Lam, E.C.; St. Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 Variants Escape Neutralization by Vaccine-Induced Humoral Immunity. Cell 2021, 184, 2372–2383.e9. [Google Scholar] [CrossRef] [PubMed]
- Fiolet, T.; Kherabi, Y.; MacDonald, C.J.; Ghosn, J.; Peiffer-Smadja, N. Comparing COVID-19 Vaccines for Their Characteristics, Efficacy and Effectiveness against SARS-CoV-2 and Variants of Concern: A Narrative Review. Clin. Microbiol. Infect. 2022, 28, 202–221. [Google Scholar] [CrossRef]
- Randolph, H.E.; Barreiro, L.B. Herd Immunity: Understanding COVID-19. Immunity 2020, 52, 737–741. [Google Scholar] [CrossRef]
- Mathieu, E.; Ritchie, H.; Ortiz-Ospina, E.; Roser, M.; Hasell, J.; Appel, C.; Giattino, C.; Rodés-Guirao, L. A Global Database of COVID-19 Vaccinations. Nat. Hum. Behav. 2021, 5, 947–953, Nat. Hum. Behav.2021, 5, 956–959. [Google Scholar] [CrossRef]
- Aksamentov, I.; Roemer, C.; Hodcroft, E.B.; Neher, R.A. Nextclade: Clade Assignment, Mutation Calling and Quality Control for Viral Genomes. J. Open Source Softw. 2021, 6, 3773. [Google Scholar] [CrossRef]
- Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 7 December 2022).
- Frampton, D.; Rampling, T.; Cross, A.; Bailey, H.; Heaney, J.; Byott, M.; Scott, R.; Sconza, R.; Price, J.; Margaritis, M.; et al. Genomic Characteristics and Clinical Effect of the Emergent SARS-CoV-2 B.1.1.7 Lineage in London, UK: A Whole-Genome Sequencing and Hospital-Based Cohort Study. Lancet Infect. Dis. 2021, 21, 1246. [Google Scholar] [CrossRef]
- Campbell, F.; Archer, B.; Laurenson-Schafer, H.; Jinnai, Y.; Konings, F.; Batra, N.; Pavlin, B.; Vandemaele, K.; van Kerkhove, M.D.; Jombart, T.; et al. Increased Transmissibility and Global Spread of SARSCoV- 2 Variants of Concern as at June 2021. Eurosurveillance 2021, 26, 2100509. [Google Scholar] [CrossRef]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated Transmissibility and Impact of SARS-CoV-2 Lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef]
- Volz, E.; Mishra, S.; Chand, M.; Barrett, J.C.; Johnson, R.; Geidelberg, L.; Hinsley, W.R.; Laydon, D.J.; Dabrera, G.; O’Toole, Á.; et al. Assessing Transmissibility of SARS-CoV-2 Lineage B.1.1.7 in England. Nature 2021, 593, 266–269. [Google Scholar] [CrossRef]
- Collier, D.A.; de Marco, A.; Ferreira, I.A.T.M.; Meng, B.; Datir, R.P.; Walls, A.C.; Kemp, S.A.; Bassi, J.; Pinto, D.; Silacci-Fregni, C.; et al. Sensitivity of SARS-CoV-2 B.1.1.7 to MRNA Vaccine-Elicited Antibodies. Nature 2021, 593, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 Variants to Neutralization by Monoclonal and Serum-Derived Polyclonal Antibodies. Nat. Med. 2021, 27, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Haas, E.J.; Angulo, F.J.; McLaughlin, J.M.; Anis, E.; Singer, S.R.; Khan, F.; Brooks, N.; Smaja, M.; Mircus, G.; Pan, K.; et al. Impact and Effectiveness of MRNA BNT162b2 Vaccine against SARS-CoV-2 Infections and COVID-19 Cases, Hospitalisations, and Deaths Following a Nationwide Vaccination Campaign in Israel: An Observational Study Using National Surveillance Data. Lancet 2021, 397, 1819–1829. [Google Scholar] [CrossRef]
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of COVID-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced Sensitivity of SARS-CoV-2 Variant Delta to Antibody Neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef]
- Liu, C.; Ginn, H.M.; Dejnirattisai, W.; Supasa, P.; Wang, B.; Tuekprakhon, A.; Nutalai, R.; Zhou, D.; Mentzer, A.J.; Zhao, Y.; et al. Reduced Neutralization of SARS-CoV-2 B.1.617 by Vaccine and Convalescent Serum. Cell 2021, 184, 4220–4236.e13. [Google Scholar] [CrossRef]
- Mlcochova, P.; Kemp, S.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.T.M.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta Variant Replication and Immune Evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef]
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital Admission and Emergency Care Attendance Risk for SARS-CoV-2 Delta (B.1.617.2) Compared with Alpha (B.1.1.7) Variants of Concern: A Cohort Study. Lancet Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef]
- Doboszewski, B.; Groaz, E.; Herdewijn, P. Synthesis of Phosphonoglycine Backbone Units for the Development of Phosphono Peptide Nucleic Acids. Eur. J. Org. Chem. 2013, 2013, 4804–4815. [Google Scholar] [CrossRef]
- Araf, Y.; Akter, F.; Tang, Y.D.; Fatemi, R.; Parvez, M.S.A.; Zheng, C.; Hossain, M.G. Omicron Variant of SARS-CoV-2: Genomics, Transmissibility, and Responses to Current COVID-19 Vaccines. J. Med. Virol. 2022, 94, 1825–1832. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Iketani, S.; Guo, Y.; Chan, J.F.W.; Wang, M.; Liu, L.; Luo, Y.; Chu, H.; Huang, Y.; Nair, M.S.; et al. Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2. Nature 2022, 602, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Dejnirattisai, W.; Shaw, R.H.; Supasa, P.; Liu, C.; Stuart, A.S.; Pollard, A.J.; Liu, X.; Lambe, T.; Crook, D.; Stuart, D.I.; et al. Reduced Neutralisation of SARS-CoV-2 Omicron B.1.1.529 Variant by Post-Immunisation Serum. Lancet 2022, 399, 234–236. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron Escapes the Majority of Existing SARS-CoV-2 Neutralizing Antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Krüger, N.; Schulz, S.; Cossmann, A.; Rocha, C.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Moldenhauer, A.S.; Winkler, M.S.; et al. The Omicron Variant Is Highly Resistant against Antibody-Mediated Neutralization: Implications for Control of the COVID-19 Pandemic. Cell 2022, 185, 447–456.e11. [Google Scholar] [CrossRef]
- Nemet, I.; Kliker, L.; Lustig, Y.; Zuckerman, N.; Erster, O.; Cohen, C.; Kreiss, Y.; Alroy-Preis, S.; Regev-Yochay, G.; Mendelson, E.; et al. Third BNT162b2 Vaccination Neutralization of SARS-CoV-2 Omicron Infection. N. Engl. J. Med. 2022, 386, 492–494. [Google Scholar] [CrossRef]
- Li, Q.; Groaz, E.; Rocha-Pereira, J.; Neyts, J.; Herdewijn, P. Anti-norovirus activity of C7-modified 4-amino-pyrrolo[2,1-f][1,2,4]triazine C-nucleosides. Eur J. Med. Chem. 2020, 195, 112198. [Google Scholar] [CrossRef]
- Ou, J.; Lan, W.; Wu, X.; Zhao, T.; Duan, B.; Yang, P.; Ren, Y.; Quan, L.; Zhao, W.; Seto, D.; et al. Tracking SARS-CoV-2 Omicron Diverse Spike Gene Mutations Identifies Multiple Inter-Variant Recombination Events. Signal Transduct. Target Ther. 2022, 7, 138. [Google Scholar] [CrossRef]
- Kimura, I.; Yamasoba, D.; Tamura, T.; Nao, N.; Suzuki, T.; Oda, Y.; Mitoma, S.; Ito, J.; Nasser, H.; Zahradnik, J.; et al. Virological Characteristics of the SARS-CoV-2 Omicron BA.2 Subvariants, Including BA.4 and BA.5. Cell 2022, 185, 3992–4007.e16. [Google Scholar] [CrossRef]
- Wilhelm, A.; Widera, M.; Grikscheit, K.; Toptan, T.; Schenk, B.; Pallas, C.; Metzler, M.; Kohmer, N.; Hoehl, S.; Marschalek, R.; et al. Limited Neutralisation of the SARS-CoV-2 Omicron Subvariants BA.1 and BA.2 by Convalescent and Vaccine Serum and Monoclonal Antibodies. EBioMedicine 2022, 82, 104158. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Zhang, L.; Rocha, C.; Sidarovich, A.; Kempf, A.; Schulz, S.; Cossmann, A.; Manger, B.; Baier, E.; Tampe, B.; et al. Comparable Neutralisation Evasion of SARS-CoV-2 Omicron Subvariants BA.1, BA.2, and BA.3. Lancet Infect. Dis. 2022, 22, 766–767. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.P.; Zeng, C.; Qu, P.; Faraone, J.; Zheng, Y.M.; Carlin, C.; Bednash, J.S.; Zhou, T.; Lozanski, G.; Mallampalli, R.; et al. Neutralization of SARS-CoV-2 Omicron Sub-Lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe 2022, 30, 1093–1102.e3. [Google Scholar] [CrossRef] [PubMed]
- Hachmann, N.P.; Miller, J.; Collier, A.Y.; Ventura, J.D.; Yu, J.; Rowe, M.; Bondzie, E.A.; Powers, O.; Surve, N.; Hall, K.; et al. Neutralization Escape by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022, 387, 86–88. [Google Scholar] [CrossRef]
- Arora, P.; Kempf, A.; Nehlmeier, I.; Schulz, S.R.; Cossmann, A.; Stankov, M.V.; Jäck, H.M.; Behrens, G.M.N.; Pöhlmann, S.; Hoffmann, M. Augmented Neutralisation Resistance of Emerging Omicron Subvariants BA.2.12.1, BA.4, and BA.5. Lancet Infect. Dis. 2022, 22, 1117–1118. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, Y.; Iketani, S.; Nair, M.S.; Li, Z.; Mohri, H.; Wang, M.; Yu, J.; Bowen, A.D.; Chang, J.Y.; et al. Antibody Evasion by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4 and BA.5. Nature 2022, 608, 603–608. [Google Scholar] [CrossRef]
- Takashita, E.; Yamayoshi, S.; Simon, V.; van Bakel, H.; Sordillo, E.M.; Pekosz, A.; Fukushi, S.; Suzuki, T.; Maeda, K.; Halfmann, P.; et al. Efficacy of Antibodies and Antiviral Drugs against Omicron BA.2.12.1, BA.4, and BA.5 Subvariants. N. Engl. J. Med. 2022, 387, 468–470. [Google Scholar] [CrossRef]
- Qu, P.; Faraone, J.; Evans, J.P.; Zou, X.; Zheng, Y.-M.; Carlin, C.; Bednash, J.S.; Lozanski, G.; Mallampalli, R.K.; Saif, L.J.; et al. Neutralization of the SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 Subvariants. N. Engl. J. Med. 2022, 386, 2526–2528. [Google Scholar] [CrossRef]
- Pavan, M.; Bassani, D.; Sturlese, M.; Moro, S. From the Wuhan-Hu-1 Strain to the XD and XE Variants: Is Targeting the SARS-CoV-2 Spike Protein Still a Pharmaceutically Relevant Option against COVID-19? J. Enzyme Inhib. Med. Chem. 2022, 37, 1704–1714. [Google Scholar] [CrossRef]
- Zheng, Z.; Groaz, E.; Snoeck, R.; De Jonghe, S.; Herdewijn, P.; Andrei, G. Influence of 4′-Substitution on the Activity of Gemcitabine and Its ProTide Against VZV and SARS-CoV-2. ACS Med. Chem. Lett. 2020, 12, 88–92. [Google Scholar] [CrossRef]
- Ullrich, S.; Nitsche, C. The SARS-CoV-2 Main Protease as Drug Target. Bioorg. Med. Chem. Lett. 2020, 30, 127377. [Google Scholar] [CrossRef]
- Anand, K.; Palm, G.J.; Mesters, J.R.; Siddell, S.G.; Ziebuhr, J.; Hilgenfeld, R. Structure of Coronavirus Main Proteinase Reveals Combination of a Chymotrypsin Fold with an Extra Alpha-Helical Domain. EMBO J. 2002, 21, 3213–3224. [Google Scholar] [CrossRef] [PubMed]
- Anand, K.; Ziebuhr, J.; Wadhwani, P.; Mesters, J.R.; Hilgenfeld, R. Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of Anti-SARS Drugs. Science 2003, 300, 1763–1767. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Yu, H.; Yang, H.; Xue, F.; Wu, Z.; Shen, W.; Li, J.; Zhou, Z.; Ding, Y.; Zhao, Q.; et al. Structures of Two Coronavirus Main Proteases: Implications for Substrate Binding and Antiviral Drug Design. J. Virol. 2008, 82, 2515–2527. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.L.; Cheng, S.C.; Shi, L.; Wang, T.Y.; Ho, K.I.; Chou, C.Y. Critical Assessment of the Important Residues Involved in the Dimerization and Catalysis of MERS Coronavirus Main Protease. PLoS ONE 2015, 10, e144865. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Verschueren, K.H.G.; Anand, K.; Shen, J.; Yang, M.; Xu, Y.; Rao, Z.; Bigalke, J.; Heisen, B.; Mesters, J.R.; et al. PH-Dependent Conformational Flexibility of the SARS-CoV Main Proteinase (M(pro)) Dimer: Molecular Dynamics Simulations and Multiple X-Ray Structure Analyses. J. Mol. Biol. 2005, 354, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Kang, X. Activation and Maturation of SARS-CoV Main Protease. Protein Cell 2011, 2, 282–290. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Snijder, E.J.; Decroly, E.; Ziebuhr, J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016, 96, 59–126. [Google Scholar] [CrossRef]
- Chen, H.; Wei, P.; Huang, C.; Tan, L.; Liu, Y.; Lai, L. Only One Protomer Is Active in the Dimer of SARS 3C-like Proteinase. J. Biol. Chem. 2006, 281, 13894–13898. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal Structure of SARS-CoV-2 Main Protease Provides a Basis for Design of Improved α-Ketoamide Inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed]
- El-Baba, T.J.; Lutomski, C.A.; Kantsadi, A.L.; Malla, T.R.; John, T.; Mikhailov, V.; Bolla, J.R.; Schofield, C.J.; Zitzmann, N.; Vakonakis, I.; et al. Allosteric Inhibition of the SARS-CoV-2 Main Protease: Insights from Mass Spectrometry Based Assays. Angew. Chem. Int. Ed. Engl. 2020, 59, 23544–23548. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.F.; Kuo, C.J.; Chang, K.T.; Chang, H.C.; Chou, C.C.; Ko, T.P.; Shr, H.L.; Chang, G.G.; Wang, A.H.J.; Liang, P.H. Mechanism of the Maturation Process of SARS-CoV 3CL Protease. J. Biol. Chem. 2005, 280, 31257–31266. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Teng, X.; Qi, Y.; Tang, B.; Shi, H.; Ma, X.; Lai, L. Conformational Flexibility of a Short Loop near the Active Site of the SARS-3CLpro Is Essential to Maintain Catalytic Activity. Sci. Rep. 2016, 6, 20918. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Song, J. The Catalysis of the SARS 3C-like Protease Is under Extensive Regulation by Its Extra Domain. FEBS J. 2006, 273, 1035–1045. [Google Scholar] [CrossRef]
- Fornasier, E.; Macchia, M.L.; Giachin, G.; Sosic, A.; Pavan, M.; Sturlese, M.; Salata, C.; Moro, S.; Gatto, B.; Bellanda, M.; et al. A New Inactive Conformation of SARS-CoV-2 Main Protease. Acta Crystallogr. D Struct. Biol. 2022, 78, 363–378. [Google Scholar] [CrossRef]
- Cheng, S.C.; Chang, G.G.; Chou, C.Y. Mutation of Glu-166 Blocks the Substrate-Induced Dimerization of SARS Coronavirus Main Protease. Biophys. J. 2010, 98, 1327–1336. [Google Scholar] [CrossRef]
- Yang, H.; Yang, M.; Ding, Y.; Liu, Y.; Lou, Z.; Zhou, Z.; Sun, L.; Mo, L.; Ye, S.; Pang, H.; et al. The Crystal Structures of Severe Acute Respiratory Syndrome Virus Main Protease and Its Complex with an Inhibitor. Proc. Natl. Acad. Sci. USA 2003, 100, 13190–13195. [Google Scholar] [CrossRef]
- Verschueren, K.H.G.; Pumpor, K.; Anemüller, S.; Chen, S.; Mesters, J.R.; Hilgenfeld, R. A Structural View of the Inactivation of the SARS Coronavirus Main Proteinase by Benzotriazole Esters. Chem. Biol. 2008, 15, 597–606. [Google Scholar] [CrossRef]
- Zhong, N.; Zhang, S.; Zou, P.; Chen, J.; Kang, X.; Li, Z.; Liang, C.; Jin, C.; Xia, B. Without Its N-Finger, the Main Protease of Severe Acute Respiratory Syndrome Coronavirus Can Form a Novel Dimer through Its C-Terminal Domain. J. Virol. 2008, 82, 4227–4234. [Google Scholar] [CrossRef]
- Kneller, D.W.; Galanie, S.; Phillips, G.; O’Neill, H.M.; Coates, L.; Kovalevsky, A. Malleability of the SARS-CoV-2 3CL Mpro Active-Site Cavity Facilitates Binding of Clinical Antivirals. Structure 2020, 28, 1313–1320.e3. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhu, Y.; Liu, X.; Jin, Z.; Duan, Y.; Zhang, Q.; Wu, C.; Feng, L.; Du, X.; Zhao, J.; et al. Structural Basis for Replicase Polyprotein Cleavage and Substrate Specificity of Main Protease from SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2022, 119, e2117142119. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kenward, C.; Worrall, L.J.; Vuckovic, M.; Gentile, F.; Ton, A.T.; Ng, M.; Cherkasov, A.; Strynadka, N.C.J.; Paetzel, M. X-Ray Crystallographic Characterization of the SARS-CoV-2 Main Protease Polyprotein Cleavage Sites Essential for Viral Processing and Maturation. Nat. Commun. 2022, 13, 5196. [Google Scholar] [CrossRef] [PubMed]
- Behnam, M.A.M. Protein Structural Heterogeneity: A Hypothesis for the Basis of Proteolytic Recognition by the Main Protease of SARS-CoV and SARS-CoV-2. Biochimie 2021, 182, 177–184. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Donchenko, A.P.; Blinov, V.M.; Koonin, E.V. Cysteine Proteases of Positive Strand RNA Viruses and Chymotrypsin-like Serine Proteases. A Distinct Protein Superfamily with a Common Structural Fold. FEBS Lett. 1989, 243, 103–114. [Google Scholar] [CrossRef]
- Perry, A.; Frey, A.D.H. Enzymatic Reaction Mechanisms; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Douangamath, A.; Fearon, D.; Gehrtz, P.; Krojer, T.; Lukacik, P.; Owen, C.D.; Resnick, E.; Strain-Damerell, C.; Aimon, A.; Ábrányi-Balogh, P.; et al. Crystallographic and Electrophilic Fragment Screening of the SARS-CoV-2 Main Protease. Nat. Commun. 2020, 11, 5047. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and Discovery of Its Inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Lee, J.; Worrall, L.J.; Vuckovic, M.; Rosell, F.I.; Gentile, F.; Ton, A.T.; Caveney, N.A.; Ban, F.; Cherkasov, A.; Paetzel, M.; et al. Crystallographic Structure of Wild-Type SARS-CoV-2 Main Protease Acyl-Enzyme Intermediate with Physiological C-Terminal Autoprocessing Site. Nat. Commun. 2020, 11, 5877. [Google Scholar] [CrossRef]
- Shi, J.; Sivaraman, J.; Song, J. Mechanism for Controlling the Dimer-Monomer Switch and Coupling Dimerization to Catalysis of the Severe Acute Respiratory Syndrome Coronavirus 3C-like Protease. J. Virol. 2008, 82, 4620–4629. [Google Scholar] [CrossRef]
- Allaire, M.; Chernaia, M.M.; Malcolm, B.A.; James, M.N.G. Picornaviral 3C Cysteine Proteinases Have a Fold Similar to Chymotrypsin-like Serine Proteinases. Nature 1994, 369, 72–76. [Google Scholar] [CrossRef]
- de Clercq, E. Strategies in the Design of Antiviral Drugs. Nat. Rev. Drug Discov. 2002, 1, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Kilianski, A.; Baker, S.C. Cell-Based Antiviral Screening against Coronaviruses: Developing Virus-Specific and Broad-Spectrum Inhibitors. Antiviral Res. 2014, 101, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.C. The Process of Structure-Based Drug Design. Chem. Biol. 2003, 10, 787–797. [Google Scholar] [CrossRef]
- Njoroge, F.G.; Chen, K.X.; Shih, N.Y.; Piwinski, J.J. Challenges in Modern Drug Discovery: A Case Study of Boceprevir, an HCV Protease Inhibitor for the Treatment of Hepatitis C Virus Infection. Acc. Chem. Res. 2008, 41, 50–59. [Google Scholar] [CrossRef]
- Pawlotsky, J.M.; Feld, J.J.; Zeuzem, S.; Hoofnagle, J.H. From Non-A, Non-B Hepatitis to Hepatitis C Virus Cure. J. Hepatol. 2015, 62, S87–S99. [Google Scholar] [CrossRef]
- Wlodawer, A.; Vondrasek, J. Inhibitors of HIV-1 Protease: A Major Success of Structure-Assisted Drug Design. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 249–284. [Google Scholar] [CrossRef]
- Palella, F.J.; Delaney, K.M.; Moorman, A.C.; Loveless, M.O.; Fuhrer, J.; Satten, G.A.; Aschman, D.J.; Holmberg, S.D. Declining Morbidity and Mortality among Patients with Advanced Human Immunodeficiency Virus Infection. N. Engl. J. Med. 1998, 338, 853–860. [Google Scholar] [CrossRef]
- Wu, C.Y.; Jan, J.T.; Ma, S.H.; Kuo, C.J.; Juan, H.F.; Cheng, Y.S.E.; Hsu, H.H.; Huang, H.C.; Wu, D.; Brik, A.; et al. Small Molecules Targeting Severe Acute Respiratory Syndrome Human Coronavirus. Proc. Natl. Acad. Sci. USA 2004, 101, 10012–10017. [Google Scholar] [CrossRef]
- Xiong, M.; Su, H.; Zhao, W.; Xie, H.; Shao, Q.; Xu, Y. What Coronavirus 3C-like Protease Tells Us: From Structure, Substrate Selectivity, to Inhibitor Design. Med. Res. Rev. 2021, 41, 1965–1998. [Google Scholar] [CrossRef]
- Poordad, F.; McCone, J.; Bacon, B.R.; Bruno, S.; Manns, M.P.; Sulkowski, M.S.; Jacobson, I.M.; Reddy, K.R.; Goodman, Z.D.; Boparai, N.; et al. Boceprevir for Untreated Chronic HCV Genotype 1 Infection. N. Engl. J. Med. 2011, 364, 1195–1206. [Google Scholar] [CrossRef] [PubMed]
- Bacon, B.R.; Gordon, S.C.; Lawitz, E.; Marcellin, P.; Vierling, J.M.; Zeuzem, S.; Poordad, F.; Goodman, Z.D.; Sings, H.L.; Boparai, N.; et al. Boceprevir for Previously Treated Chronic HCV Genotype 1 Infection. N. Engl. J. Med. 2011, 364, 1207–1217. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Kim, Y.; Liu, H.; Galasiti Kankanamalage, A.C.; Eckstrand, C.; Groutas, W.C.; Bannasch, M.; Meadows, J.M.; Chang, K.O. Efficacy of a 3C-like Protease Inhibitor in Treating Various Forms of acquired Feline Infectious Peritonitis. J. Feline Med. Surg. 2018, 20, 378. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Sacco, M.D.; Hurst, B.; Townsend, J.A.; Hu, Y.; Szeto, T.; Zhang, X.; Tarbet, B.; Marty, M.T.; Chen, Y.; et al. Boceprevir, GC-376, and Calpain Inhibitors II, XII Inhibit SARS-CoV-2 Viral Replication by Targeting the Viral Main Protease. Cell Res. 2020, 30, 678–692. [Google Scholar] [CrossRef] [PubMed]
- Vuong, W.; Khan, M.B.; Fischer, C.; Arutyunova, E.; Lamer, T.; Shields, J.; Saffran, H.A.; McKay, R.T.; van Belkum, M.J.; Joyce, M.A.; et al. Feline Coronavirus Drug Inhibits the Main Protease of SARS-CoV-2 and Blocks Virus Replication. Nat. Commun. 2020, 11, 4282. [Google Scholar] [CrossRef]
- Singh, J.; Petter, R.C.; Baillie, T.A.; Whitty, A. The Resurgence of Covalent Drugs. Nat. Rev. Drug Discov. 2011, 10, 307–317. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V.; Hayashi, Y.; Jung, S.H. An Overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL Protease Inhibitors: Peptidomimetics and Small Molecule Chemotherapy. J. Med. Chem. 2016, 59, 6595–6628. [Google Scholar] [CrossRef]
- Hoffman, R.L.; Kania, R.S.; Brothers, M.A.; Davies, J.F.; Ferre, R.A.; Gajiwala, K.S.; He, M.; Hogan, R.J.; Kozminski, K.; Li, L.Y.; et al. Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19. J. Med. Chem. 2020, 63, 12725–12747. [Google Scholar] [CrossRef]
- Boras, B.; Jones, R.M.; Anson, B.J.; Arenson, D.; Aschenbrenner, L.; Bakowski, M.A.; Beutler, N.; Binder, J.; Chen, E.; Eng, H.; et al. Preclinical Characterization of an Intravenous Coronavirus 3CL Protease Inhibitor for the Potential Treatment of COVID19. Nat. Commun. 2021, 12, 6055. [Google Scholar] [CrossRef]
- De Vries, M.; Mohamed, A.S.; Prescott, R.A.; Valero-Jimenez, A.M.; Desvignes, L.; O’Connor, R.; Steppan, C.; Devlin, J.C.; Ivanova, E.; Herrera, A.; et al. A Comparative Analysis of SARS-CoV-2 Antivirals Characterizes 3CLpro Inhibitor PF-00835231 as a Potential New Treatment for COVID-19. J. Virol. 2021, 95, e01819-20. [Google Scholar] [CrossRef]
- Vandyck, K.; Deval, J. Considerations for the Discovery and Development of 3-Chymotrypsin-like Cysteine Protease Inhibitors Targeting SARS-CoV-2 Infection. Curr. Opin. Virol. 2021, 49, 36. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.R.; Allerton, C.M.N.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An Oral SARS-CoV-2 M pro Inhibitor Clinical Candidate for the Treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Lead- and Drug-like Compounds: The Rule-of-Five Revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- How Pfizer Scientists Transformed an Old Drug Lead into a COVID-19 Antiviral. Available online: https://cen.acs.org/pharmaceuticals/drug-discovery/How-Pfizer-scientists-transformed-an-old-drug-lead-into-a-COVID-19-antiviral/100/i3 (accessed on 14 December 2022).
- Pfizer Unveils Its Oral SARS-CoV-2 Inhibitor. Available online: https://cen.acs.org/acs-news/acs-meeting-news/Pfizer-unveils-oral-SARS-CoV/99/i13 (accessed on 14 December 2022).
- Lamb, Y.N. Nirmatrelvir Plus Ritonavir: First Approval. Drugs 2022, 82, 585–591. [Google Scholar] [CrossRef]
- Mahase, E. COVID-19: Pfizer’s Paxlovid Is 89% Effective in Patients at Risk of Serious Illness, Company Reports. BMJ 2021, 375, n2713. [Google Scholar] [CrossRef]
- Hung, Y.P.; Lee, J.C.; Chiu, C.W.; Lee, C.C.; Tsai, P.J.; Hsu, I.L.; Ko, W.C. Oral Nirmatrelvir/Ritonavir Therapy for COVID-19: The Dawn in the Dark? Antibiotics 2022, 11, 220. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, S.; Ekanayake, K.B.; Otting, G.; Nitsche, C. Main Protease Mutants of SARS-CoV-2 Variants Remain Susceptible to Nirmatrelvir. Bioorg. Med. Chem. Lett. 2022, 62, 128629. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, E.; Costacurta, F.; Moghadasi, S.A.; Ye, C.; Pavan, M.; Bassani, D.; Volland, A.; Ascher, C.; Weiss, A.K.H.; Bante, D.; et al. SARS-CoV-2 3CLpro Mutations Selected in a VSV-Based System Confer Resistance to Nirmatrelvir, Ensitrelvir, and GC376. Sci. Transl. Med. 2022, 15, eabq7360. [Google Scholar] [CrossRef]
- McCarthy, M.W. Ensitrelvir as a Potential Treatment for COVID-19. Expert Opin. Pharmacother. 2022, 23, 1995–1998. [Google Scholar] [CrossRef]
- Shimizu, R.; Sonoyama, T.; Fukuhara, T.; Kuwata, A.; Matsuo, Y.; Kubota, R. Safety, Tolerability, and Pharmacokinetics of the Novel Antiviral Agent Ensitrelvir Fumaric Acid, a SARS-CoV-2 3CL Protease Inhibitor, in Healthy Adults. Antimicrob. Agents Chemother. 2022, 66, e00632-22. [Google Scholar] [CrossRef] [PubMed]
- Mukae, H.; Yotsuyanagi, H.; Ohmagari, N.; Doi, Y.; Imamura, T.; Sonoyama, T.; Fukuhara, T.; Ichihashi, G.; Sanaki, T.; Baba, K.; et al. A Randomized Phase 2/3 Study of Ensitrelvir, a Novel Oral SARS-CoV-2 3C-Like Protease Inhibitor, in Japanese Patients with Mild-to-Moderate COVID-19 or Asymptomatic SARS-CoV-2 Infection: Results of the Phase 2a Part. Antimicrob. Agents Chemother. 2022, 66, e00697-22. [Google Scholar] [CrossRef] [PubMed]
- Anighoro, A.; Bajorath, J.; Rastelli, G. Polypharmacology: Challenges and Opportunities in Drug Discovery. J. Med. Chem. 2014, 57, 7874–7887. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.U. Polypharmacology—Foe or Friend? J. Med. Chem. 2013, 56, 8955–8971. [Google Scholar] [CrossRef]
- Reddy, A.S.; Zhang, S. Polypharmacology: Drug Discovery for the Future. Expert Rev. Clin. Pharmacol. 2013, 6, 41–47. [Google Scholar] [CrossRef]
- Nasab, M.G.; Saghazadeh, A.; Rezaei, N. SARS-CoV-2-A Tough Opponent for the Immune System. Arch. Med. Res. 2020, 51, 589–592. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Lubin, J.H.; Zardecki, C.; Dolan, E.M.; Lu, C.; Shen, Z.; Dutta, S.; Westbrook, J.D.; Hudson, B.P.; Goodsell, D.S.; Williams, J.K.; et al. Evolution of the SARS-CoV-2 Proteome in Three Dimensions (3D) during the First 6 Months of the COVID-19 Pandemic. Proteins: Struct. Funct. Bioinform. 2022, 90, 1054–1080. [Google Scholar] [CrossRef]
- Osipiuk, J.; Azizi, S.A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G.; et al. Structure of Papain-like Protease from SARS-CoV-2 and Its Complexes with Non-Covalent Inhibitors. Nat. Commun. 2021, 12, 743. [Google Scholar] [CrossRef]
- Rut, W.; Lv, Z.; Zmudzinski, M.; Patchett, S.; Nayak, D.; Snipas, S.J.; El Oualid, F.; Huang, T.T.; Bekes, M.; Drag, M.; et al. Activity Profiling and Crystal Structures of Inhibitor-Bound SARS-CoV-2 Papain-like Protease: A Framework for Anti–COVID-19 Drug Design. Sci. Adv. 2020, 6, 4596–4612. [Google Scholar] [CrossRef]
- Klemm, T.; Ebert, G.; Calleja, D.J.; Allison, C.C.; Richardson, L.W.; Bernardini, J.P.; Lu, B.G.; Kuchel, N.W.; Grohmann, C.; Shibata, Y.; et al. Mechanism and Inhibition of the Papain-like Protease, PLpro, of SARS-CoV-2. EMBO J. 2020, 39, e106275. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like Protease Regulates SARS-CoV-2 Viral Spread and Innate Immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-Dependent RNA Polymerase from COVID-19 Virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef] [PubMed]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of Replicating SARS-CoV-2 Polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, J.; Wang, H.; Gao, Y.; Liu, Q.; Mu, A.; Ji, W.; Yan, L.; Zhu, Y.; Zhu, C.; et al. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell 2020, 182, 417–428.e13. [Google Scholar] [CrossRef]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural Basis for Inhibition of the RNA-Dependent RNA Polymerase from SARS-CoV-2 by Remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef]
- Kokic, G.; Hillen, H.S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Höbartner, C.; Cramer, P. Mechanism of SARS-CoV-2 Polymerase Stalling by Remdesivir. Nat. Commun. 2021, 12, 279. [Google Scholar] [CrossRef]
- Chen, J.; Malone, B.; Llewellyn, E.; Grasso, M.; Shelton, P.M.M.; Olinares, P.D.B.; Maruthi, K.; Eng, E.T.; Vatandaslar, H.; Chait, B.T.; et al. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell 2020, 182, 1560–1573.e13. [Google Scholar] [CrossRef]
- Awan, F.M.; Yang, B.B.; Naz, A.; Hanif, A.; Ikram, A.; Obaid, A.; Malik, A.; Janjua, H.A.; Ali, A.; Sharif, S. The Emerging Role and Significance of Circular RNAs in Viral Infections and Antiviral Immune Responses: Possible Implication as Theranostic Agents. RNA Biol. 2021, 18, 1–15. [Google Scholar] [CrossRef]
- Adedeji, A.O.; Marchand, B.; te Velthuis, A.J.W.; Snijder, E.J.; Weiss, S.; Eoff, R.L.; Singh, K.; Sarafianos, S.G. Mechanism of Nucleic Acid Unwinding by SARS-CoV Helicase. PLoS ONE 2012, 7, e36521. [Google Scholar] [CrossRef]
- Mickolajczyk, K.J.; Shelton, P.M.M.; Grasso, M.; Cao, X.; Warrington, S.E.; Aher, A.; Liu, S.; Kapoor, T.M. Force-Dependent Stimulation of RNA Unwinding by SARS-CoV-2 Nsp13 Helicase. Biophys. J. 2021, 120, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.A.; Douangamath, A.; Yadzani, S.; Yosaatmadja, Y.; Aimon, A.; Brandão-Neto, J.; Dunnett, L.; Gorrie-stone, T.; Skyner, R.; Fearon, D.; et al. Structure, Mechanism and Crystallographic Fragment Screening of the SARS-CoV-2 NSP13 Helicase. Nat. Commun. 2021, 12, 4848. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, K.A.; Thiel, V.; Dobbe, J.C.; van der Meer, Y.; Snijder, E.J.; Ziebuhr, J. Multiple Enzymatic Activities Associated with Severe Acute Respiratory Syndrome Coronavirus Helicase. J. Virol. 2004, 78, 5619–5632. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Su, C.; Ke, M.; Jin, X.; Xu, L.; Zhang, Z.; Wu, A.; Sun, Y.; Yang, Z.; Tien, P.; et al. Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by Nsp16/Nsp10 Protein Complex. PLoS Pathog. 2011, 7, e1002294. [Google Scholar] [CrossRef]
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells 2020, 9, 1267. [Google Scholar] [CrossRef]
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic Characterization of a Novel SARS-CoV-2. Gene Rep. 2020, 19, 100682. [Google Scholar] [CrossRef]
- Ogando, N.S.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Bredenbeek, P.J.; Posthuma, C.C.; Snijder, E.J. The Enzymatic Activity of the Nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020, 94, e01246-20. [Google Scholar] [CrossRef]
- Ma, Y.; Wu, L.; Shaw, N.; Gao, Y.; Wang, J.; Sun, Y.; Lou, Z.; Yan, L.; Zhang, R.; Rao, Z. Structural Basis and Functional Analysis of the SARS Coronavirus Nsp14-Nsp10 Complex. Proc. Natl. Acad. Sci. USA 2015, 112, 9436–9441. [Google Scholar] [CrossRef]
- Lin, S.; Chen, H.; Chen, Z.; Yang, F.; Ye, F.; Zheng, Y.; Yang, J.; Lin, X.; Sun, H.; Wang, L.; et al. Crystal Structure of SARS-CoV-2 Nsp10 Bound to Nsp14-ExoN Domain Reveals an Exoribonuclease with Both Structural and Functional Integrity. Nucleic Acids Res. 2021, 49, 5382–5392. [Google Scholar] [CrossRef]
- Eckerle, L.D.; Becker, M.M.; Halpin, R.A.; Li, K.; Venter, E.; Lu, X.; Scherbakova, S.; Graham, R.L.; Baric, R.S.; Stockwell, T.B.; et al. Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing. PLoS Pathog. 2010, 6, e1000896. [Google Scholar] [CrossRef] [PubMed]
- Czarna, A.; Plewka, J.; Kresik, L.; Matsuda, A.; Karim, A.; Robinson, C.; O’Byrne, S.; Cunningham, F.; Georgiou, I.; Wilk, P.; et al. Refolding of Lid Subdomain of SARS-CoV-2 Nsp14 upon Nsp10 Interaction Releases Exonuclease Activity. Structure 2022, 30, 1050–1054.e2. [Google Scholar] [CrossRef] [PubMed]
- Wilamowski, M.; Sherrell, D.A.; Minasov, G.; Kim, Y.; Shuvalova, L.; Lavens, A.; Chard, R.; Maltseva, N.; Jedrzejczak, R.; Rosas-Lemus, M.; et al. 2′-O Methylation of RNA Cap in SARS-CoV-2 Captured by Serial Crystallography. Proc. Natl. Acad. Sci. USA 2021, 118, e2100170118. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Chen, H.; Ye, F.; Chen, Z.; Yang, F.; Zheng, Y.; Cao, Y.; Qiao, J.; Yang, S.; Lu, G. Crystal Structure of SARS-CoV-2 Nsp10/Nsp16 2′-O-Methylase and Its Implication on Antiviral Drug Design. Signal Transduct. Target. Ther. 2020, 5, 131. [Google Scholar] [CrossRef]
- Rosas-Lemus, M.; Minasov, G.; Shuvalova, L.; Inniss, N.L.; Kiryukhina, O.; Brunzelle, J.; Satchell, K.J.F. High-Resolution Structures of the SARS-CoV-2 2′-O-Methyltransferase Reveal Strategies for Structure-Based Inhibitor Design. Sci. Signal 2020, 13, 1202. [Google Scholar] [CrossRef]
- Yoshimoto, F.K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 2020, 39, 198–216. [Google Scholar] [CrossRef]
- Frazier, M.N.; Wilson, I.M.; Krahn, J.M.; Butay, K.J.; Dillard, L.B.; Borgnia, M.J.; Stanley, R.E. Flipped over U: Structural Basis for DsRNA Cleavage by the SARS-CoV-2 Endoribonuclease. Nucleic Acids Res. 2022, 50, 8290–8301. [Google Scholar] [CrossRef]
- Pillon, M.C.; Frazier, M.N.; Dillard, L.B.; Williams, J.G.; Kocaman, S.; Krahn, J.M.; Perera, L.; Hayne, C.K.; Gordon, J.; Stewart, Z.D.; et al. Cryo-EM Structures of the SARS-CoV-2 Endoribonuclease Nsp15 Reveal Insight into Nuclease Specificity and Dynamics. Nat. Commun. 2021, 12, 636. [Google Scholar] [CrossRef]
- Kim, Y.; Jedrzejczak, R.; Maltseva, N.I.; Wilamowski, M.; Endres, M.; Godzik, A.; Michalska, K.; Joachimiak, A. Crystal Structure of Nsp15 Endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020, 29, 1596–1605. [Google Scholar] [CrossRef]
- Frazier, M.N.; Dillard, L.B.; Krahn, J.M.; Perera, L.; Williams, J.G.; Wilson, I.M.; Stewart, Z.D.; Pillon, M.C.; Deterding, L.J.; Borgnia, M.J.; et al. Characterization of SARS2 Nsp15 Nuclease Activity Reveals It’s Mad about U. Nucleic Acids Res. 2021, 49, 10136–10149. [Google Scholar] [CrossRef]
- Kim, Y.; Wower, J.; Maltseva, N.; Chang, C.; Jedrzejczak, R.; Wilamowski, M.; Kang, S.; Nicolaescu, V.; Randall, G.; Michalska, K.; et al. Tipiracil Binds to Uridine Site and Inhibits Nsp15 Endoribonuclease NendoU from SARS-CoV-2. Commun. Biol. 2021, 4, 193. [Google Scholar] [CrossRef] [PubMed]
- Littler, D.R.; MacLachlan, B.J.; Watson, G.M.; Vivian, J.P.; Gully, B.S. A Pocket Guide on How to Structure SARS-CoV-2 Drugs and Therapies. Biochem. Soc. Trans. 2020, 48, 2625–2641. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.R.; Yin, W.C.; Jiang, Y.; Xu, H.E. Structure Genomics of SARS-CoV-2 and Its Omicron Variant: Drug Design Templates for COVID-19. Acta Pharmacol. Sin. 2022, 43, 3021–3033. [Google Scholar] [CrossRef] [PubMed]
- Cragg, G.M.; Newman, D.J. Natural Products: A Continuing Source of Novel Drug Leads. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [PubMed]
- Gurib-Fakim, A. Medicinal Plants: Traditions of Yesterday and Drugs of Tomorrow. Mol. Asp. Med. 2006, 27, 1–93. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L. Natural Products in Drug Discovery. Drug Discov. Today 2008, 13, 894–901. [Google Scholar] [CrossRef]
- Ban, T.A. The Role of Serendipity in Drug Discovery. Dialogues Clin. Neurosci. 2006, 8, 335–344. [Google Scholar] [CrossRef]
- Morphy, R.; Kay, C.; Rankovic, Z. From Magic Bullets to Designed Multiple Ligands. Drug Discov. Today 2004, 9, 641–651. [Google Scholar] [CrossRef]
- Jaskolski, M.; Dauter, Z.; Wlodawer, A. A Brief History of Macromolecular Crystallography, Illustrated by a Family Tree and Its Nobel Fruits. FEBS J. 2014, 281, 3985–4009. [Google Scholar] [CrossRef]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W. Computational Methods in Drug Discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef]
- Leelananda, S.P.; Lindert, S. Computational Methods in Drug Discovery. Beilstein J. Org. Chem. 2016, 12, 2694–2718. [Google Scholar] [CrossRef] [PubMed]
- Kapetanovic, I.M. Computer-Aided Drug Discovery and Development (CADDD): In Silico-Chemico-Biological Approach. Chem. Biol. Interact. 2008, 171, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Macalino, S.J.Y.; Gosu, V.; Hong, S.; Choi, S. Role of Computer-Aided Drug Design in Modern Drug Discovery. Arch. Pharm. Res. 2015, 38, 1686–1701. [Google Scholar] [CrossRef]
- Yu, W.; Mackerell, A.D. Computer-Aided Drug Design Methods. Methods Mol. Biol. 2017, 1520, 85–106. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.C.; McMullan, G.; Scheres, S.H.W. How Cryo-EM Is Revolutionizing Structural Biology. Trends Biochem. Sci. 2015, 40, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.S.; Martin, J.H.J. X Ray Crystallography. Mol. Pathol. 2000, 53, 8. [Google Scholar] [CrossRef] [PubMed]
- Markwick, P.R.L.; Malliavin, T.; Nilges, M. Structural Biology by NMR: Structure, Dynamics, and Interactions. PLoS Comput. Biol. 2008, 4, e1000168. [Google Scholar] [CrossRef]
- Nwanochie, E.; Uversky, V.N. Structure Determination by Single-Particle Cryo-Electron Microscopy: Only the Sky (and Intrinsic Disorder) Is the Limit. Int. J. Mol. Sci. 2019, 20, 4186. [Google Scholar] [CrossRef]
- Kuhlman, B.; Bradley, P. Advances in Protein Structure Prediction and Design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. [Google Scholar] [CrossRef]
- Martí-Renom, M.A.; Stuart, A.C.; Fiser, A.; Sánchez, R.; Melo, F.; Šali, A. Comparative Protein Structure Modeling of Genes and Genomes. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 291–325. [Google Scholar] [CrossRef] [PubMed]
- Cavasotto, C.N.; Phatak, S.S. Homology Modeling in Drug Discovery: Current Trends and Applications. Drug Discov. Today 2009, 14, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, R.; Strauss, C.E.M.; Rohl, C.A.; Chivian, D.; Bradley, P.; Malmströ, L.; Robertson, T.; Baker, D. De Novo Prediction of Three-Dimensional Structures for Major Protein Families. J. Mol. Biol. 2002, 322, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Protein Structure Prediction Using Multiple Deep Neural Networks in the 13th Critical Assessment of Protein Structure Prediction (CASP13). Proteins Struct. Funct. Bioinform. 2019, 87, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.; Simpkin, A.J.; Hartmann, M.D.; Rigden, D.J.; Keegan, R.M.; Lupas, A.N. High-Accuracy Protein Structure Prediction in CASP14. Proteins 2021, 89, 1687–1699. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Applying and Improving AlphaFold at CASP14. Proteins Struct. Funct. Bioinform. 2021, 89, 1711–1721. [Google Scholar] [CrossRef]
- Kwon, S.; Won, J.; Kryshtafovych, A.; Seok, C. Assessment of Protein Model Structure Accuracy Estimation in CASP14: Old and New Challenges. Proteins Struct. Funct. Bioinform. 2021, 89, 1940–1948. [Google Scholar] [CrossRef]
- Dong, S.; Sun, J.; Mao, Z.; Wang, L.; Lu, Y.L.; Li, J. A Guideline for Homology Modeling of the Proteins from Newly Discovered Betacoronavirus, 2019 Novel Coronavirus (2019-NCoV). J. Med. Virol. 2020, 92, 1542–1548. [Google Scholar] [CrossRef]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of Therapeutic Targets for SARS-CoV-2 and Discovery of Potential Drugs by Computational Methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef]
- Bassani, D.; Ragazzi, E.; Lapolla, A.; Sartore, G.; Moro, S. Omicron Variant of SARS-CoV-2 Virus: In Silico Evaluation of the Possible Impact on People Affected by Diabetes Mellitus. Front. Endocrinol. 2022, 13, 284. [Google Scholar] [CrossRef]
- Gan, H.H.; Twaddle, A.; Marchand, B.; Gunsalus, K.C. Structural Modeling of the SARS-CoV-2 Spike/Human ACE2 Complex Interface Can Identify High-Affinity Variants Associated with Increased Transmissibility. J. Mol. Biol. 2021, 433, 167051. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Praissman, J.L.; Grant, O.C.; Cai, Y.; Xiao, T.; Rosenbalm, K.E.; Aoki, K.; Kellman, B.P.; Bridger, R.; Barouch, D.H.; et al. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell Host Microbe 2020, 28, 586–601.e6. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Wang, J.; Chen, G.; Zhang, H.; An, K.; Xu, P.; Du, Y.; Ye, R.D.; Saha, A.; Zhang, A.; et al. Predicting Mutational Effects on Receptor Binding of the Spike Protein of SARS-CoV-2 Variants. J. Am. Chem. Soc. 2021, 143, 17646–17654. [Google Scholar] [CrossRef] [PubMed]
- Shahhosseini, N.; Babuadze, G.; Wong, G.; Kobinger, G.P. Mutation Signatures and in Silico Docking of Novel SARS-CoV-2 Variants of Concern. Microorganisms 2021, 9, 926. [Google Scholar] [CrossRef] [PubMed]
- Pavan, M.; Bassani, D.; Sturlese, M.; Moro, S. Bat Coronaviruses Related to SARS-CoV-2: What about Their 3CL Proteases (MPro)? J. Enzym. Inhib. Med. Chem. 2022, 37, 1077–1082. [Google Scholar] [CrossRef]
- Martin, R.W.; Butts, C.T.; Cross, T.J.; Takahashi, G.R.; Diessner, E.M.; Crosby, M.G.; Farahmand, V.; Zhuang, S. Sequence Characterization and Molecular Modeling of Clinically Relevant Variants of the SARS-CoV-2 Main Protease. Biochemistry 2020, 59, 3741–3756. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, C.; Pearce, R.; Omenn, G.S.; Zhang, Y. Identifying the Zoonotic Origin of SARS-CoV-2 by Modeling the Binding Affinity between the Spike Receptor-Binding Domain and Host ACE2. J. Proteome Res. 2020, 19, 4844–4856. [Google Scholar] [CrossRef]
- Rodrigues, J.P.G.L.M.; Barrera-Vilarmau, S.; Teixeira, J.M.C.; Sorokina, M.; Seckel, E.; Kastritis, P.L.; Levitt, M. Insights on Cross-Species Transmission of SARS-CoV-2 from Structural Modeling. PLoS Comput. Biol. 2020, 16, e1008449. [Google Scholar] [CrossRef]
- Piplani, S.; Singh, P.K.; Winkler, D.A.; Petrovsky, N. In Silico Comparison of SARS-CoV-2 Spike Protein-ACE2 Binding Affinities across Species and Implications for Virus Origin. Sci. Rep. 2021, 11, 13063. [Google Scholar] [CrossRef]
- Sharma, P.; Kumar, M.; Tripathi, M.K.; Gupta, D.; Vishwakarma, P.; Das, U.; Kaur, P. Genomic and Structural Mechanistic Insight to Reveal the Differential Infectivity of Omicron and Other Variants of Concern. Comput. Biol. Med. 2022, 150, 106129. [Google Scholar] [CrossRef]
- Jacob, J.J.; Vasudevan, K.; Pragasam, A.K.; Gunasekaran, K.; Veeraraghavan, B.; Mutreja, A. Evolutionary Tracking of SARS-CoV-2 Genetic Variants Highlights an Intricate Balance of Stabilizing and Destabilizing Mutations. mBio 2021, 12, e01188-21. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Delaunay-Moisan, A.; Timmis, K.; Danchin, A. SARS-CoV-2 Biology and Variants: Anticipation of Viral Evolution and What Needs to Be Done. Environ. Microbiol. 2021, 23, 2339–2363. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Brindisi, M.; Shahabi, D.; Chapman, M.E.; Mesecar, A.D. Drug Development and Medicinal Chemistry Efforts toward SARS-Coronavirus and COVID-19 Therapeutics. ChemMedChem 2020, 15, 907–932. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Mishevich, J.L.; Mesecar, A.; Mitsuya, H. Recent Drug Development and Medicinal Chemistry Approaches for the Treatment of SARS-CoV-2 Infection and COVID-19. ChemMedChem 2022, 17, e202200440. [Google Scholar] [CrossRef]
- Tiwari, V.; Beer, J.C.; Sankaranarayanan, N.V.; Swanson-Mungerson, M.; Desai, U.R. Discovering Small-Molecule Therapeutics against SARS-CoV-2. Drug Discov. Today 2020, 25, 1535–1544. [Google Scholar] [CrossRef]
- Adamson, C.S.; Chibale, K.; Goss, R.J.M.; Jaspars, M.; Newman, D.J.; Dorrington, R.A. Antiviral Drug Discovery: Preparing for the next Pandemic. Chem. Soc. Rev. 2021, 50, 3647–3655. [Google Scholar] [CrossRef]
- Consortium, T.C.M.; Achdout, H.; Aimon, A.; Bar-David, E.; Barr, H.; Ben-Shmuel, A.; Bennett, J.; Bilenko, V.A.; Bilenko, V.A.; Boby, M.L.; et al. Open Science Discovery of Oral Non-Covalent SARS-CoV-2 Main Protease Inhibitor Therapeutics. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kuntz, I.D. Structure-Based Strategies for Drug Design and Discovery. Science 1992, 257, 1078–1082. [Google Scholar] [CrossRef]
- Forli, S. Charting a Path to Success in Virtual Screening. Molecules 2015, 20, 18732–18758. [Google Scholar] [CrossRef]
- Halgren, T.A. Identifying and Characterizing Binding Sites and Assessing Druggability. J. Chem. Inf. Model. 2009, 49, 377–389. [Google Scholar] [CrossRef]
- Liang, J.; Edelsbrunner, H.; Woodward, C. Anatomy of Protein Pockets and Cavities: Measurement of Binding Site Geometry and Implications for Ligand Design. Protein Sci. 1998, 7, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and Scoring in Virtual Screening for Drug Discovery: Methods and Applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Kuntz, I.D.; Blaney, J.M.; Oatley, S.J.; Langridge, R.; Ferrin, T.E. A Geometric Approach to Macromolecule-Ligand Interactions. J. Mol. Biol. 1982, 161, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Current Computer Aided-Drug Design 2012, 7, 146–157. [Google Scholar] [CrossRef]
- Salmaso, V.; Moro, S. Bridging Molecular Docking to Molecular Dynamics in Exploring Ligand-Protein Recognition Process: An Overview. Front. Pharmacol. 2018, 9, 923. [Google Scholar] [CrossRef]
- Halperin, I.; Ma, B.; Wolfson, H.; Nussinov, R. Principles of Docking: An Overview of Search Algorithms and a Guide to Scoring Functions. Proteins Struct. Funct. Genet. 2002, 47, 409–443. [Google Scholar] [CrossRef]
- Warren, G.L.; Andrews, C.W.; Capelli, A.M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A Critical Assessment of Docking Programs and Scoring Functions. J. Med. Chem. 2006, 49, 5912–5931. [Google Scholar] [CrossRef]
- Bassani, D.; Pavan, M.; Bolcato, G.; Sturlese, M.; Moro, S. Re-Exploring the Ability of Common Docking Programs to Correctly Reproduce the Binding Modes of Non-Covalent Inhibitors of SARS-CoV-2 Protease Mpro. Pharmaceuticals 2022, 15, 180. [Google Scholar] [CrossRef]
- Ton, A.T.; Gentile, F.; Hsing, M.; Ban, F.; Cherkasov, A. Rapid Identification of Potential Inhibitors of SARS-CoV-2 Main Protease by Deep Docking of 1.3 Billion Compounds. Mol. Inform. 2020, 39, 2000028. [Google Scholar] [CrossRef]
- Acharya, A.; Agarwal, R.; Baker, M.B.; Baudry, J.; Bhowmik, D.; Boehm, S.; Byler, K.G.; Chen, S.Y.; Coates, L.; Cooper, C.J.; et al. Supercomputer-Based Ensemble Docking Drug Discovery Pipeline with Application to COVID-19. J. Chem. Inf. Model. 2020, 60, 5832–5852. [Google Scholar] [CrossRef]
- Gorgulla, C.; Padmanabha Das, K.M.; Leigh, K.E.; Cespugli, M.; Fischer, P.D.; Wang, Z.F.; Tesseyre, G.; Pandita, S.; Shnapir, A.; Calderaio, A.; et al. A Multi-Pronged Approach Targeting SARS-CoV-2 Proteins Using Ultra-Large Virtual Screening. iScience 2021, 24, 102021. [Google Scholar] [CrossRef] [PubMed]
- Manelfi, C.; Gossen, J.; Gervasoni, S.; Talarico, C.; Albani, S.; Philipp, B.J.; Musiani, F.; Vistoli, G.; Rossetti, G.; Beccari, A.R.; et al. Combining Different Docking Engines and Consensus Strategies to Design and Validate Optimized Virtual Screening Protocols for the SARS-CoV-2 3CL Protease. Molecules 2021, 26, 797. [Google Scholar] [CrossRef] [PubMed]
- Corona, A.; Wycisk, K.; Talarico, C.; Manelfi, C.; Milia, J.; Cannalire, R.; Esposito, F.; Gribbon, P.; Zaliani, A.; Iaconis, D.; et al. Natural Compounds Inhibit SARS-CoV-2 Nsp13 Unwinding and ATPase Enzyme Activities. ACS Pharmacol. Transl. Sci. 2022, 5, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved Protein-Ligand Docking Using GOLD. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Kolarič, A.; Jukič, M.; Bren, U. Novel Small-Molecule Inhibitors of the SARS-CoV-2 Spike Protein Binding to Neuropilin 1. Pharmaceuticals 2022, 15, 165. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785. [Google Scholar] [CrossRef]
- Vatansever, E.C.; Yang, K.S.; Drelich, A.K.; Kratch, K.C.; Cho, C.C.; Kempaiah, K.R.; Hsu, J.C.; Mellott, D.M.; Xu, S.; Tseng, C.T.K.; et al. Bepridil Is Potent against SARS-CoV-2 in Vitro. Proc. Natl. Acad. Sci. USA 2021, 118, e2012201118. [Google Scholar] [CrossRef]
- Neves, M.A.C.; Totrov, M.; Abagyan, R. Docking and Scoring with ICM: The Benchmarking Results and Strategies for Improvement. J. Comput. Aided Mol. Des. 2012, 26, 675–686. [Google Scholar] [CrossRef]
- Lam, P.C.H.; Abagyan, R.; Totrov, M. Ligand-Biased Ensemble Receptor Docking (LigBEnD): A Hybrid Ligand/Receptor Structure-Based Approach. J. Comput. Aided Mol. Des. 2018, 32, 187–198. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J. Comput. Chem. 2010, 31, 455. [Google Scholar] [CrossRef]
- Kao, H.T.; Orry, A.; Palfreyman, M.G.; Porton, B. Synergistic Interactions of Repurposed Drugs That Inhibit Nsp1, a Major Virulence Factor for COVID-19. Sci. Rep. 2022, 12, 10174. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, H.; Hu, X.; Wang, Q.; Zhong, F.; Zhou, X.; Lin, C.; Yang, Y.; Wei, J.; Du, W.; et al. Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus. J. Virol. 2022, 96, 1253–1274. [Google Scholar] [CrossRef] [PubMed]
- Huff, S.; Kummetha, I.R.; Tiwari, S.K.; Huante, M.B.; Clark, A.E.; Wang, S.; Bray, W.; Smith, D.; Carlin, A.F.; Endsley, M.; et al. Discovery and Mechanism of SARS-CoV-2 Main Protease Inhibitors. J. Med. Chem. 2022, 65, 2866–2879. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zou, R.; Cui, W.; Li, M.; Wang, X.; Dong, J.; Li, H.; Li, H.; Wang, P.; Shao, X.; et al. Clinical HDAC Inhibitors Are Effective Drugs to Prevent the Entry of SARS-CoV2. ACS Pharmacol. Transl. Sci. 2020, 3, 1361–1370. [Google Scholar] [CrossRef]
- Rao, S.N.; Head, M.S.; Kulkarni, A.; LaLonde, J.M. Validation Studies of the Site-Directed Docking Program LibDock. J. Chem. Inf. Model. 2007, 47, 2159–2171. [Google Scholar] [CrossRef]
- Wang, G.; Yang, M.L.; Duan, Z.L.; Liu, F.L.; Jin, L.; Long, C.B.; Zhang, M.; Tang, X.P.; Xu, L.; Li, Y.C.; et al. Dalbavancin Binds ACE2 to Block Its Interaction with SARS-CoV-2 Spike Protein and Is Effective in Inhibiting SARS-CoV-2 Infection in Animal Models. Cell Res. 2020, 31, 17–24. [Google Scholar] [CrossRef]
- Luttens, A.; Gullberg, H.; Abdurakhmanov, E.; Vo, D.D.; Akaberi, D.; Talibov, V.O.; Nekhotiaeva, N.; Vangeel, L.; de Jonghe, S.; Jochmans, D.; et al. Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses. J. Am. Chem. Soc 2022, 144, 2905–2920. [Google Scholar] [CrossRef]
- Cross, S.S.J. Improved FlexX Docking Using FlexS-Determined Base Fragment Placement. J. Chem. Inf. Model. 2005, 45, 993–1001. [Google Scholar] [CrossRef]
- Welker, A.; Kersten, C.; Müller, C.; Madhugiri, R.; Zimmer, C.; Müller, P.; Zimmermann, R.; Hammerschmidt, S.; Maus, H.; Ziebuhr, J.; et al. Structure-Activity Relationships of Benzamides and Isoindolines Designed as SARS-CoV Protease Inhibitors Effective against SARS-CoV-2. ChemMedChem 2021, 16, 340–354. [Google Scholar] [CrossRef]
- Otava, T.; Šála, M.; Li, F.; Fanfrlík, J.; Devkota, K.; Perveen, S.; Chau, I.; Pakarian, P.; Hobza, P.; Vedadi, M.; et al. The Structure-Based Design of SARS-CoV-2 Nsp14 Methyltransferase Ligands Yields Nanomolar Inhibitors. ACS Infect Dis. 2021, 7, 2214–2220. [Google Scholar] [CrossRef]
- Wang, Y.T.; Long, X.Y.; Ding, X.; Fan, S.R.; Cai, J.Y.; Yang, B.J.; Zhang, X.F.; Luo, R.H.; Yang, L.; Ruan, T.; et al. Novel Nucleocapsid Protein-Targeting Phenanthridine Inhibitors of SARS-CoV-2. Eur. J. Med. Chem. 2022, 227, 113966. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C. Beware of Docking! Trends Pharmacol. Sci. 2015, 36, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Llanos, M.A.; Gantner, M.E.; Rodriguez, S.; Alberca, L.N.; Bellera, C.L.; Talevi, A.; Gavernet, L. Strengths and Weaknesses of Docking Simulations in the SARS-CoV-2 Era: The Main Protease (Mpro) Case Study. J. Chem. Inf. Model. 2021, 61, 3758–3770. [Google Scholar] [CrossRef] [PubMed]
- Chaput, L.; Mouawad, L. Efficient Conformational Sampling and Weak Scoring in Docking Programs? Strategy of the Wisdom of Crowds. J. Cheminform. 2017, 9, 37. [Google Scholar] [CrossRef]
- Neves, B.J.; Mottin, M.; Moreira-Filho, J.T.; de Paula Sousa, B.K.; Mendonca, S.S.; Andrade, C.H. Best Practices for Docking-Based Virtual Screening. Mol. Docking Comput. Aided Drug Des. 2021, 2021, 75–98. [Google Scholar] [CrossRef]
- Cerón-Carrasco, J.P. When Virtual Screening Yields Inactive Drugs: Dealing with False Theoretical Friends. ChemMedChem 2022, 17, e202200278. [Google Scholar] [CrossRef]
- Scior, T.; Bender, A.; Tresadern, G.; Medina-Franco, J.L.; Martínez-Mayorga, K.; Langer, T.; Cuanalo-Contreras, K.; Agrafiotis, D.K. Recognizing Pitfalls in Virtual Screening: A Critical Review. J. Chem. Inf. Model. 2012, 52, 867–881. [Google Scholar] [CrossRef]
- Alonso, H.; Bliznyuk, A.A.; Gready, J.E. Combining Docking and Molecular Dynamic Simulations in Drug Design. Med. Res. Rev. 2006, 26, 531–568. [Google Scholar] [CrossRef]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular Dynamics Simulations of Biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Durrant, J.D.; McCammon, J.A. Molecular Dynamics Simulations and Drug Discovery. BMC Biol. 2011, 9, 1–9. [Google Scholar] [CrossRef]
- Ferreira, L.G.; dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular Docking and Structure-Based Drug Design Strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Tan, L.; Batista, J.; Bajorath, J. Computational Methodologies for Compound Database Searching That Utilize Experimental Protein-Ligand Interaction Information. Chem. Biol. Drug Des. 2010, 76, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Peach, M.L.; Nicklaus, M.C. Combining Docking with Pharmacophore Filtering for Improved Virtual Screening. J. Cheminform. 2009, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Muthas, D.; Sabnis, Y.A.; Lundborg, M.; Karlén, A. Is It Possible to Increase Hit Rates in Structure-Based Virtual Screening by Pharmacophore Filtering? An Investigation of the Advantages and Pitfalls of Post-Filtering. J. Mol. Graph. Model. 2008, 26, 1237–1251. [Google Scholar] [CrossRef]
- Rácz, A.; Bajusz, D.; Héberger, K. Life beyond the Tanimoto Coefficient: Similarity Measures for Interaction Fingerprints. J. Cheminform. 2018, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Pavan, M.; Menin, S.; Bassani, D.; Sturlese, M.; Moro, S. Implementing a Scoring Function Based on Interaction Fingerprint for Autogrow4: Protein Kinase CK1δ as a Case Study. Front. Mol. Biosci. 2022, 9, 629. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wen, J.; Yang, Y.; Liu, H.; Wang, S.; Ding, X.; Zhou, C.; Zhang, X. Identification of Highly Effective Inhibitors against SARS-CoV-2 Main Protease: From Virtual Screening to in Vitro Study. Front. Pharmacol. 2022, 13, 4934. [Google Scholar] [CrossRef]
- Tian, X.; Zhao, Q.; Chen, X.; Peng, Z.; Tan, X.; Wang, Q.; Chen, L.; Yang, Y. Discovery of Novel and Highly Potent Inhibitors of SARS-CoV-2 Papain-Like Protease Through Structure-Based Pharmacophore Modeling, Virtual Screening, Molecular Docking, Molecular Dynamics Simulations, and Biological Evaluation. Front. Pharmacol. 2022, 13, 16. [Google Scholar] [CrossRef]
- Yin, S.; Mei, S.; Li, Z.; Xu, Z.; Wu, Y.; Chen, X.; Liu, D.; Niu, M.-M.; Li, J. Non-Covalent Cyclic Peptides Simultaneously Targeting Mpro and NRP1 Are Highly Effective against Omicron BA.2.75. Front. Pharmacol. 2022, 13, 4723. [Google Scholar] [CrossRef]
- Gossen, J.; Albani, S.; Hanke, A.; Joseph, B.P.; Bergh, C.; Kuzikov, M.; Costanzi, E.; Manelfi, C.; Storici, P.; Gribbon, P.; et al. A Blueprint for High Affinity SARS-CoV-2 Mpro Inhibitors from Activity-Based Compound Library Screening Guided by Analysis of Protein Dynamics. ACS Pharmacol. Transl. Sci. 2021, 4, 1079–1095. [Google Scholar] [CrossRef]
- Hu, X.; Chen, C.Z.; Xu, M.; Hu, Z.; Guo, H.; Itkin, Z.; Shinn, P.; Ivin, P.; Leek, M.; Liang, T.J.; et al. Discovery of Small Molecule Entry Inhibitors Targeting the Fusion Peptide of SARS-CoV-2 Spike Protein. ACS Med. Chem. Lett. 2021, 12, 1267–1274. [Google Scholar] [CrossRef]
- Jang, W.D.; Jeon, S.; Kim, S.; Lee, S.Y. Drugs Repurposed for COVID-19 by Virtual Screening of 6,218 Drugs and Cell-Based Assay. Proc. Natl. Acad. Sci. USA 2021, 118, e2024302118. [Google Scholar] [CrossRef]
- McGovern, S.L.; Shoichet, B.K. Information Decay in Molecular Docking Screens against Holo, Apo, and Modeled Conformations of Enzymes. J. Med. Chem. 2003, 46, 2895–2907. [Google Scholar] [CrossRef]
- Salmaso, V.; Sturlese, M.; Cuzzolin, A.; Moro, S. Combining Self- and Cross-Docking as Benchmark Tools: The Performance of DockBench in the D3R Grand Challenge 2. J. Comput. Aided Mol. Des. 2018, 32, 251–264. [Google Scholar] [CrossRef]
- Korb, O.; Olsson, T.S.G.; Bowden, S.J.; Hall, R.J.; Verdonk, M.L.; Liebeschuetz, J.W.; Cole, J.C. Potential and Limitations of Ensemble Docking. J. Chem. Inf. Model. 2012, 52, 1262–1274. [Google Scholar] [CrossRef] [PubMed]
- Knegtel, R.M.A.; Kuntz, I.D.; Oshiro, C.M. Molecular Docking to Ensembles of Protein Structures. J. Mol. Biol. 1997, 266, 424–440. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Y.; Zou, X. Ensemble Docking of Multiple Protein Structures: Considering Protein Structural Variations in Molecular Docking. Proteins Struct. Funct. Genet. 2007, 66, 399–421. [Google Scholar] [CrossRef]
- Wang, R.; Wang, S. How Does Consensus Scoring Work for Virtual Library Screening? An Idealized Computer Experiment. J. Chem. Inf. Comput. Sci. 2001, 41, 1422–1426. [Google Scholar] [CrossRef] [PubMed]
- Charifson, P.S.; Corkery, J.J.; Murcko, M.A.; Walters, W.P. Consensus Scoring: A Method for Obtaining Improved Hit Rates from Docking Databases of Three-Dimensional Structures into Proteins. J. Med. Chem. 1999, 42, 5100–5109. [Google Scholar] [CrossRef]
- Bissantz, C.; Folkers, G.; Rognan, D. Protein-Based Virtual Screening of Chemical Databases. 1. Evaluation of Different Docking/Scoring Combinations. J. Med. Chem. 2000, 43, 4759–4767. [Google Scholar] [CrossRef] [PubMed]
- McGann, M. FRED and HYBRID Docking Performance on Standardized Datasets. J. Comput. Aided Mol. Des. 2012, 26, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Gimeno, A.; Mestres-Truyol, J.; Ojeda-Montes, M.J.; Macip, G.; Saldivar-Espinoza, B.; Cereto-Massagué, A.; Pujadas, G.; Garcia-Vallvé, S. Prediction of Novel Inhibitors of the Main Protease (M-pro) of SARS-CoV-2 through Consensus Docking and Drug Reposition. Int. J. Mol. Sci. 2020, 21, 3793. [Google Scholar] [CrossRef]
- Yang, J.; Lin, X.; Xing, N.; Zhang, Z.; Zhang, H.; Wu, H.; Xue, W. Structure-Based Discovery of Novel Nonpeptide Inhibitors Targeting SARS-CoV-2 Mpro. J. Chem. Inf. Model. 2021, 61, 3917–3926. [Google Scholar] [CrossRef]
- Alhossary, A.; Handoko, S.D.; Mu, Y.; Kwoh, C.K. Fast, Accurate, and Reliable Molecular Docking with QuickVina 2. Bioinformatics 2015, 31, 2214–2216. [Google Scholar] [CrossRef]
- Rubio-Martínez, J.; Jiménez-Alesanco, A.; Ceballos-Laita, L.; Ortega-Alarcón, D.; Vega, S.; Calvo, C.; Benítez, C.; Abian, O.; Velázquez-Campoy, A.; Thomson, T.M.; et al. Discovery of Diverse Natural Products as Inhibitors of SARS-CoV-2 MproProtease through Virtual Screening. J. Chem. Inf. Model. 2021, 61, 6094–6106. [Google Scholar] [CrossRef]
- Clyde, A.; Galanie, S.; Kneller, D.W.; Ma, H.; Babuji, Y.; Blaiszik, B.; Brace, A.; Brettin, T.; Chard, K.; Chard, R.; et al. High-Throughput Virtual Screening and Validation of a SARS-CoV-2 Main Protease Noncovalent Inhibitor. J. Chem. Inf. Model. 2022, 62, 116–128. [Google Scholar] [CrossRef]
- Glaab, E.; Manoharan, G.B.; Abankwa, D. Pharmacophore Model for SARS-CoV-2 3CLpro Small-Molecule Inhibitors and in Vitro Experimental Validation of Computationally Screened Inhibitors. J. Chem. Inf. Model. 2021, 61, 4082–4096. [Google Scholar] [CrossRef]
- Ghahremanpour, M.M.; Tirado-Rives, J.; Deshmukh, M.; Ippolito, J.A.; Zhang, C.H.; Cabeza De Vaca, I.; Liosi, M.E.; Anderson, K.S.; Jorgensen, W.L. Identification of 14 Known Drugs as Inhibitors of the Main Protease of SARS-CoV-2. ACS Med. Chem. Lett. 2020, 11, 2526–2533. [Google Scholar] [CrossRef]
- Wang, L.; Wu, Y.; Deng, Y.; Kim, B.; Pierce, L.; Krilov, G.; Lupyan, D.; Robinson, S.; Dahlgren, M.K.; Greenwood, J.; et al. Accurate and Reliable Prediction of Relative Ligand Binding Potency in Prospective Drug Discovery by Way of a Modern Free-Energy Calculation Protocol and Force Field. J. Am. Chem. Soc. 2015, 137, 2695–2703. [Google Scholar] [CrossRef]
- Rastelli, G.; Degliesposti, G.; del Rio, A.; Sgobba, M. Binding Estimation after Refinement, a New Automated Procedure for the Refinement and Rescoring of Docked Ligands in Virtual Screening. Chem. Biol. Drug Des. 2009, 73, 283–286. [Google Scholar] [CrossRef]
- Jespers, W.; Åqvist, J.; Gutiérrez-de-Terán, H. Free Energy Calculations for Protein–Ligand Binding Prediction. Methods Mol. Biol. 2021, 2266, 203–226. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the Performance of the Molecular Mechanics/Poisson Boltzmann Surface Area and Molecular Mechanics/Generalized Born Surface Area Methods. II. The Accuracy of Ranking Poses Generated from Docking. J. Comput. Chem. 2011, 32, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Stone, E.A.; Deshmukh, M.; Ippolito, J.A.; Ghahremanpour, M.M.; Tirado-Rives, J.; Spasov, K.A.; Zhang, S.; Takeo, Y.; Kudalkar, S.N.; et al. Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations. ACS Cent. Sci. 2021, 7, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, X.; Huang, Y.Y.; Wu, Y.; Liu, R.; Zhou, L.; Lin, Y.; Wu, D.; Zhang, L.; Liu, H.; et al. Identify Potent SARS-CoV-2 Main Protease Inhibitors via Accelerated Free Energy Perturbation-Based Virtual Screening of Existing Drugs. Proc. Natl. Acad. Sci. USA 2020, 117, 27381–27387. [Google Scholar] [CrossRef]
- Ngo, S.T.; Tam, N.M.; Pham, M.Q.; Nguyen, T.H. Benchmark of Popular Free Energy Approaches Revealing the Inhibitors Binding to SARS-CoV-2 Mpro. J. Chem. Inf. Model. 2021, 61, 2302–2312. [Google Scholar] [CrossRef]
- Sherman, W.; Beard, H.S.; Farid, R. Use of an Induced Fit Receptor Structure in Virtual Screening. Chem. Biol. Drug Des. 2006, 67, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449. [Google Scholar] [CrossRef]
- Ibrahim, I.M.; Elfiky, A.A.; Fathy, M.M.; Mahmoud, S.H.; ElHefnawi, M. Targeting SARS-CoV-2 Endoribonuclease: A Structure-Based Virtual Screening Supported by in Vitro Analysis. Sci. Rep. 2022, 12, 13337. [Google Scholar] [CrossRef]
- Copeland, R.A.; Pompliano, D.L.; Meek, T.D. Drug-Target Residence Time and Its Implications for Lead Optimization. Nat. Rev. Drug Discov. 2006, 5, 730–739. [Google Scholar] [CrossRef] [PubMed]
- Bernetti, M.; Masetti, M.; Rocchia, W.; Cavalli, A. Kinetics of Drug Binding and Residence Time. Annu. Rev. Phys. Chem. 2019, 70, 143–171. [Google Scholar] [CrossRef] [PubMed]
- Pavan, M.; Menin, S.; Bassani, D.; Sturlese, M.; Moro, S. Qualitative Estimation of Protein–Ligand Complex Stability through Thermal Titration Molecular Dynamics Simulations. J. Chem. Inf. Model. 2022, 62, 5715–5728. [Google Scholar] [CrossRef] [PubMed]
- Zaidman, D.; Gehrtz, P.; Filep, M.; Fearon, D.; Gabizon, R.; Douangamath, A.; Prilusky, J.; Duberstein, S.; Cohen, G.; Owen, C.D.; et al. An Automatic Pipeline for the Design of Irreversible Derivatives Identifies a Potent SARS-CoV-2 Mpro Inhibitor. Cell Chem. Biol. 2021, 28, 1795–1806.e5. [Google Scholar] [CrossRef]
- Valiente, P.A.; Wen, H.; Nim, S.; Lee, J.; Kim, H.J.; Kim, J.; Perez-Riba, A.; Paudel, Y.P.; Hwang, I.; Kim, K.-D.; et al. Computational Design of Potent D-Peptide Inhibitors of SARS-CoV-2. J. Med. Chem. 2021, 64, 14955–14967. [Google Scholar] [CrossRef]
- Dehouck, Y.; Kwasigroch, J.M.; Rooman, M.; Gilis, D. BeAtMuSiC: Prediction of Changes in Protein-Protein Binding Affinity on Mutations. Nucleic Acids Res. 2013, 41, W333–W339. [Google Scholar] [CrossRef]
- Kandeel, M.; Yamamoto, M.; Tani, H.; Kobayashi, A.; Gohda, J.; Kawaguchi, Y.; Park, B.K.; Kwon, H.J.; Inoue, J.I.; Alkattan, A. Discovery of New Fusion Inhibitor Peptides against SARS-CoV-2 by Targeting the Spike S2 Subunit. Biomol. Ther. 2021, 29, 282–289. [Google Scholar] [CrossRef]
- Leman, J.K.; Weitzner, B.D.; Lewis, S.M.; Adolf-Bryfogle, J.; Alam, N.; Alford, R.F.; Aprahamian, M.; Baker, D.; Barlow, K.A.; Barth, P.; et al. Macromolecular Modeling and Design in Rosetta: Recent Methods and Frameworks. Nat. Methods 2020, 17, 665–680. [Google Scholar] [CrossRef]
- Jeong, B.S.; Cha, J.S.; Hwang, I.; Kim, U.; Adolf-Bryfogle, J.; Coventry, B.; Cho, H.S.; Kim, K.D.; Oh, B.H. Computational Design of a Neutralizing Antibody with Picomolar Binding Affinity for All Concerning SARS-CoV-2 Variants. MAbs 2022, 14, 2021601. [Google Scholar] [CrossRef]
- Sun, M.; Liu, S.; Wei, X.; Wan, S.; Huang, M.; Song, T.; Lu, Y.; Weng, X.; Lin, Z.; Chen, H.; et al. Aptamer Blocking Strategy Inhibits SARS-CoV-2 Virus Infection. Angew. Chem. Int. Ed. 2021, 60, 10266–10272. [Google Scholar] [CrossRef]
- Dou, J.; Vorobieva, A.A.; Sheffler, W.; Doyle, L.A.; Park, H.; Bick, M.J.; Mao, B.; Foight, G.W.; Lee, M.Y.; Gagnon, L.A.; et al. De Novo Design of a Fluorescence-Activating β-Barrel. Nature 2018, 561, 485–491. [Google Scholar] [CrossRef]
- Cao, L.; Goreshnik, I.; Coventry, B.; Case, J.B.; Miller, L.; Kozodoy, L.; Chen, R.E.; Carter, L.; Walls, A.C.; Park, Y.J.; et al. De Novo Design of Picomolar SARS-CoV-2 Miniprotein Inhibitors. Science 2020, 370, 426–431. [Google Scholar] [CrossRef]
- Kortemme, T.; Kim, D.E.; Baker, D. Computational Alanine Scanning of Protein-Protein Interfaces. Sci. STKE 2004, 2004, pl2. [Google Scholar] [CrossRef]
- Kortemme, T.; Baker, D. A Simple Physical Model for Binding Energy Hot Spots in Protein-Protein Complexes. Proc. Natl. Acad. Sci. USA 2002, 99, 14116–14121. [Google Scholar] [CrossRef]
- Glasgow, A.; Glasgow, J.; Limonta, D.; Solomon, P.; Lui, I.; Zhang, Y.; Nix, M.A.; Rettko, N.J.; Zha, S.; Yamin, R.; et al. Engineered ACE2 Receptor Traps Potently Neutralize SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 28046–28055. [Google Scholar] [CrossRef]
- de Vivo, M.; Masetti, M.; Bottegoni, G.; Cavalli, A. Role of Molecular Dynamics and Related Methods in Drug Discovery. J. Med. Chem. 2016, 59, 4035–4061. [Google Scholar] [CrossRef]
- Ferrari, F.; Bissaro, M.; Fabbian, S.; de Almeida Roger, J.; Mammi, S.; Moro, S.; Bellanda, M.; Sturlese, M. HT-SuMD: Making Molecular Dynamics Simulations Suitable for Fragment-Based Screening. a Comparative Study with NMR. J. Enzym. Inhib. Med. Chem. 2020, 36, 1–14. [Google Scholar] [CrossRef]
- Verdonk, M.L.; Giangreco, I.; Hall, R.J.; Korb, O.; Mortenson, P.N.; Murray, C.W. Docking Performance of Fragments and Druglike Compounds. J. Med. Chem. 2011, 54, 5422–5431. [Google Scholar] [CrossRef]
- Bissaro, M.; Bolcato, G.; Pavan, M.; Bassani, D.; Sturlese, M.; Moro, S. Inspecting the Mechanism of Fragment Hits Binding on SARS-CoV-2 M pro by Using Supervised Molecular Dynamics (SuMD) Simulations. ChemMedChem 2021, 16, 2075–2081. [Google Scholar] [CrossRef]
- Sabbadin, D.; Moro, S. Supervised Molecular Dynamics (SuMD) as a Helpful Tool to Depict GPCR-Ligand Recognition Pathway in a Nanosecond Time Scale. J. Chem. Inf. Model. 2014, 54, 372–376. [Google Scholar] [CrossRef]
- Cuzzolin, A.; Sturlese, M.; Deganutti, G.; Salmaso, V.; Sabbadin, D.; Ciancetta, A.; Moro, S. Deciphering the Complexity of Ligand-Protein Recognition Pathways Using Supervised Molecular Dynamics (SuMD) Simulations. J. Chem. Inf. Model. 2016, 56, 687–705. [Google Scholar] [CrossRef]
- Pavan, M.; Bolcato, G.; Bassani, D.; Sturlese, M.; Moro, S. Supervised Molecular Dynamics (SuMD) Insights into the Mechanism of Action of SARS-CoV-2 Main Protease Inhibitor PF-07321332. J. Enzym. Inhib. Med. Chem. 2021, 36, 1646–1650. [Google Scholar] [CrossRef]
- Pavan, M.; Bassani, D.; Sturlese, M.; Moro, S. Investigating RNA–Protein Recognition Mechanisms through Supervised Molecular Dynamics (SuMD) Simulations. NAR Genom. Bioinform. 2022, 4, lqac088. [Google Scholar] [CrossRef]
- Mathieu, E.; Ritchie, H.; Rodés-Guirao, L.; Appel, C.; Giattino, C.; Hasell, J.; Macdonald, B.; Dattani, S.; Beltekian, D.; Ortiz-Ospina, E.; et al. Coronavirus Pandemic (COVID-19). Our World Data 2020. [Google Scholar]
- Sallam, M. COVID-19 Vaccine Hesitancy Worldwide: A Concise Systematic Review of Vaccine Acceptance Rates. Vaccines 2021, 9, 160. [Google Scholar] [CrossRef]
- Fisher, K.A.; Bloomstone, S.J.; Walder, J.; Crawford, S.; Fouayzi, H.; Mazor, K.M. Attitudes toward a Potential SARS-CoV-2 Vaccine: A Survey of U.S. Adults. Ann. Intern. Med. 2020, 173, 964–973. [Google Scholar] [CrossRef]
- SARS-CoV-2 Variants of Concern as of 9 February 2023. Available online: https://www.ecdc.europa.eu/en/covid-19/variants-concern (accessed on 17 February 2023).
- Hall, V.; Foulkes, S.; Insalata, F.; Kirwan, P.; Saei, A.; Atti, A.; Wellington, E.; Khawam, J.; Munro, K.; Cole, M.; et al. Protection against SARS-CoV-2 after COVID-19 Vaccination and Previous Infection. N. Engl. J. Med. 2022, 386, 1207–1220. [Google Scholar] [CrossRef]
- Coronavirus Disease (COVID-19) Situation Reports. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 17 February 2023).
- Miller, J.K.; Elenberg, K.; Dubrawski, A. Forecasting Emergence of COVID-19 Variants of Concern. PLoS ONE 2022, 17, e0264198. [Google Scholar] [CrossRef]
- Magiorkinis, G. On the Evolution of SARS-CoV-2 and the Emergence of Variants of Concern. Trends Microbiol. 2023, 31, 5–8. [Google Scholar] [CrossRef]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and Ecology of Influenza A Viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [CrossRef]
- Carrat, F.; Flahault, A. Influenza Vaccine: The Challenge of Antigenic Drift. Vaccine 2007, 25, 6852–6862. [Google Scholar] [CrossRef]
- Martin, R.; Löchel, H.F.; Welzel, M.; Hattab, G.; Hauschild, A.C.; Heider, D. CORDITE: The Curated CORona Drug InTERactions Database for SARS-CoV-2. iScience 2020, 23, 101297. [Google Scholar] [CrossRef]
- Crook, H.; Raza, S.; Nowell, J.; Young, M.; Edison, P. Long COVID—Mechanisms, Risk Factors, and Management. BMJ 2021, 374, n1648. [Google Scholar] [CrossRef] [PubMed]
- Hijikata, A.; Shionyu, C.; Nakae, S.; Shionyu, M.; Ota, M.; Kanaya, S.; Shirai, T. Current Status of Structure-Based Drug Repurposing against COVID-19 by Targeting SARS-CoV-2 Proteins. Biophys. Physicobiol. 2021, 18, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Gupta, Y.; Savytskyi, O.V.; Coban, M.; Venugopal, A.; Pleqi, V.; Weber, C.A.; Chitale, R.; Durvasula, R.; Hopkins, C.; Kempaiah, P.; et al. Protein Structure-Based in-Silico Approaches to Drug Discovery: Guide to COVID-19 Therapeutics. Mol. Asp. Med. 2023, 91, 101151. [Google Scholar] [CrossRef] [PubMed]
- Gurung, A.B.; Ali, M.A.; Lee, J.; Farah, M.A.; Al-Anazi, K.M. An Updated Review of Computer-Aided Drug Design and Its Application to COVID-19. Biomed. Res. Int. 2021, 2021, 8853056. [Google Scholar] [CrossRef] [PubMed]
- Floresta, G.; Zagni, C.; Gentile, D.; Patamia, V.; Rescifina, A. Artificial Intelligence Technologies for COVID-19 De Novo Drug Design. Int. J. Mol. Sci. 2022, 23, 3261. [Google Scholar] [CrossRef] [PubMed]
- Marani, M.; Katul, G.G.; Pan, W.K.; Parolari, A.J. Intensity and Frequency of Extreme Novel Epidemics. Proc. Natl. Acad. Sci. USA 2021, 118, e2105482118. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).