Intracellular Nitric Oxide and cAMP Are Involved in Cellulolytic Enzyme Production in Neurospora crassa
Abstract
:1. Introduction
2. Results
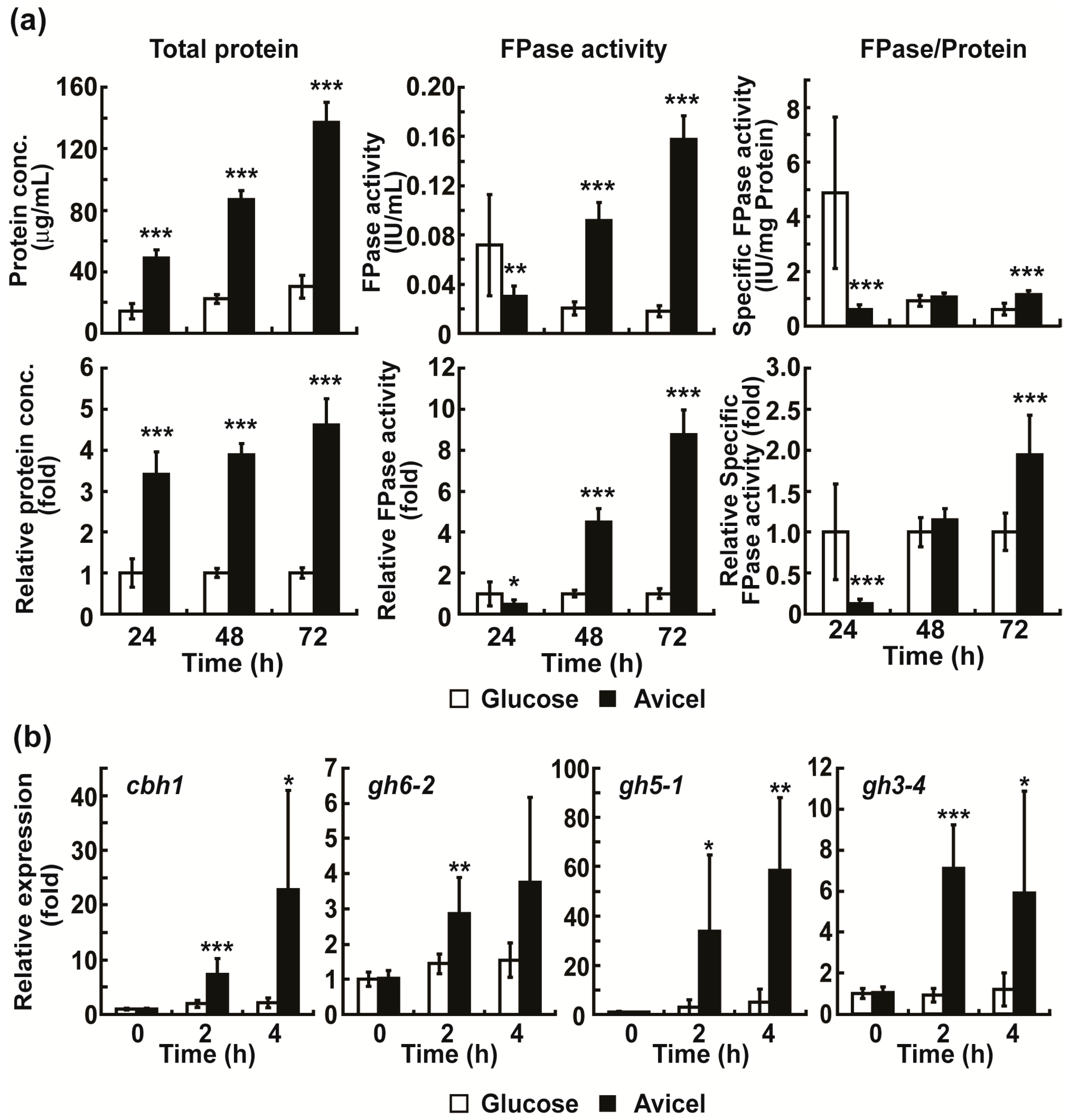
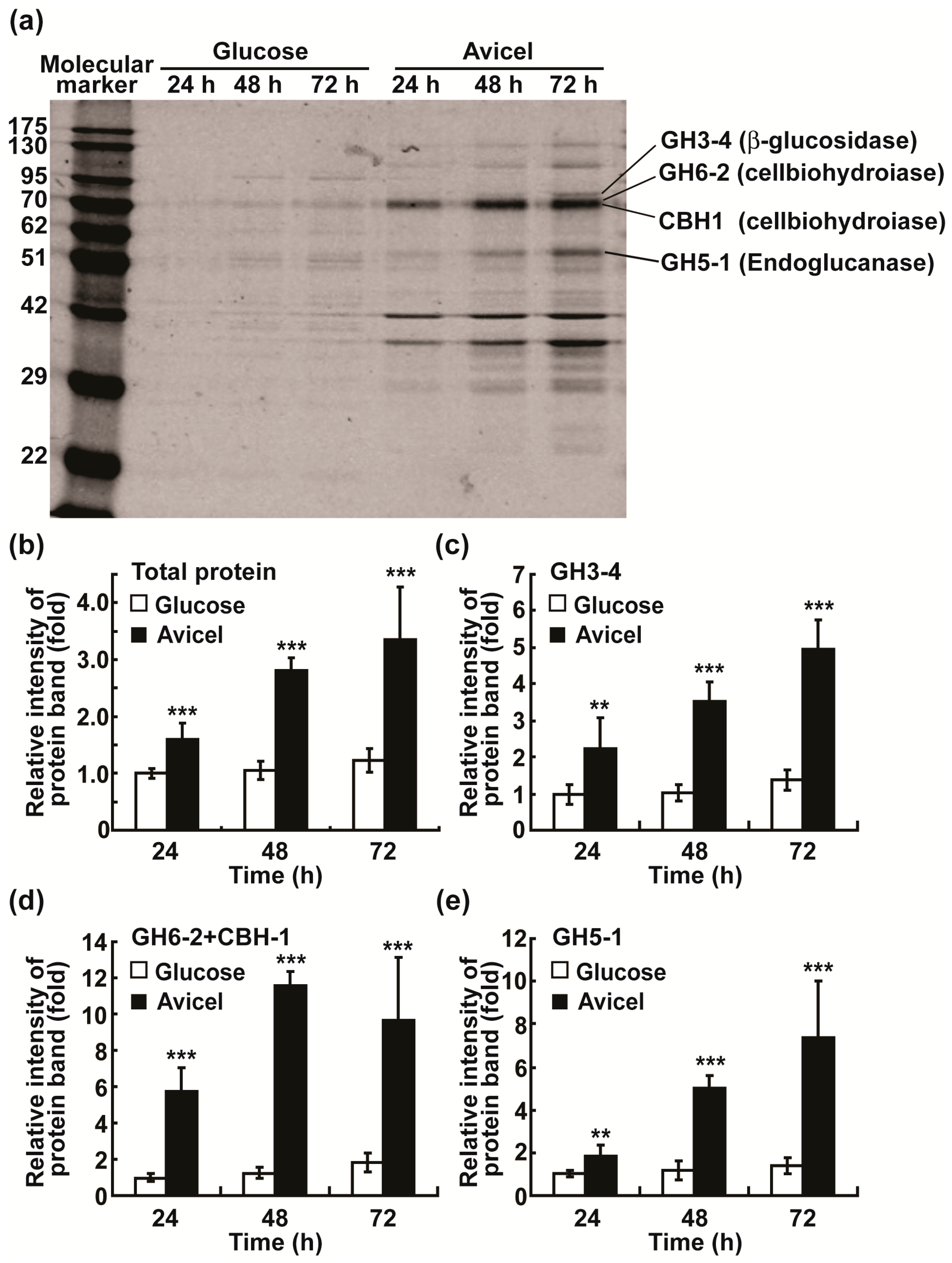
2.1. Avicel Induces Cellulase Production in N. crassa
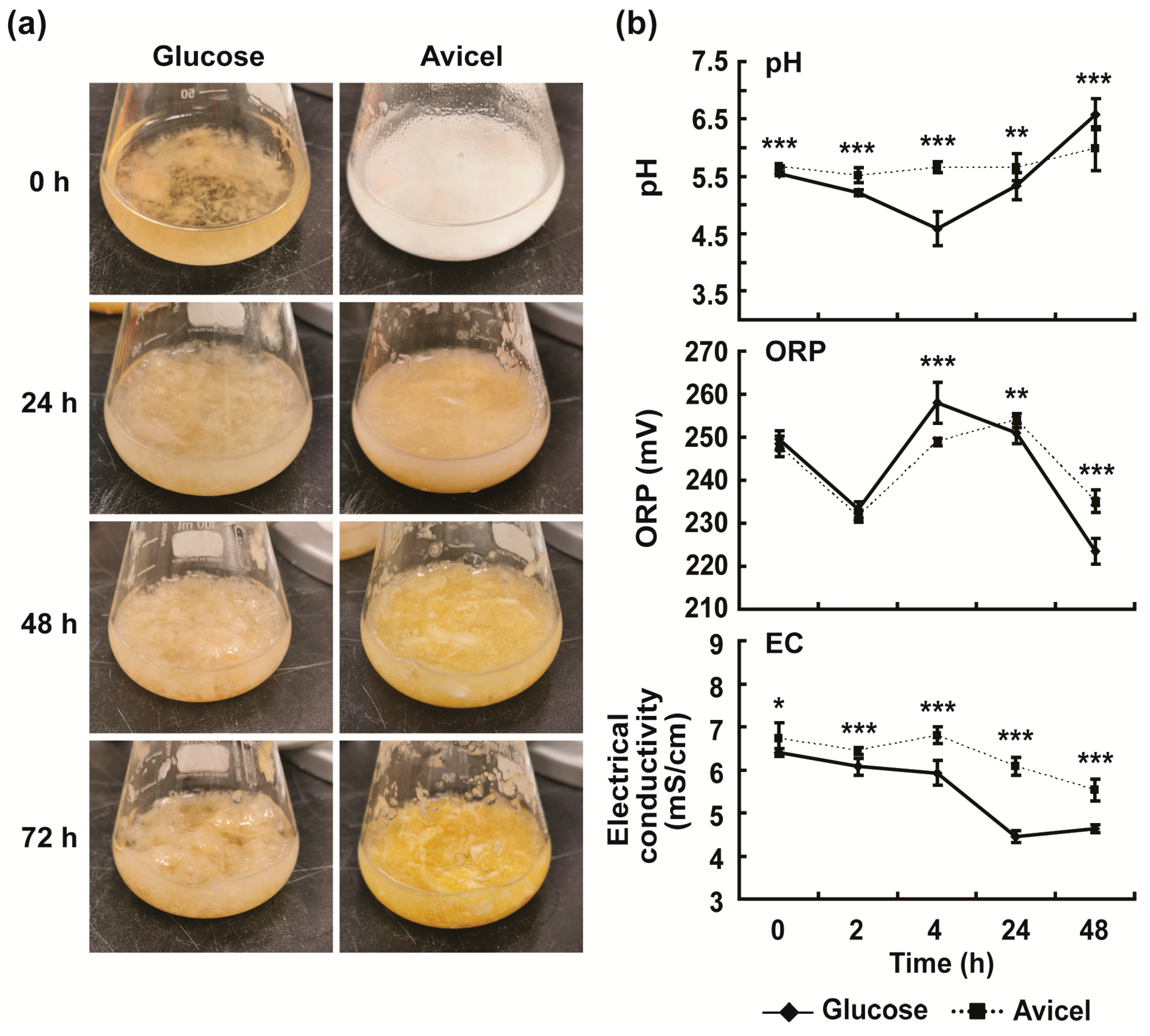
2.2. Physicochemical Properties Were Different between Glucose and Avicel Media
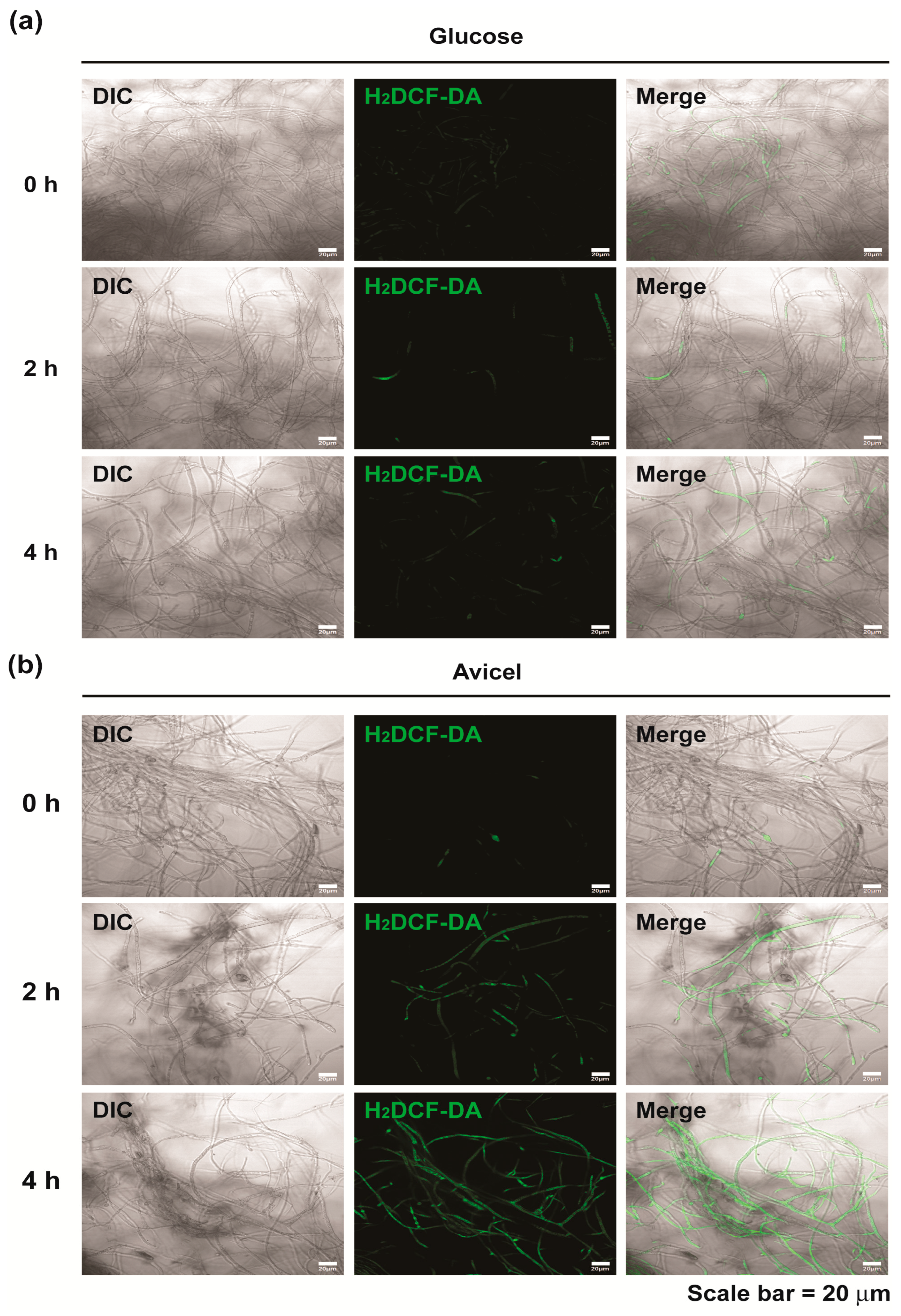
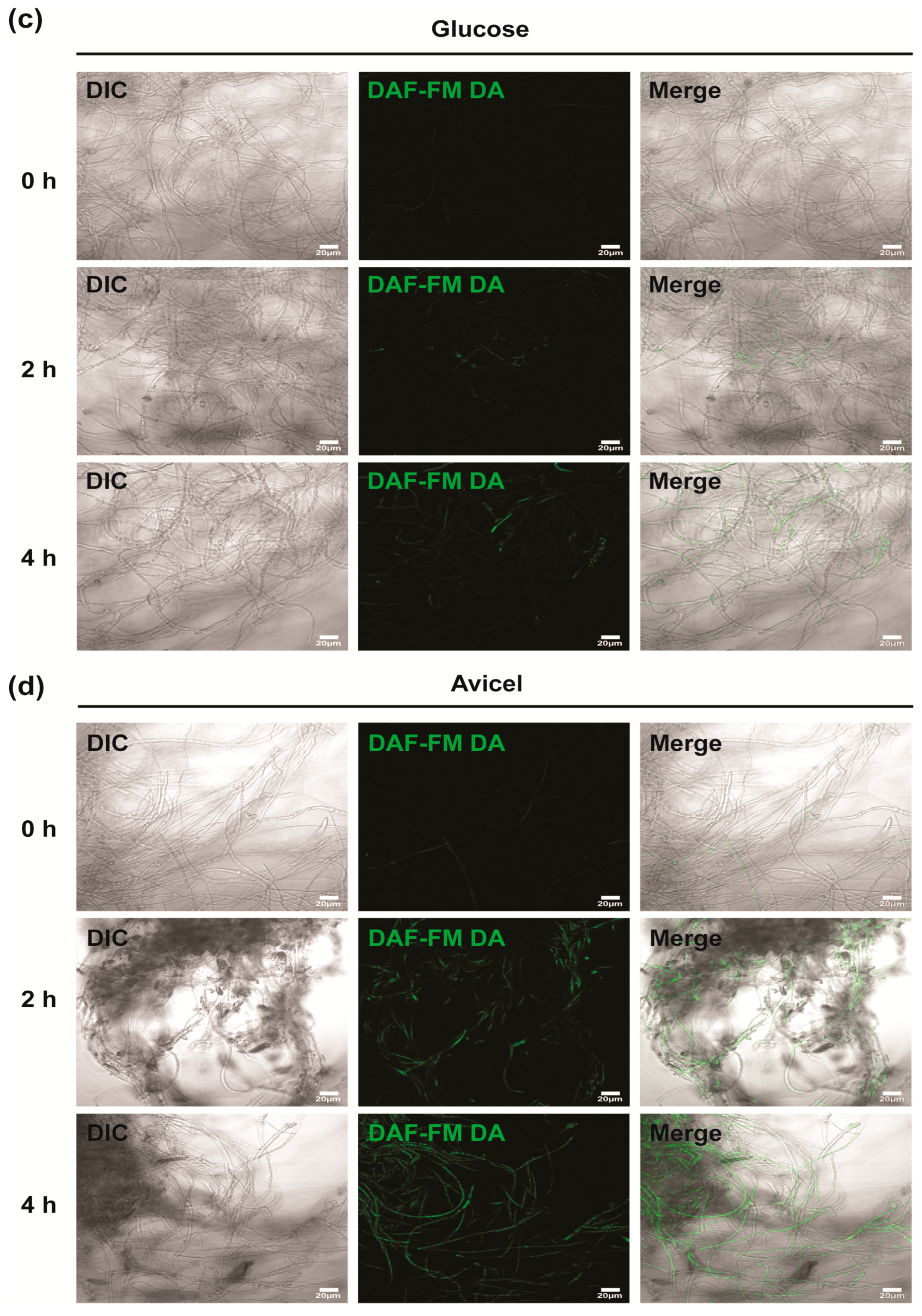
2.3. Intracellular NO and ROS Levels Significantly Increased in N. crassa Cultured in Avicel Medium
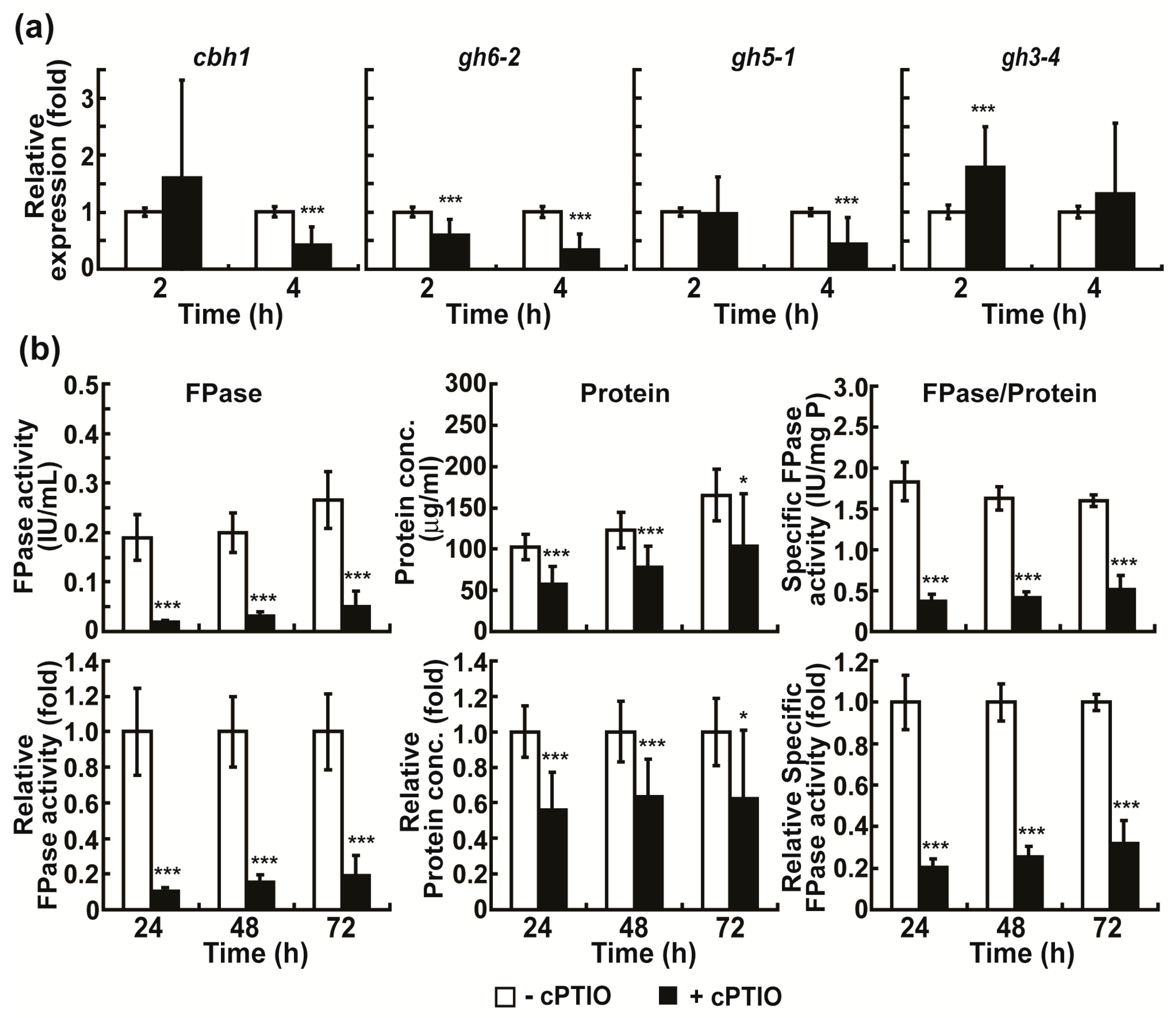
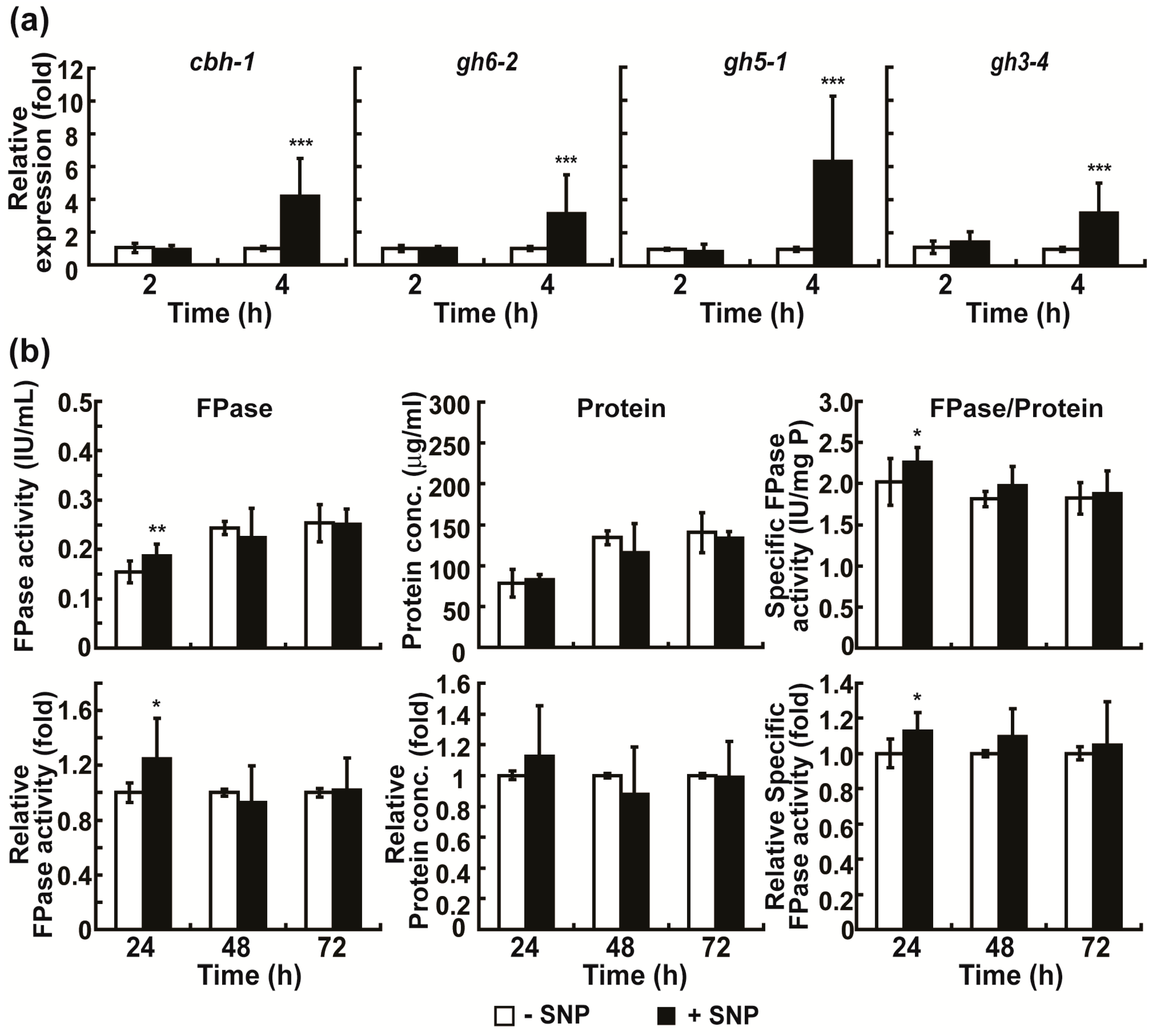
2.4. Intracellular NO Is Involved in the Regulation of Cellulolytic Enzyme Production in Avicel Medium
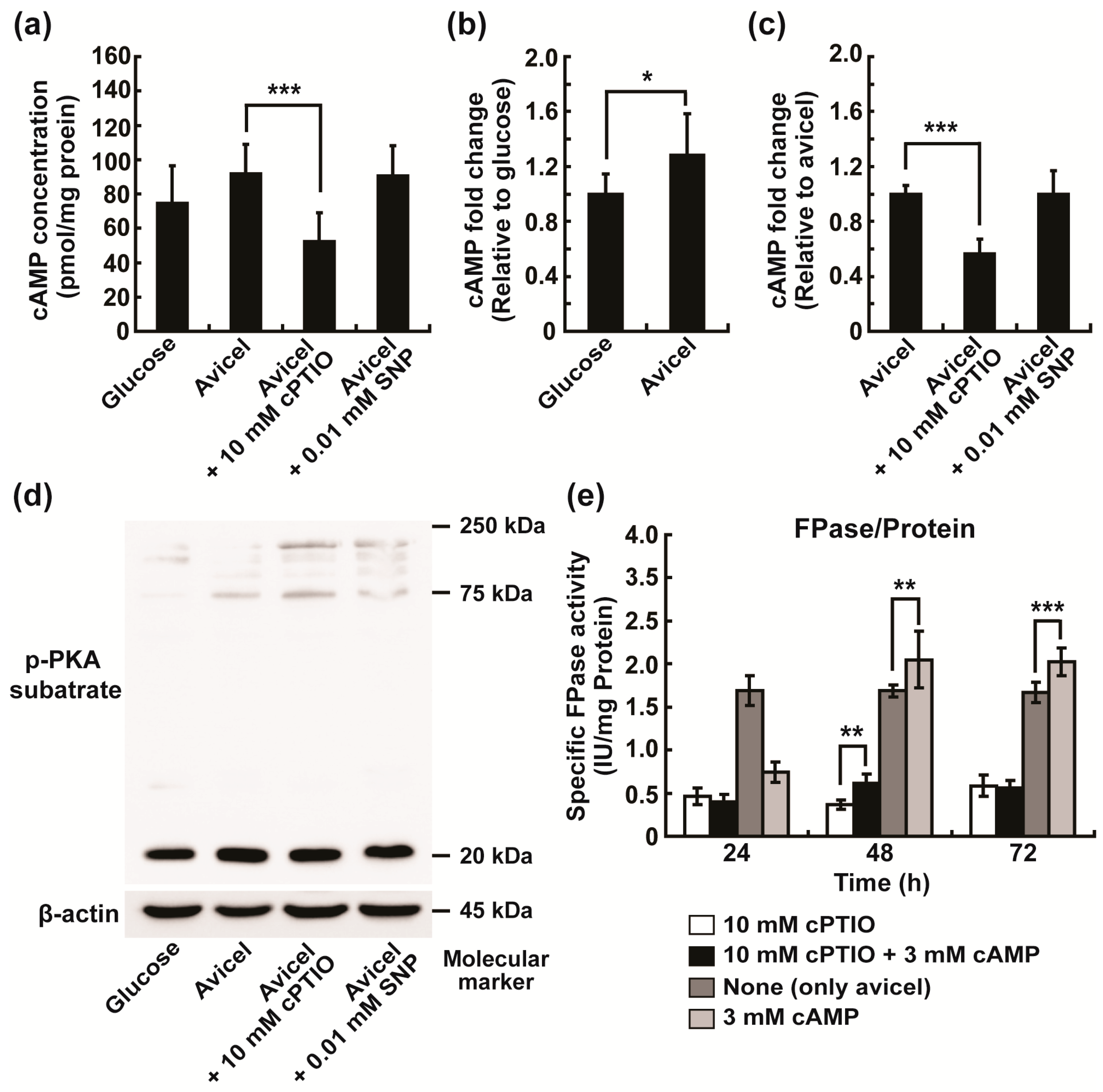
2.5. Intracellular NO May Be Closely Associated with cAMP Signaling in the Regulation of Cellulolytic Enzyme Production
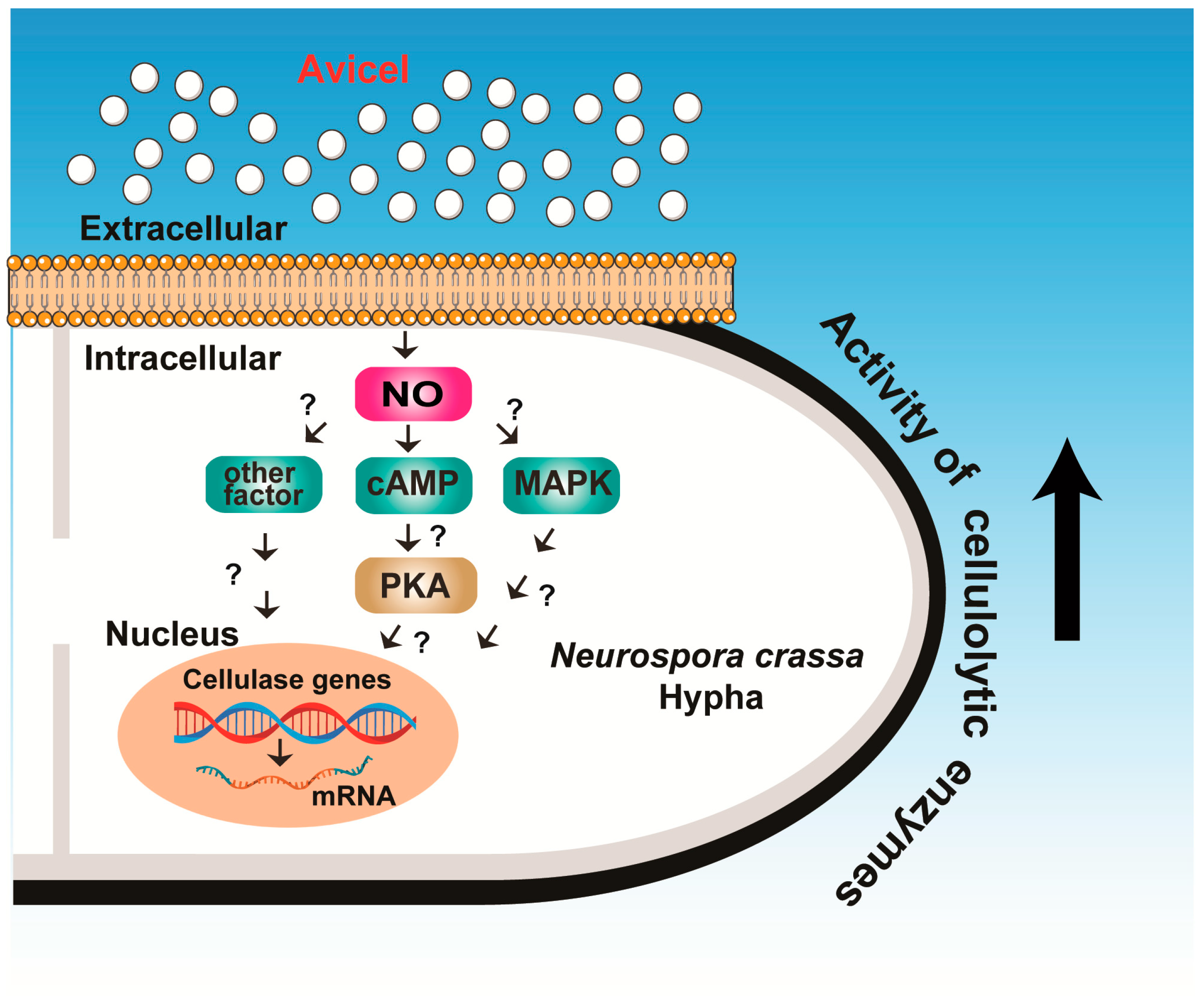
3. Discussion
4. Materials and Methods
4.1. Fungal Strain and Culture Conditions
4.2. Measurement of EC, ORP, and pH in the Media
4.3. Assay for Enzymatic Activities and Protein Concentration
4.4. Protein Gel Electrophoresis and Western Blotting
4.5. Quantitative Real-Time PCR Analysis
4.6. Fluorescence Staining
4.7. cPTIO (NO Scavenger), SNP (NO Donor), and cAMP Treatment
4.8. cAMP Concentration Measurement
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reimer, M.; Zollfrank, C. Cellulose for Light Manipulation: Methods, Applications, and Prospects. Adv. Energy Mater. 2021, 11, 2003866. [Google Scholar] [CrossRef]
- Jayasekara, S.; Ratnayake, R. Microbial Cellulases: An overview and applications, cellulose. In Cellulose; Pascual, A.R., María, E., Martín, E., Eds.; IntechOpen: London, UK, 2019. [Google Scholar]
- Bhardwaj, N.; Kumar, B.; Agrawal, K.; Verma, P. Current perspective on production and applications of microbial cellulases: A review. Bioresour. Bioprocess. 2021, 8, 95. [Google Scholar] [CrossRef]
- Li, J.X.; Zhang, F.; Jiang, D.D.; Li, J.; Wang, F.L.; Zhang, Z.; Wang, W.; Zhao, X.Q. Diversity of Cellulase-Producing Filamentous Fungi From Tibet and Transcriptomic Analysis of a Superior Cellulase Producer Trichoderma harzianum LZ117. Front. Microbiol. 2020, 11, 1617. [Google Scholar] [CrossRef]
- Imran, M.; Anwar, Z.; Irshad, M.; Asad, M.J.; Ashfaq, H. Cellulase Production from Species of Fungi and Bacteria from Agricultural Wastes and Its Utilization in Industry: A Review. Adv. Enzym. Res. 2016, 4, 44–55. [Google Scholar] [CrossRef] [Green Version]
- Lubeck, M.; Lubeck, P.S. Fungal Cell Factories for Efficient and Sustainable Production of Proteins and Peptides. Microorganisms 2022, 10, 753. [Google Scholar] [CrossRef]
- Zhao, X.-Q.; Zhang, X.-Y.; Zhang, F.; Zhang, R.; Jiang, B.-J.; Bai, F.-W. Metabolic engineering of fungal strains for efficient production of cellulolytic enzymes. In Fungal Cellulolytic Enzymes; Springer: New York, NY, USA, 2018; pp. 27–41. [Google Scholar]
- Znameroski, E.A.; Coradetti, S.T.; Roche, C.M.; Tsai, J.C.; Iavarone, A.T.; Cate, J.H.D.; Glass, N.L. Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. Proc. Natl. Acad. Sci. USA 2012, 109, 6012–6017. [Google Scholar] [CrossRef] [Green Version]
- Martzy, R.; Mello-de-Sousa, T.M.; Mach, R.L.; Yaver, D.; Mach-Aigner, A.R. The phenomenon of degeneration of industrial Trichoderma reesei strains. Biotechnol. Biofuels 2021, 14, 193. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.M.; Iavarone, A.T.; Marletta, M.A. Quantitative Proteomic Approach for Cellulose Degradation by Neurospora crassa. J. Proteome Res. 2011, 10, 4177–4185. [Google Scholar] [CrossRef]
- Lin, L.C.; Wang, S.S.; Li, X.L.; He, Q.; Benz, J.P.; Tian, C.G. STK-12 act as a transcriptional brake to control the expression of cellulase-encoding genes in Neurospora crassa. PLoS Genet. 2019, 15, e1008510. [Google Scholar] [CrossRef] [Green Version]
- Mattam, A.J.; Chaudhari, Y.B.; Velankar, H.R. Factors regulating cellulolytic gene expression in filamentous fungi: An overview. Microbial Cell Factories 2022, 21, 44. [Google Scholar] [CrossRef] [PubMed]
- Znameroski, E.A.; Glass, N.L. Using a model filamentous fungus to unravel mechanisms of lignocellulose deconstruction. Biotechnol. Biofuels 2013, 6, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Znameroski, E.A.; Li, X.; Tsai, J.C.; Galazka, J.M.; Glass, N.L.; Cate, J.H.D. Evidence for Transceptor Function of Cellodextrin Transporters in Neurospora crassa. J. Biol. Chem. 2014, 289, 2610–2619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collier, L.A.; Ghosh, A.; Borkovich, K.A. Heterotrimeric G-Protein Signaling Is Required for Cellulose Degradation in Neurospora crassa. Mbio 2020, 11, e02419–e02420. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, Q.; Yang, J.; Jiang, B.; Wang, F.; Liu, K.; Fang, X. A Mitogen-Activated Protein Kinase Tmk3 Participates in High Osmolarity Resistance, Cell Wall Integrity Maintenance and Cellulase Production Regulation in Trichoderma reesei. PLoS ONE 2013, 8, e72189. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Zou, G.; Wang, J.; Wang, J.; Liu, R.; Jiang, Y.; Zhao, G.; Zhou, Z. Characterization of the Ca2+-responsive signaling pathway in regulating the expression and secretion of cellulases in Trichoderma reesei Rut-C30. Mol. Microbiol. 2016, 100, 560–575. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Liu, Y.J.; Hao, X.R.; Wang, D.; Akhberdi, O.; Xiang, B.Y.; Zhu, X.D. Regulation of the G alpha-cAMP/PKA signaling pathway in cellulose utilization of Chaetomium globosum. Microb. Cell Factories 2018, 17, 160. [Google Scholar] [CrossRef]
- de Paula, R.G.; Antoniêto, A.C.C.; Carraro, C.B.; Lopes, D.C.B.; Persinoti, G.F.; Peres, N.T.A.; Martinez-Rossi, N.M.; Silva-Rocha, R.; Silva, R.N. The Duality of the MAPK Signaling Pathway in the Control of Metabolic Processes and Cellulase Production in Trichoderma reesei. Sci. Rep. 2018, 8, 14931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins-Santana, L.; de Paula, R.G.; Silva, A.G.; Lopes, D.C.B.; Silva, R.D.; Silva-Rocha, R. CRZ1 regulator and calcium cooperatively modulate holocellulases gene expression in Trichoderma reesei QM6a. Genet. Mol. Biol. 2020, 43, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Cheng, S.; Li, Z.; Men, Y.; Wu, J. Impacts of Cellulase and Amylase on Enzymatic Hydrolysis and Methane Production in the Anaerobic Digestion of Corn Straw. Sustainability 2020, 12, 5453. [Google Scholar] [CrossRef]
- Chen, Y.; Fan, X.; Zhao, X.; Shen, Y.; Xu, X.; Wei, L.; Wang, W.; Wei, D. cAMP activates calcium signalling via phospholipase C to regulate cellulase production in the filamentous fungus Trichoderma reesei. Biotechnol. Biofuels 2021, 14, 62. [Google Scholar] [CrossRef]
- Ma, B.; Ning, Y.-N.; Li, C.-X.; Tian, D.; Guo, H.; Pang, X.-M.; Luo, X.-M.; Zhao, S.; Feng, J.-X. A mitogen-activated protein kinase PoxMK1 mediates regulation of the production of plant-biomass-degrading enzymes, vegetative growth, and pigment biosynthesis in Penicillium oxalicum. Appl. Microbiol. Biotechnol. 2021, 105, 661–678. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Xu, Y.; Yu, X.-W. From induction to secretion: A complicated route for cellulase production in Trichoderma reesei. Bioresour. Bioprocess. 2021, 8, 107. [Google Scholar] [CrossRef]
- Lian, L.D.; Shi, L.Y.; Zhu, J.; Liu, R.; Shi, L.; Ren, A.; Yu, H.S.; Zhao, M.W. GlSwi6 Positively Regulates Cellulase and Xylanase Activities through Intracellular Ca2+ Signaling in Ganoderma lucidum. J. Fungi 2022, 8, 187. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.T.; Wei, L.F.; Wang, Y.F.; Li, J.Y.; Liu, X.; Zhou, Y.H.; Du, P.P.; Fang, H.; Liesche, J.; Wei, Y.H.; et al. Parallel proteomic and phosphoproteomic analyses reveal cellobiose-dependent regulation of lignocellulase secretion in the filamentous fungus Neurospora crassa. Glob. Change Biol. Bioenergy 2021, 13, 1372–1387. [Google Scholar] [CrossRef]
- Li, Q.X.; Ng, W.T.; Wu, J.C. Isolation, characterization and application of a cellulose-degrading strain Neurospora crassa S1 from oil palm empty fruit bunch. Microb. Cell Factories 2014, 13, 157. [Google Scholar] [CrossRef]
- Adnan, M.; Zheng, W.H.; Islam, W.; Arif, M.; Abubakar, Y.S.; Wang, Z.H.; Lu, G.D. Carbon Catabolite Repression in Filamentous Fungi. Int. J. Mol. Sci. 2018, 19, 48. [Google Scholar] [CrossRef] [Green Version]
- Coradetti, S.T.; Xiong, Y.; Glass, N.L. Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. MicrobiologyOpen 2013, 2, 595–609. [Google Scholar] [CrossRef]
- Vongvichiankul, C.; Deebao, J.; Khongnakorn, W. Relationship between pH, Oxidation Reduction Potential (ORP) and Biogas Production in Mesophilic Screw Anaerobic Digester. Energy Procedia 2017, 138, 877–882. [Google Scholar] [CrossRef]
- Shcherbakov, V.V.; Artemkina, Y.M.; Akimova, I.A.; Artemkina, I.M. Dielectric Characteristics, Electrical Conductivity and Solvation of Ions in Electrolyte Solutions. Materials 2021, 14, 5617. [Google Scholar] [CrossRef]
- Jones, A.R.; Meshulam, T.; Oliveira, M.F.; Burritt, N.; Corkey, B.E. Extracellular Redox Regulation of Intracellular Reactive Oxygen Generation, Mitochondrial Function and Lipid Turnover in Cultured Human Adipocytes. PLoS ONE 2016, 11, e0164011. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, Y.; Sawa, Y.; Nishimura, M.; Fukuyama, N.; Ichikawa, H.; Ohtake, S.; Nakazawa, H.; Matsuda, H. Peroxynitrite, a product between nitric oxide and superoxide anion, plays a cytotoxic role in the development of post-bypass systemic inflammatory response. Eur. J. Cardiothorac. Surg. 2004, 26, 276–280. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.M.; Shen, Y.L.; Wang, W.; Wei, D.Z. Mn2+ modulates the expression of cellulase genes in Trichoderma reesei Rut-C30 via calcium signaling. Biotechnol. Biofuels 2018, 11, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.Y.; Zhang, M.L.; Li, L.; Dong, Y.M.; Jiang, Y.; Liu, K.M.; Zhang, R.Q.; Jiang, B.J.; Niu, K.L.; Fang, X. Role of Trichoderma reesei mitogen-activated protein kinases (MAPKs) in cellulase formation. Biotechnol. Biofuels 2017, 10, 99. [Google Scholar] [CrossRef]
- Wang, Z.X.; An, N.; Xu, W.Q.; Zhang, W.X.; Meng, X.F.; Chen, G.J.; Liu, W.F. Functional characterization of the upstream components of the Hog1-like kinase cascade in hyperosmotic and carbon sensing in Trichoderma reesei. Biotechnol. Biofuels 2018, 11, 97. [Google Scholar] [CrossRef] [Green Version]
- Wiberth, C.-C.; Casandra, A.-Z.C.; Zhiliang, F.; Gabriela, H. Oxidative enzymes activity and hydrogen peroxide production in white-rot fungi and soil-borne micromycetes co-cultures. Ann. Microbiol. 2019, 69, 171–181. [Google Scholar] [CrossRef]
- Li, N.; Zeng, Y.; Chen, Y.; Shen, Y.; Wang, W. Induction of cellulase production by Sr2+ in Trichoderma reesei via calcium signaling transduction. Bioresour. Bioprocess. 2022, 9, 96. [Google Scholar] [CrossRef]
- Kimbrough, D.; Kouame, Y.; Moheban, P.; Springthorpe, S. The effect of electrolysis and oxidation-reduction potential on microbial survival, growth, and disinfection. Int. J. Environ. Pollut. 2006, 27, 211–221. [Google Scholar] [CrossRef]
- Essien, J.P.; Itah, A.Y.; Eduok, S.I. Influence of electrical conductivity on microorganisms and rate of crude oil mineralization in Niger Delta Ultisol. Glob. J. Pure Appl. Sci. 2003, 9, 199–204. [Google Scholar] [CrossRef] [Green Version]
- Schuster, A.; Tisch, D.; Seidl-Seiboth, V.; Kubicek, C.P.; Schmoll, M. Roles of protein kinase A and adenylate cyclase in light-modulated cellulase regulation in Trichoderma reesei. Appl. Env. Microbiol. 2012, 78, 2168–2178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, R.; Ma, L.; Li, C.; Jia, W.; Li, D.; Zhang, D.; Chen, S. Trpac1, a pH response transcription regulator, is involved in cellulase gene expression in Trichoderma reesei. Enzym. Microb. Technol. 2014, 67, 17–26. [Google Scholar] [CrossRef]
- Antonieto, A.C.C.; Pedersoli, W.R.; Castro, L.D.; Santos, R.D.; Cruz, A.H.D.; Nogueira, K.M.V.; Silva-Rocha, R.; Rossi, A.; Silva, R.N. Deletion of pH Regulator pac-3 Affects Cellulase and Xylanase Activity during Sugarcane Bagasse Degradation by Neurospora crassa. PLoS ONE 2017, 12, e0169796. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Genes | Primer Sequences |
|---|---|
| β-actin | Forward- 5′-TGA TCT TAC CGA CTA CCT-3′ |
| Reverse- 5′-CAG AGC TTC TCC TTG ATG-3′ | |
| cbh-1 | Forward- 5′-ATC TGG GAA GCG AAC AAA G-3′ |
| Reverse- 5′-TAG CGG TCG TCG GAA TAG-3′ | |
| gh6-2 | Forward- 5′-CCC ATC ACC ACT ACT ACC-3′ |
| Reverse- 5′-CCA GCC CTG AAC ACC AAG-3′ | |
| gh5-1 | Forward- 5′-GAG TTC ACA TTC CCT GAC A-3′ |
| Reverse- 5′-CGA AGC CAA CAC GGAAGA-3′ | |
| gh3-4 | Forward- 5′-AAC AAG GTC AAC GGT ACG TGG-3′ |
| Reverse- 5′-TCG TCA TAT CCA TAC CAC TGT TTG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, N.-N.; Ketya, W.; Park, G. Intracellular Nitric Oxide and cAMP Are Involved in Cellulolytic Enzyme Production in Neurospora crassa. Int. J. Mol. Sci. 2023, 24, 4503. https://doi.org/10.3390/ijms24054503
Yu N-N, Ketya W, Park G. Intracellular Nitric Oxide and cAMP Are Involved in Cellulolytic Enzyme Production in Neurospora crassa. International Journal of Molecular Sciences. 2023; 24(5):4503. https://doi.org/10.3390/ijms24054503
Chicago/Turabian StyleYu, Nan-Nan, Wirinthip Ketya, and Gyungsoon Park. 2023. "Intracellular Nitric Oxide and cAMP Are Involved in Cellulolytic Enzyme Production in Neurospora crassa" International Journal of Molecular Sciences 24, no. 5: 4503. https://doi.org/10.3390/ijms24054503
APA StyleYu, N. -N., Ketya, W., & Park, G. (2023). Intracellular Nitric Oxide and cAMP Are Involved in Cellulolytic Enzyme Production in Neurospora crassa. International Journal of Molecular Sciences, 24(5), 4503. https://doi.org/10.3390/ijms24054503







