Psoriatic Resolved Skin Epidermal Keratinocytes Retain Disease-Residual Transcriptomic and Epigenomic Profiles
Abstract
1. Introduction
2. Results
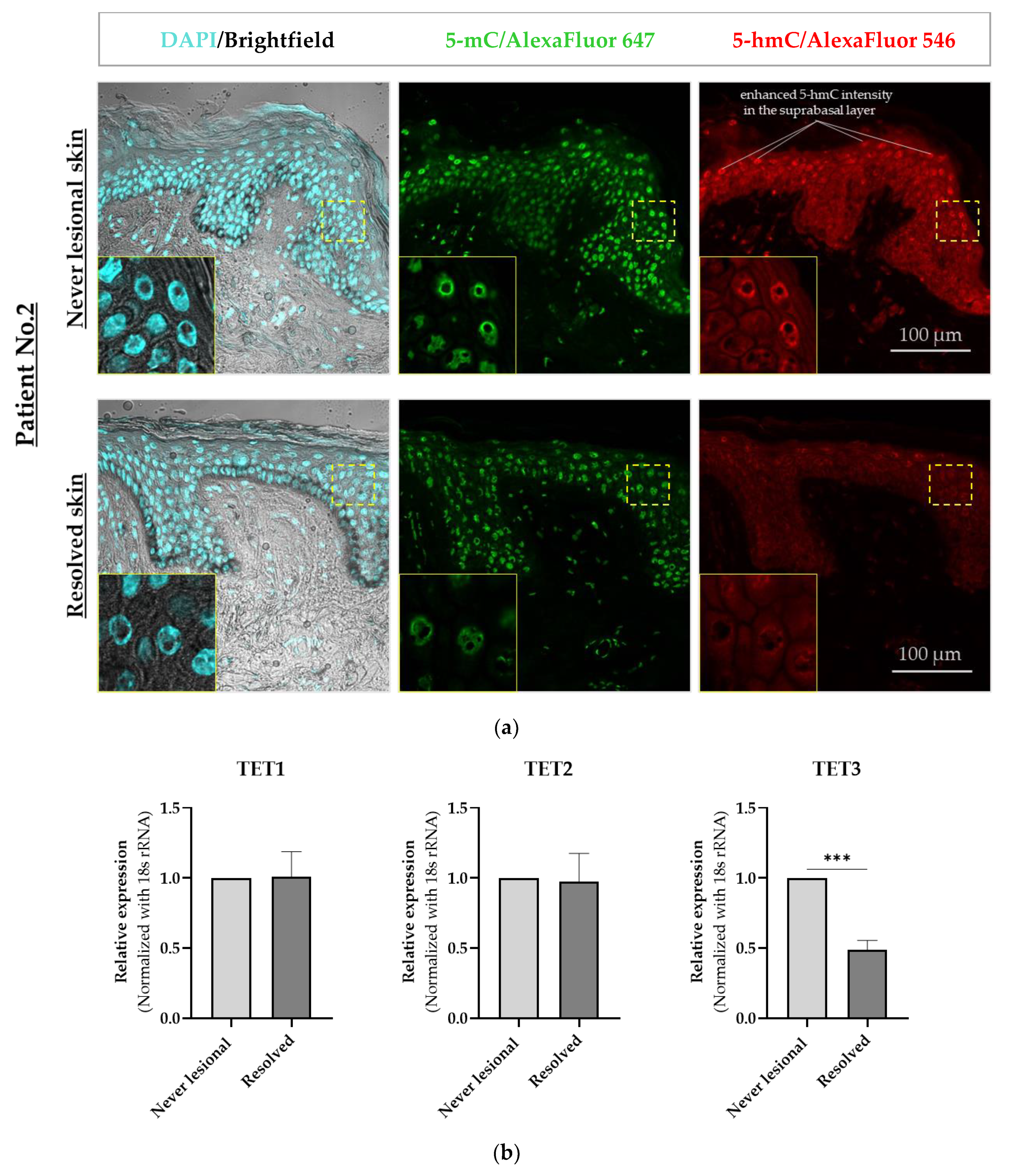
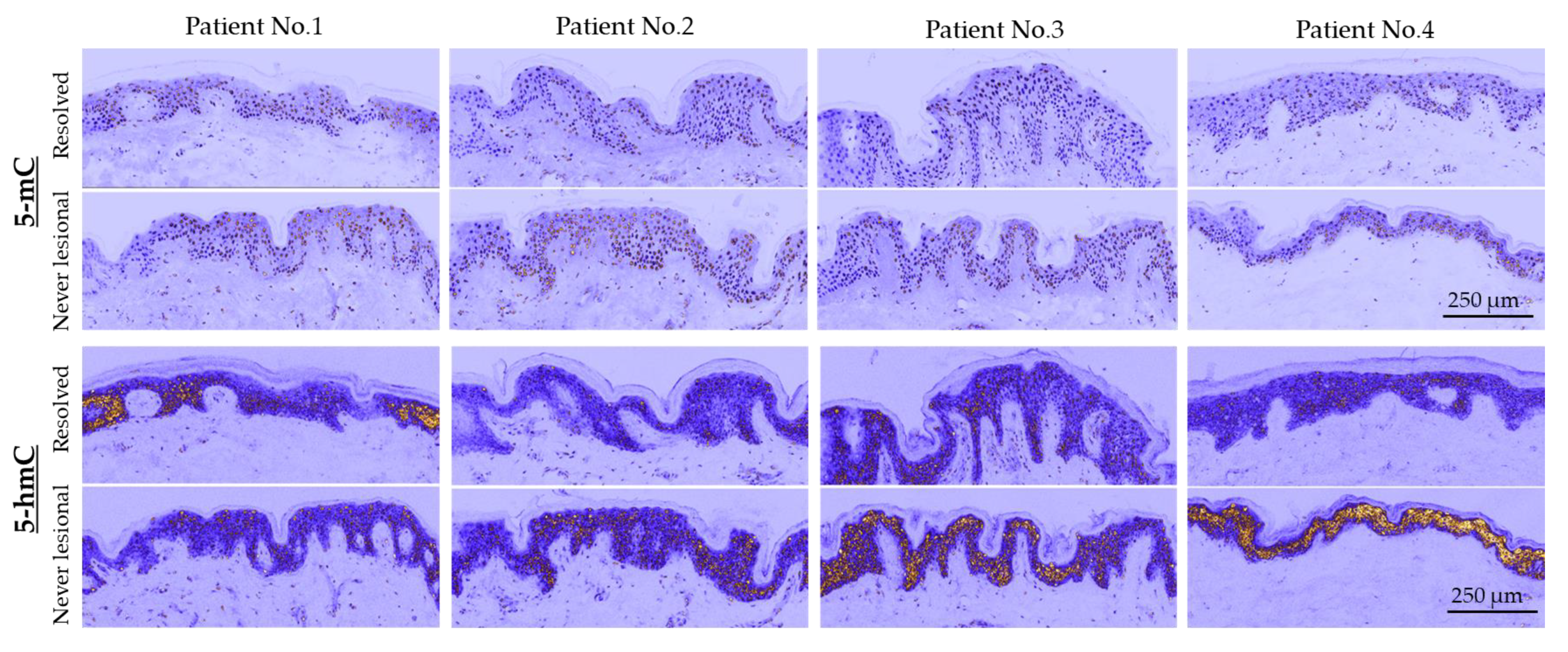
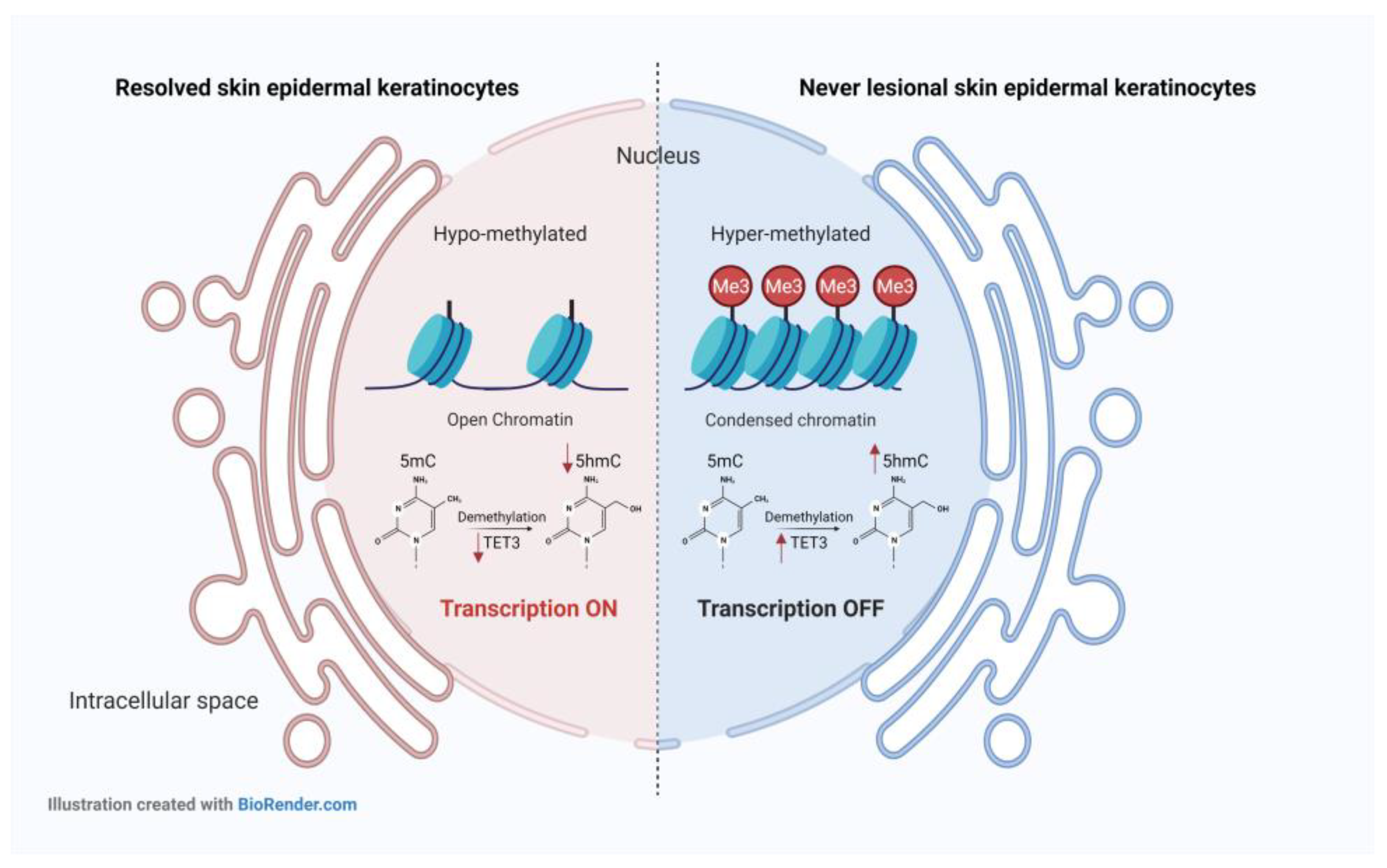
2.1. 5-mC and 5-hmC Amounts Were Overall Lower in Resolved Epidermis as Compared to Never-Lesional Epidermis
2.2. 5-hmC Weaker Intensity Was Accompanied by Decreased TET3 mRNA Expression Level in Resolved Epidermis
2.3. Transcriptome Profiling of the Resolved Epidermis vs. Never-Lesional Epidermis
2.3.1. Analysis of Differentially Expressed Genes (DEGs)
2.3.2. The Most Down-Regulated DEGs in Resolved Epidermis vs. Never-Lesional Epidermis (|FC| ≥ 2.5)
2.3.3. The Most Up-Regulated DEGs in Resolved Epidermis vs. Never-Lesional Epidermis (|FC| ≥ 2.5)
2.3.4. The Most Down-Regulated DEGs in Resolved Dermis vs. Never-Lesional Dermis (|FC| > 10)
2.3.5. The Most Up-Regulated DEGs in Resolved Dermis vs. Never-Lesional Dermis (|FC| > 10)
2.3.6. Identifying Biological Processes (BPs), Molecular Functions (MFs), and Cellular Components (CCs) Enriched among the DEGs
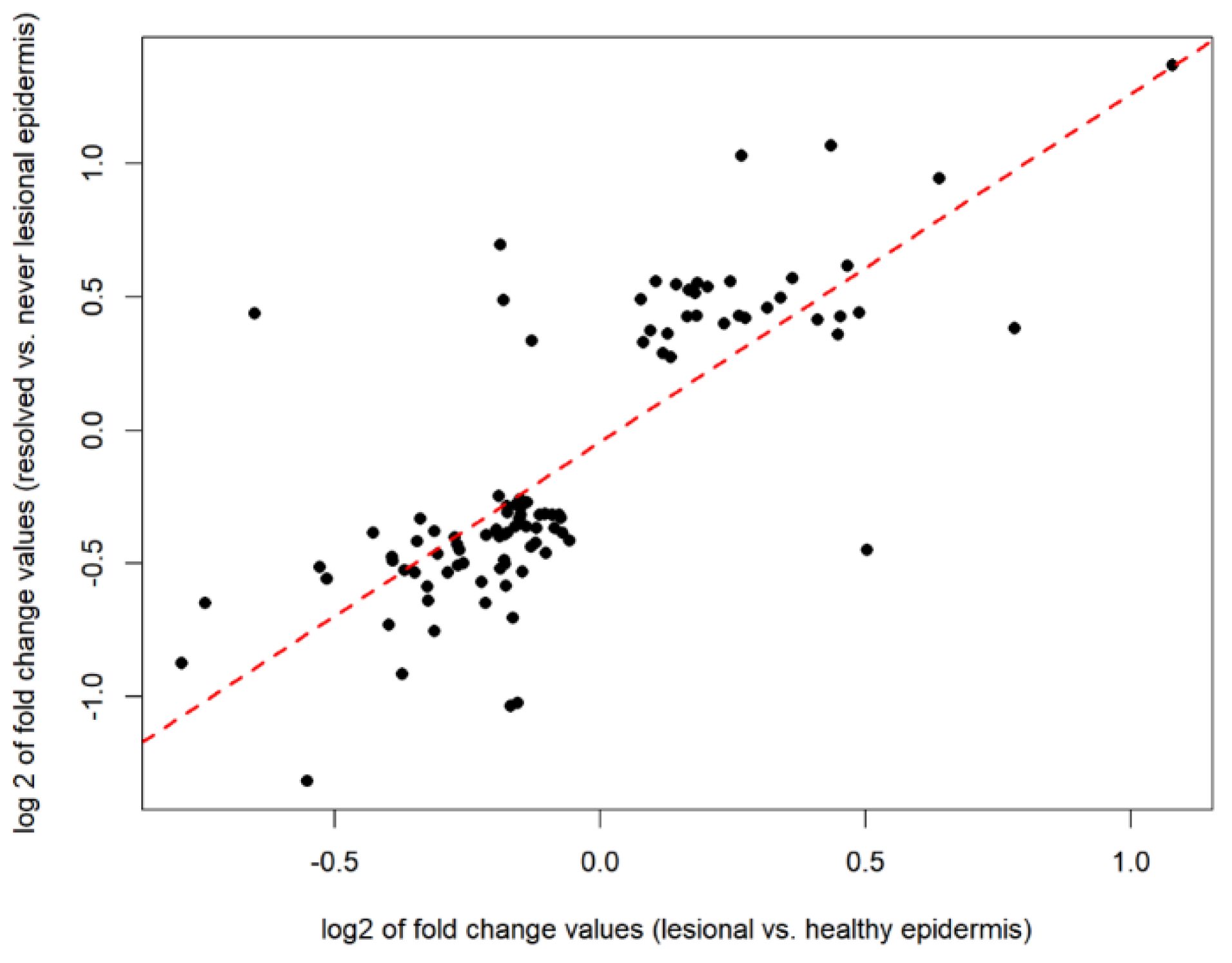
2.4. Defining the Disease-Residual Transcriptomic Profile (DRTP)
3. Discussion
4. Materials and Methods
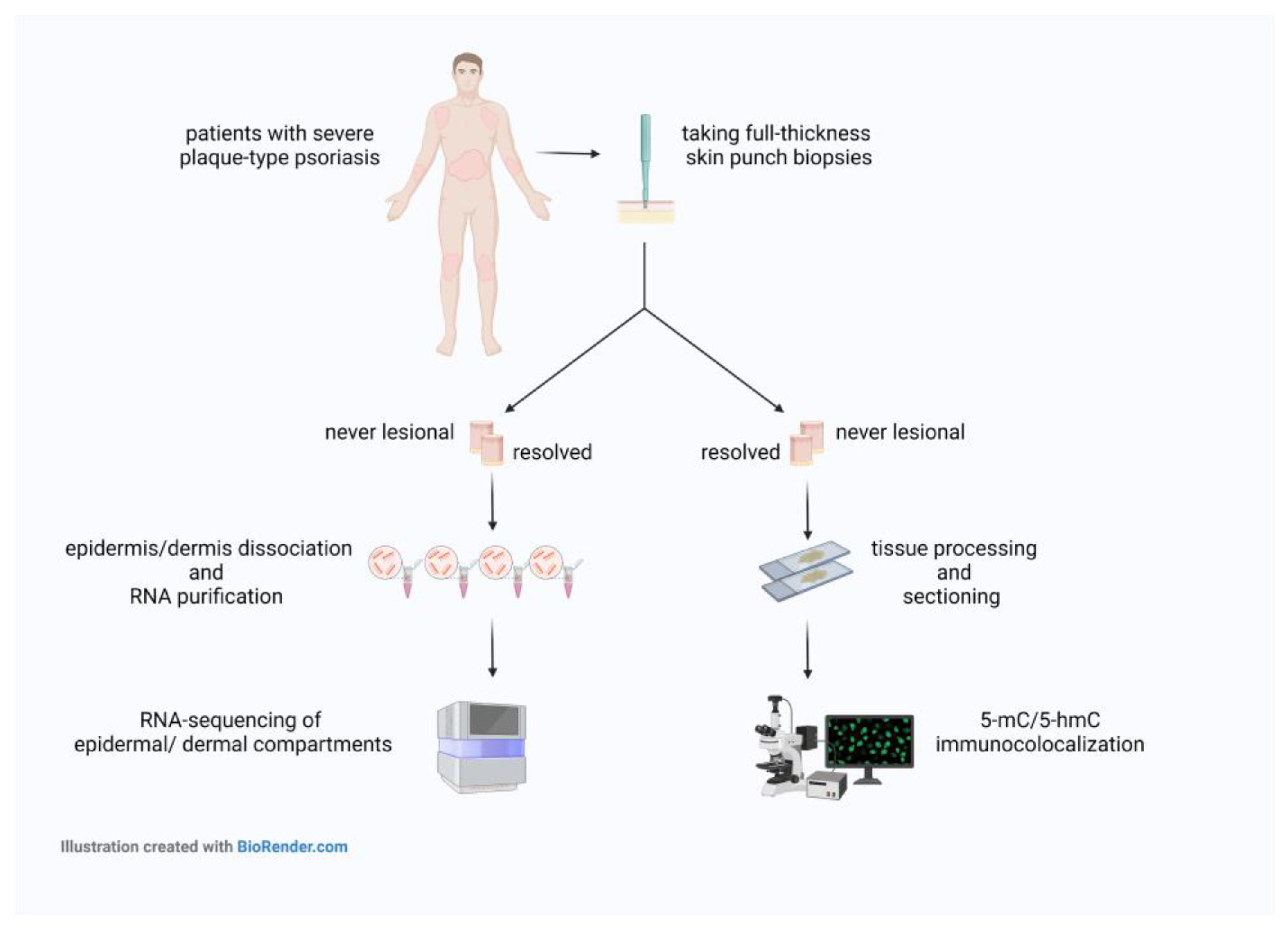
4.1. Psoriatic Skin Samples Collection and Ethics
4.2. Co-Localization of 5-mC and 5-hmC Epigenetic Marks in Psoriatic Never-Lesional and Resolved Skin Sections
4.3. Microscopy
4.4. RNA Extraction and Real-Time RT-PCR
4.5. High-Throughput mRNA Sequencing
4.5.1. RNA Sequencing Data Analysis
4.5.2. Pathway Analyses
4.6. Examining the Overlap between Resolved vs. Never-Lesional DEGs and Lesional vs. Healthy DEGs
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Billi, A.C.; Gudjonsson, J.E.; Voorhees, J.J. Psoriasis: Past, Present, and Future. J. Investig. Dermatol. 2019, 139, e133–e142. [Google Scholar] [CrossRef] [PubMed]
- Owczarek, W.; Dzik, M.; Narbutt, J.; Walecka, I.; Kowalczyk, M. Real-world evidence on time to relapse of plaque psoriasis after discontinuation of biologic treatment in Poland. Dermatol. Ther. 2021, 34, e15052. [Google Scholar] [CrossRef] [PubMed]
- Bu, J.; Ding, R.; Zhou, L.; Chen, X.; Shen, E. Epidemiology of Psoriasis and Comorbid Diseases: A Narrative Review. Front. Immunol. 2022, 13, 880201. [Google Scholar] [CrossRef] [PubMed]
- Chamian, F.; Lowes, M.A.; Lin, S.-L.; Lee, E.; Kikuchi, T.; Gilleaudeau, P.; Sullivan-Whalen, M.; Cardinale, I.; Khatcherian, A.; Novitskaya, I.; et al. Alefacept reduces infiltrating T cells, activated dendritic cells, and inflammatory genes in psoriasis vulgaris. Proc. Natl. Acad. Sci. USA 2005, 102, 2075–2080. [Google Scholar] [CrossRef]
- Zaba, L.C.; Cardinale, I.; Gilleaudeau, P.; Sullivan-Whalen, M.; Suárez-Fariñas, M.; Suárez Fariñas, M.; Fuentes-Duculan, J.; Novitskaya, I.; Khatcherian, A.; Bluth, M.J.; et al. Amelioration of epidermal hyperplasia by TNF inhibition is associated with reduced Th17 responses. J. Exp. Med. 2007, 204, 3183–3194. [Google Scholar] [CrossRef]
- Dattola, A.; Silvestri, M.; Bennardo, L.; Passante, M.; Rizzuto, F.; Dastoli, S.; Patruno, C.; Bianchi, L.; Nisticò, S.P. A novel vehicle for the treatment of psoriasis. Dermatol. Ther. 2020, 33, e13185. [Google Scholar] [CrossRef]
- Nast, A.; Gisondi, P.; Ormerod, A.D.; Saiag, P.; Smith, C.; Spuls, P.I.; Arenberger, P.; Bachelez, H.; Barker, J.; Dauden, E.; et al. European S3-Guidelines on the systemic treatment of psoriasis vulgaris–Update 2015–Short version–EDF in cooperation with EADV and IPC. J. Eur. Acad. Dermatol. Venereol. JEADV 2015, 29, 2277–2294. [Google Scholar] [CrossRef]
- Langley, R.G.B.; Krueger, G.G.; Griffiths, C.E.M. Psoriasis: Epidemiology, clinical features, and quality of life. Ann. Rheum. Dis. 2005, 64 (Suppl. S2), ii18–i23; discussion ii24–ii25. [Google Scholar] [CrossRef]
- Szabó, K.; Bata-Csörgő, Z.; Dallos, A.; Bebes, A.; Francziszti, L.; Dobozy, A.; Kemény, L.; Széll, M. Regulatory networks contributing to psoriasis susceptibility. Acta Derm. Venereol. 2014, 94, 380–385. [Google Scholar] [CrossRef]
- Bozó, R.; Danis, J.; Flink, L.B.; Vidács, D.L.; Kemény, L.; Bata-Csörgő, Z. Stress-Related Regulation Is Abnormal in the Psoriatic Uninvolved Skin. Life 2021, 11, 599. [Google Scholar] [CrossRef]
- Bozó, R.; Szél, E.; Danis, J.; Gubán, B.; Bata-Csörgő, Z.; Szabó, K.; Kemény, L.; Groma, G. Cartilage Oligomeric Matrix Protein Negatively Influences Keratinocyte Proliferation via α5β1-Integrin: Potential Relevance of Altered Cartilage Oligomeric Matrix Protein Expression in Psoriasis. J. Investig. Dermatol. 2020, 140, 1733–1742.e7. [Google Scholar] [CrossRef]
- Bata-Csorgo, Z.; Hammerberg, C.; Voorhees, J.J.; Cooper, K.D. Kinetics and regulation of human keratinocyte stem cell growth in short-term primary ex vivo culture. Cooperative growth factors from psoriatic lesional T lymphocytes stimulate proliferation among psoriatic uninvolved, but not normal, stem keratinocytes. J. Clin. Investig. 1995, 95, 317–327. [Google Scholar] [CrossRef]
- Széll, M.; Bata-Csörgo, Z.; Koreck, A.; Pivarcsi, A.; Polyánka, H.; Szeg, C.; Gaál, M.; Dobozy, A.; Kemény, L. Proliferating keratinocytes are putative sources of the psoriasis susceptibility-related EDA+ (extra domain A of fibronectin) oncofetal fibronectin. J. Investig. Dermatol. 2004, 123, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Fariñas, M.; Fuentes-Duculan, J.; Lowes, M.A.; Krueger, J.G. Resolved psoriasis lesions retain expression of a subset of disease-related genes. J. Investig. Dermatol. 2011, 131, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Brodmerkel, C.; Li, K.; Garcet, S.; Hayden, K.; Chiricozzi, A.; Novitskaya, I.; Fuentes-Duculan, J.; Suarez-Farinas, M.; Campbell, K.; Krueger, J.G. Modulation of inflammatory gene transcripts in psoriasis vulgaris: Differences between ustekinumab and etanercept. J. Allergy Clin. Immunol. 2019, 143, 1965–1969. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, S.; Baum, P.; Vinisko, R.; Schmid, R.; Flack, M.; Lalovic, B.; Kleiner, O.; Fuentes-Duculan, J.; Garcet, S.; Davis, J.W.; et al. Psoriatic skin molecular and histopathologic profiles after treatment with risankizumab versus ustekinumab. J. Allergy Clin. Immunol. 2019, 143, 2158–2169. [Google Scholar] [CrossRef]
- Sofen, H.; Smith, S.; Matheson, R.T.; Leonardi, C.L.; Calderon, C.; Brodmerkel, C.; Li, K.; Campbell, K.; Marciniak, S.J.; Wasfi, Y.; et al. Guselkumab (an IL-23-specific mAb) demonstrates clinical and molecular response in patients with moderate-to-severe psoriasis. J. Allergy Clin. Immunol. 2014, 133, 1032–1040. [Google Scholar] [CrossRef]
- Kulig, P.; Musiol, S.; Freiberger, S.N.; Schreiner, B.; Gyülveszi, G.; Russo, G.; Pantelyushin, S.; Kishihara, K.; Alessandrini, F.; Kündig, T.; et al. IL-12 protects from psoriasiform skin inflammation. Nat. Commun. 2016, 7, 13466. [Google Scholar] [CrossRef]
- Ruchusatsawat, K.; Wongpiyabovorn, J.; Shuangshoti, S.; Hirankarn, N.; Mutirangura, A. SHP-1 promoter 2 methylation in normal epithelial tissues and demethylation in psoriasis. J. Mol. Med. Berl. Ger. 2006, 84, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chen, Z.-Q.; Cui, P.-G.; Yao, X.; Li, Y.-M.; Li, A.-S.; Gong, J.-Q.; Cao, Y.-H. The methylation pattern of p16INK4a gene promoter in psoriatic epidermis and its clinical significance. Br. J. Dermatol. 2008, 158, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Liu, Z.; Xu, Z.; Ke, F.; Zhang, L.; Zhu, H.; Lou, F.; Wang, H.; Fei, Y.; Shi, Y.-L.; et al. Epigenetic downregulation of SFRP4 contributes to epidermal hyperplasia in psoriasis. J. Immunol. 2015, 194, 4185–4198. [Google Scholar] [CrossRef]
- Ruchusatsawat, K.; Wongpiyabovorn, J.; Protjaroen, P.; Chaipipat, M.; Shuangshoti, S.; Thorner, P.S.; Mutirangura, A. Parakeratosis in skin is associated with loss of inhibitor of differentiation 4 via promoter methylation. Hum. Pathol. 2011, 42, 1878–1887. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Boldrup, L.; Coates, P.J.; Fahraeus, R.; Nylander, E.; Loizou, C.; Olofsson, K.; Norberg-Spaak, L.; Gärskog, O.; Nylander, K. Epigenetic regulation of OAS2 shows disease-specific DNA methylation profiles at individual CpG sites. Sci. Rep. 2016, 6, 32579. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Su, Y.; Chen, H.; Zhao, M.; Lu, Q. Abnormal DNA methylation in skin lesions and PBMCs of patients with psoriasis vulgaris. J. Dermatol. Sci. 2010, 60, 40–42. [Google Scholar] [CrossRef] [PubMed]
- Yooyongsatit, S.; Ruchusatsawat, K.; Noppakun, N.; Hirankarn, N.; Mutirangura, A.; Wongpiyabovorn, J. Patterns and functional roles of LINE-1 and Alu methylation in the keratinocyte from patients with psoriasis vulgaris. J. Hum. Genet. 2015, 60, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Roberson, E.D.O.; Liu, Y.; Ryan, C.; Joyce, C.E.; Duan, S.; Cao, L.; Martin, A.; Liao, W.; Menter, A.; Bowcock, A.M. A subset of methylated CpG sites differentiate psoriatic from normal skin. J. Investig. Dermatol. 2012, 132, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhao, M.; Liang, G.; Yin, G.; Huang, D.; Su, F.; Zhai, H.; Wang, L.; Su, Y.; Lu, Q. Whole-genome DNA methylation in skin lesions from patients with psoriasis vulgaris. J. Autoimmun. 2013, 41, 17–24. [Google Scholar] [CrossRef]
- Li, F.; Yuan, C.W.; Xu, S.; Zu, T.; Woappi, Y.; Lee, C.A.A.; Abarzua, P.; Wells, M.; Ramsey, M.R.; Frank, N.Y.; et al. Loss of the Epigenetic Mark 5-hmC in Psoriasis: Implications for Epidermal Stem Cell Dysregulation. J. Investig. Dermatol. 2020, 140, 1266–1275.e3. [Google Scholar] [CrossRef]
- Gu, X.; Nylander, E.; Coates, P.J.; Fahraeus, R.; Nylander, K. Correlation between Reversal of DNA Methylation and Clinical Symptoms in Psoriatic Epidermis Following Narrow-Band UVB Phototherapy. J. Investig. Dermatol. 2015, 135, 2077–2083. [Google Scholar] [CrossRef]
- Zhou, F.; Wang, W.; Shen, C.; Li, H.; Zuo, X.; Zheng, X.; Yue, M.; Zhang, C.; Yu, L.; Chen, M.; et al. Epigenome-Wide Association Analysis Identified Nine Skin DNA Methylation Loci for Psoriasis. J. Investig. Dermatol. 2016, 136, 779–787. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011, 25, 2436–2452. [Google Scholar] [CrossRef]
- Guo, J.U.; Su, Y.; Zhong, C.; Ming, G.; Song, H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle Georget. Tex 2011, 10, 2662–2668. [Google Scholar] [CrossRef]
- Carpenter, S.; Aiello, D.; Atianand, M.K.; Ricci, E.P.; Gandhi, P.; Hall, L.L.; Byron, M.; Monks, B.; Henry-Bezy, M.; Lawrence, J.B.; et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013, 341, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Prinz, F.; Kapeller, A.; Pichler, M.; Klec, C. The Implications of the Long Non-Coding RNA NEAT1 in Non-Cancerous Diseases. Int. J. Mol. Sci. 2019, 20, 627. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Cao, L.; Zhou, R.; Yang, X.; Wu, M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat. Commun. 2019, 10, 1495. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Ahn, R.; Lai, K.; Mullins, E.; Debbaneh, M.; Dimon, M.; Arron, S.; Liao, W. Landscape of Long Noncoding RNAs in Psoriatic and Healthy Skin. J. Investig. Dermatol. 2016, 136, 603–609. [Google Scholar] [CrossRef]
- Lee, E.J.; Seo, J.H.; Park, J.-H.; Vo, T.T.L.; An, S.; Bae, S.-J.; Le, H.; Lee, H.S.; Wee, H.-J.; Lee, D.; et al. SAMHD1 acetylation enhances its deoxynucleotide triphosphohydrolase activity and promotes cancer cell proliferation. Oncotarget 2017, 8, 68517–68529. [Google Scholar] [CrossRef] [PubMed]
- Raposo, R.A.; Gupta, R.; Abdel-Mohsen, M.; Dimon, M.; Debbaneh, M.; Jiang, W.; York, V.A.; Leadabrand, K.S.; Brown, G.; Malakouti, M.; et al. Antiviral gene expression in psoriasis. J. Eur. Acad. Dermatol. Venereol. JEADV 2015, 29, 1951–1957. [Google Scholar] [CrossRef]
- Sumantran, V.N.; Mishra, P.; Bera, R.; Sudhakar, N. Microarray Analysis of Differentially-Expressed Genes Encoding CYP450 and Phase II Drug Metabolizing Enzymes in Psoriasis and Melanoma. Pharmaceutics 2016, 8, 4. [Google Scholar] [CrossRef]
- Gudjonsson, J.E.; Ding, J.; Johnston, A.; Tejasvi, T.; Guzman, A.M.; Nair, R.P.; Voorhees, J.J.; Abecasis, G.R.; Elder, J.T. Assessment of the psoriatic transcriptome in a large sample: Additional regulated genes and comparisons with in vitro models. J. Investig. Dermatol. 2010, 130, 1829–1840. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yi, X.; Ding, Y. Combined Transcriptomic Analysis Revealed AKR1B10 Played an Important Role in Psoriasis through the Dysregulated Lipid Pathway and Overproliferation of Keratinocyte. BioMed Res. Int. 2017, 2017, 8717369. [Google Scholar] [CrossRef]
- Cao, D.; Fan, S.T.; Chung, S.S. Identification and characterization of a novel human aldose reductase-like gene. J. Biol. Chem. 1998, 273, 11429–11435. [Google Scholar] [CrossRef] [PubMed]
- Ramana, K.V.; Bhatnagar, A.; Srivastava, S.K. Inhibition of aldose reductase attenuates TNF-alpha-induced expression of adhesion molecules in endothelial cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2004, 18, 1209–1218. [Google Scholar] [CrossRef]
- Yadav, U.C.S.; Mishra, R.; Aguilera-Aguirre, L.; Sur, S.; Bolodgh, I.; Ramana, K.V.; Srivatsava, S.K. Prevention of allergic rhinitis by aldose reductase inhibition in a murine model. Inflamm. Allergy Drug Targets 2013, 12, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Pladzyk, A.; Reddy, A.B.M.; Yadav, U.C.S.; Tammali, R.; Ramana, K.V.; Srivastava, S.K. Inhibition of aldose reductase prevents lipopolysaccharide-induced inflammatory response in human lens epithelial cells. Investig. Ophthalmol. Vis. Sci. 2006, 47, 5395–5403. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Yadav, U.C.S.; Reddy, A.B.M.; Saxena, A.; Tammali, R.; Shoeb, M.; Ansari, N.H.; Bhatnagar, A.; Petrash, M.J.; Srivastava, S.; et al. Aldose reductase inhibition suppresses oxidative stress-induced inflammatory disorders. Chem. Biol. Interact. 2011, 191, 330–338. [Google Scholar] [CrossRef]
- Yadav, U.C.S.; Ramana, K.V.; Srivastava, S.K. Aldose reductase inhibition suppresses airway inflammation. Chem. Biol. Interact. 2011, 191, 339–345. [Google Scholar] [CrossRef]
- Yang, M.; Tang, M.; Ma, X.; Yang, L.; He, J.; Peng, X.; Guo, G.; Zhou, L.; Luo, N.; Yuan, Z.; et al. AP-57/C10orf99 is a new type of multifunctional antimicrobial peptide. Biochem. Biophys. Res. Commun. 2015, 457, 347–352. [Google Scholar] [CrossRef]
- Suply, T.; Hannedouche, S.; Carte, N.; Li, J.; Grosshans, B.; Schaefer, M.; Raad, L.; Beck, V.; Vidal, S.; Hiou-Feige, A.; et al. A natural ligand for the orphan receptor GPR15 modulates lymphocyte recruitment to epithelia. Sci. Signal. 2017, 10, eaal0180. [Google Scholar] [CrossRef]
- Ocón, B.; Pan, J.; Dinh, T.T.; Chen, W.; Ballet, R.; Bscheider, M.; Habtezion, A.; Tu, H.; Zabel, B.A.; Butcher, E.C. A Mucosal and Cutaneous Chemokine Ligand for the Lymphocyte Chemoattractant Receptor GPR15. Front. Immunol. 2017, 8, 1111. [Google Scholar] [CrossRef]
- Lahl, K.; Sweere, J.; Pan, J.; Butcher, E. Orphan chemoattractant receptor GPR15 mediates dendritic epidermal T-cell recruitment to the skin. Eur. J. Immunol. 2014, 44, 2577–2581. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Fariñas, M.; Li, K.; Fuentes-Duculan, J.; Hayden, K.; Brodmerkel, C.; Krueger, J.G. Expanding the psoriasis disease profile: Interrogation of the skin and serum of patients with moderate-to-severe psoriasis. J. Investig. Dermatol. 2012, 132, 2552–2564. [Google Scholar] [CrossRef]
- Rønholt, K.; Nielsen, A.L.-L.; Johansen, C.; Vestergaard, C.; Fauerbye, A.; López-Vales, R.; Dinarello, C.A.; Iversen, L. IL-37 Expression Is Downregulated in Lesional Psoriasis Skin. ImmunoHorizons 2020, 4, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, S.; Wolf, C.; König, N.; Staroske, W.; Guck, J.; Häusler, M.; Luksch, H.; Nguyen, L.A.; Kim, B.; Alexopoulou, D.; et al. SAMHD1 prevents autoimmunity by maintaining genome stability. Ann. Rheum. Dis. 2015, 74, e17. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Luo, Y.; Mai, G.; Zhang, M.; Wang, G.; Zhao, M.; Gao, L.; Li, F.; Zhou, F. Gene expression profile based classification models of psoriasis. Genomics 2014, 103, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Tseng, P.-Y.; Hoon, M.A. GPR15L is an epithelial inflammation-derived pruritogen. Sci. Adv. 2022, 8, eabm7342. [Google Scholar] [CrossRef]
- Ghosh, D.; Ding, L.; Sivaprasad, U.; Geh, E.; Biagini Myers, J.; Bernstein, J.A.; Khurana Hershey, G.K.; Mersha, T.B. Multiple Transcriptome Data Analysis Reveals Biologically Relevant Atopic Dermatitis Signature Genes and Pathways. PLoS ONE 2015, 10, e0144316. [Google Scholar] [CrossRef] [PubMed]
- Jumper, N.; Hodgkinson, T.; Arscott, G.; Har-Shai, Y.; Paus, R.; Bayat, A. The Aldo-Keto Reductase AKR1B10 Is Up-Regulated in Keloid Epidermis, Implicating Retinoic Acid Pathway Dysregulation in the Pathogenesis of Keloid Disease. J. Investig. Dermatol. 2016, 136, 1500–1512. [Google Scholar] [CrossRef]
- Ruiz, F.X.; Porté, S.; Parés, X.; Farrés, J. Biological role of aldo-keto reductases in retinoic Acid biosynthesis and signaling. Front. Pharmacol. 2012, 3, 58. [Google Scholar] [CrossRef]
- Geiger, J.M. Efficacy of acitretin in severe psoriasis. Skin Ther. Lett. 2003, 8, 1–3, 7. [Google Scholar]
- Huang, L.; He, R.; Luo, W.; Zhu, Y.-S.; Li, J.; Tan, T.; Zhang, X.; Hu, Z.; Luo, D. Aldo-Keto Reductase Family 1 Member B10 Inhibitors: Potential Drugs for Cancer Treatment. Recent Pat. Anticancer Drug Discov. 2016, 11, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.G.; Xu, Y.; Ceol, C.; Wu, F.; Larson, A.; Dresser, K.; Xu, W.; Tan, L.; Hu, Y.; Zhan, Q.; et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 2012, 150, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Ficz, G.; Gribben, J.G. Loss of 5-hydroxymethylcytosine in cancer: Cause or consequence? Genomics 2014, 104, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, M. DNA methylation in cancer: Too much, but also too little. Oncogene 2002, 21, 5400–5413. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, C.M.; Mäbert, K.; Rauen, T.; Tsokos, G.C. DNA methylation in systemic lupus erythematosus. Epigenomics 2017, 9, 505–525. [Google Scholar] [CrossRef]
- Zhou, Y.; Lu, Q. DNA methylation in T cells from idiopathic lupus and drug-induced lupus patients. Autoimmun. Rev. 2008, 7, 376–383. [Google Scholar] [CrossRef]
- Karouzakis, E.; Gay, R.E.; Michel, B.A.; Gay, S.; Neidhart, M. DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2009, 60, 3613–3622. [Google Scholar] [CrossRef]
- Moscarello, M.A.; Brady, G.W.; Fein, D.B.; Wood, D.D.; Cruz, T.F. The role of charge microheterogeneity of basic protein in the formation and maintenance of the multilayered structure of myelin: A possible role in multiple sclerosis. J. Neurosci. Res. 1986, 15, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Moscarello, M.A.; Wood, D.D.; Ackerley, C.; Boulias, C. Myelin in multiple sclerosis is developmentally immature. J. Clin. Investig. 1994, 94, 146–154. [Google Scholar] [CrossRef]
- Gerdes, S.; Körber, A.; Biermann, M.; Karnthaler, C.; Reinhardt, M. Absolute and relative psoriasis area and severity index (PASI) treatment goals and their association with health-related quality of life. J. Dermatol. Treat. 2020, 31, 470–475. [Google Scholar] [CrossRef]
- Singh, R.K.; Diaz, P.E.; Binette, F.; Nasonkin, I.O. Immunohistochemical Detection of 5-Methylcytosine and 5-Hydroxymethylcytosine in Developing and Postmitotic Mouse Retina. J. Vis. Exp. JoVE 2018, 138, e58274. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Szabad, G.; Kormos, B.; Pivarcsi, A.; Széll, M.; Kis, K.; Kenderessy Szabó, A.; Dobozy, A.; Kemény, L.; Bata-Csörgo, Z. Human adult epidermal melanocytes cultured without chemical mitogens express the EGF receptor and respond to EGF. Arch. Dermatol. Res. 2007, 299, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Andrianne, M.; Assabban, A.; La, C.; Mogilenko, D.; Salle, D.S.; Fleury, S.; Doumont, G.; Van Simaeys, G.; Nedospasov, S.A.; Blackshear, P.J.; et al. Tristetraprolin expression by keratinocytes controls local and systemic inflammation. JCI Insight 2017, 2, 92979. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Eden, E.; Lipson, D.; Yogev, S.; Yakhini, Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 2007, 3, e39. [Google Scholar] [CrossRef]



| The 25 Most Down-Regulated and Up-Regulated Resolved vs. Never-Lesional DEGs in Epidermal and Dermal Compartments | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Epidermis (n = 4) | Dermis (n = 3) | ||||||||
| Down-Regulated | Up-Regulated | Down-Regulated | Up-Regulated | ||||||
| Number | Gene Symbol | FC | Gene Symbol | FC | Number | Gene Symbol | FC | Gene Symbol | FC |
| 1 | NEAT1_3 | −4.66 | STAC2 | 5.35 | 1 | SPRR4 | −32.61 | IGHV3-7 | 12.68 |
| 2 | SAMHD1 | −2.57 | AQP5 | 4.38 | 2 | ATP12A | −25.92 | IGHV3-33 | 10.90 |
| 3 | HOXB2 | −2.50 | FAM25C | 3.36 | 3 | CST6 | −17.10 | IGHJ4 | 10.56 |
| 4 | CACNA2D1 | −2.49 | ELOVL3 | 3.31 | 4 | SPRR1A | −17.00 | IGKV3-20 | 7.39 |
| 5 | FHL1 | −2.22 | C10orf99 | 2.75 | 5 | CRCT1 | −14.41 | IGKV3-15 | 7.01 |
| 6 | RPL24P7 | −2.14 | AKR1B10 | 2.57 | 6 | MSMB | −12.93 | IGLV1-44 | 6.40 |
| 7 | SNORA9 | −2.11 | FAM25G | 2.42 | 7 | KRT34 | −10.56 | POU6F2 | 6.10 |
| 8 | CYP4B1 | −2.05 | CRNN | 2.41 | 8 | GJB4 | −10.54 | IGKV1D-12 | 5.72 |
| 9 | ODF3L1 | −2.04 | RPL31P63 | 2.28 | 9 | RNF222 | −9.99 | LINC00619 | 5.61 |
| 10 | WNT2 | −2.03 | FABP5P2 | 2.28 | 10 | CTSV | −9.46 | ADH4 | 5.57 |
| 11 | KLHL11 | −1.94 | FABP5P10 | 2.26 | 11 | KRTAP3-2 | −9.37 | IGKV1-17 | 5.03 |
| 12 | TMEM256 | −1.93 | LGR6 | 2.20 | 12 | KRT33A | −8.82 | IGKV1-12 | 4.87 |
| 13 | PELI2 | −1.88 | IL1F10 | 2.19 | 13 | PLA2G2F | −8.56 | IGHV2-70 | 4.69 |
| 14 | WDR82P2 | −1.88 | FABP5P1 | 2.12 | 14 | SLC15A1 | −8.40 | ITIH1 | 4.49 |
| 15 | LIF | −1.88 | FABP5 | 2.09 | 15 | KRT31 | −8.30 | IGKV1-16 | 4.38 |
| 16 | TPPP | −1.83 | WISP3 | 2.09 | 16 | RPP21 | −8.29 | ADAM20P1 | 4.33 |
| 17 | RCAN2 | −1.77 | FABP5P7 | 2.09 | 17 | TRIM39-RPP21 | −7.85 | SAA2 | 4.27 |
| 18 | PKIB | −1.74 | NPM1P25 | 2.07 | 18 | SPRR1B | −7.48 | IGHV1-18 | 4.16 |
| 19 | MIR23A | −1.71 | MRPS10P1 | 2.07 | 19 | B4GALNT2 | −7.48 | RHPN1-AS1 | 4.10 |
| 20 | KLF9 | −1.68 | PYDC1 | 2.03 | 20 | CASP1P2 | −7.15 | ANP32BP3 | 3.99 |
| 21 | LRRC8C | −1.68 | IGFBP2 | 2.00 | 21 | BPIFC | −6.94 | RHOXF2 | 3.96 |
| 22 | SLC47A1 | −1.67 | RPS26P39 | 2.00 | 22 | KRT38 | −6.89 | IGHV1-69 | 3.93 |
| 23 | CNTNAP3P2 | −1.66 | ENTPD3 | 1.98 | 23 | LIPK | −6.87 | OLAH | 3.89 |
| 24 | IKZF2 | −1.66 | KRT2 | 1.94 | 24 | CARD18 | −6.43 | IGLV1-47 | 3.83 |
| 25 | WDR45BP1 | −1.66 | WNT5A | 1.92 | 25 | PCSK1N | −6.14 | MT3 | 3.79 |
| The KEGG Enrichment Analysis of the 102 Overlapping Genes between Resolved and Lesional Epidermis | |||||||
|---|---|---|---|---|---|---|---|
| Gene Set | Description | Size | Expect | Ratio | p Value | FDR | |
| 1 | hsa05217 | Basal cell carcinoma | 59 | 0.38756 | 12.901 | 3.78 × 10−5 | 0.0122 |
| 2 | hsa04550 | Signaling pathways regulating pluripotency of stem cells | 132 | 0.86709 | 5.7664 | 1.63 × 10−3 | 0.1296 |
| 3 | hsa04310 | WNT signaling pathway | 140 | 0.91964 | 5.4369 | 2.12 × 10−3 | 0.1296 |
| 4 | hsa05224 | Breast cancer | 142 | 0.93277 | 5.3604 | 2.25 × 10−3 | 0.1296 |
| 5 | hsa05226 | Gastric cancer | 142 | 0.93277 | 5.3604 | 2.25 × 10−3 | 0.1296 |
| 6 | hsa04150 | mTOR signaling pathway | 144 | 0.94591 | 5.2859 | 2.39 × 10−3 | 0.1296 |
| 7 | hsa04916 | Melanogenesis | 95 | 0.62404 | 6.4099 | 3.36 × 10−3 | 0.1534 |
| 8 | hsa05225 | Hepatocellular carcinoma | 160 | 1.051 | 4.7573 | 3.78 × 10−3 | 0.1534 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghaffarinia, A.; Ayaydin, F.; Póliska, S.; Manczinger, M.; Bolla, B.S.; Flink, L.B.; Balogh, F.; Veréb, Z.; Bozó, R.; Szabó, K.; et al. Psoriatic Resolved Skin Epidermal Keratinocytes Retain Disease-Residual Transcriptomic and Epigenomic Profiles. Int. J. Mol. Sci. 2023, 24, 4556. https://doi.org/10.3390/ijms24054556
Ghaffarinia A, Ayaydin F, Póliska S, Manczinger M, Bolla BS, Flink LB, Balogh F, Veréb Z, Bozó R, Szabó K, et al. Psoriatic Resolved Skin Epidermal Keratinocytes Retain Disease-Residual Transcriptomic and Epigenomic Profiles. International Journal of Molecular Sciences. 2023; 24(5):4556. https://doi.org/10.3390/ijms24054556
Chicago/Turabian StyleGhaffarinia, Ameneh, Ferhan Ayaydin, Szilárd Póliska, Máté Manczinger, Beáta Szilvia Bolla, Lili Borbála Flink, Fanni Balogh, Zoltán Veréb, Renáta Bozó, Kornélia Szabó, and et al. 2023. "Psoriatic Resolved Skin Epidermal Keratinocytes Retain Disease-Residual Transcriptomic and Epigenomic Profiles" International Journal of Molecular Sciences 24, no. 5: 4556. https://doi.org/10.3390/ijms24054556
APA StyleGhaffarinia, A., Ayaydin, F., Póliska, S., Manczinger, M., Bolla, B. S., Flink, L. B., Balogh, F., Veréb, Z., Bozó, R., Szabó, K., Bata-Csörgő, Z., & Kemény, L. (2023). Psoriatic Resolved Skin Epidermal Keratinocytes Retain Disease-Residual Transcriptomic and Epigenomic Profiles. International Journal of Molecular Sciences, 24(5), 4556. https://doi.org/10.3390/ijms24054556










