R-Propranolol Has Broad-Spectrum Anti-Coronavirus Activity and Suppresses Factors Involved in Pathogenic Angiogenesis
Abstract
:1. Introduction
2. Results
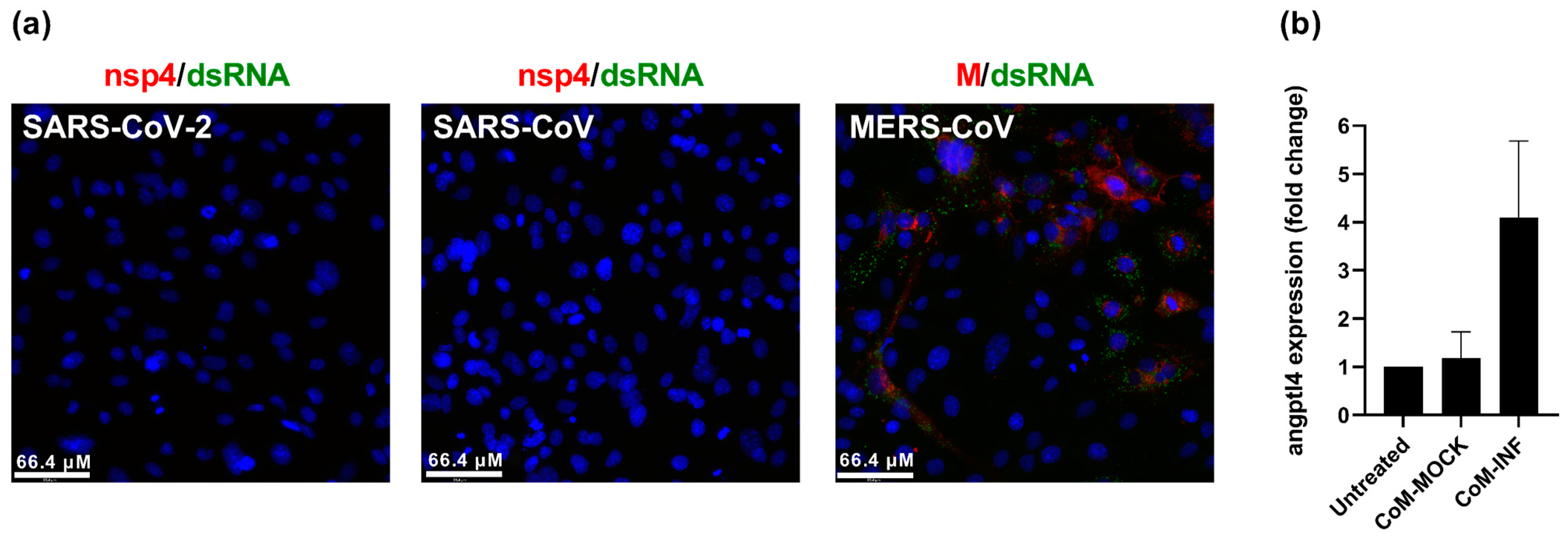
2.1. SARS-CoV-2 Infection Upregulates Angiogenic Factor angptl4 in Endothelial Cells
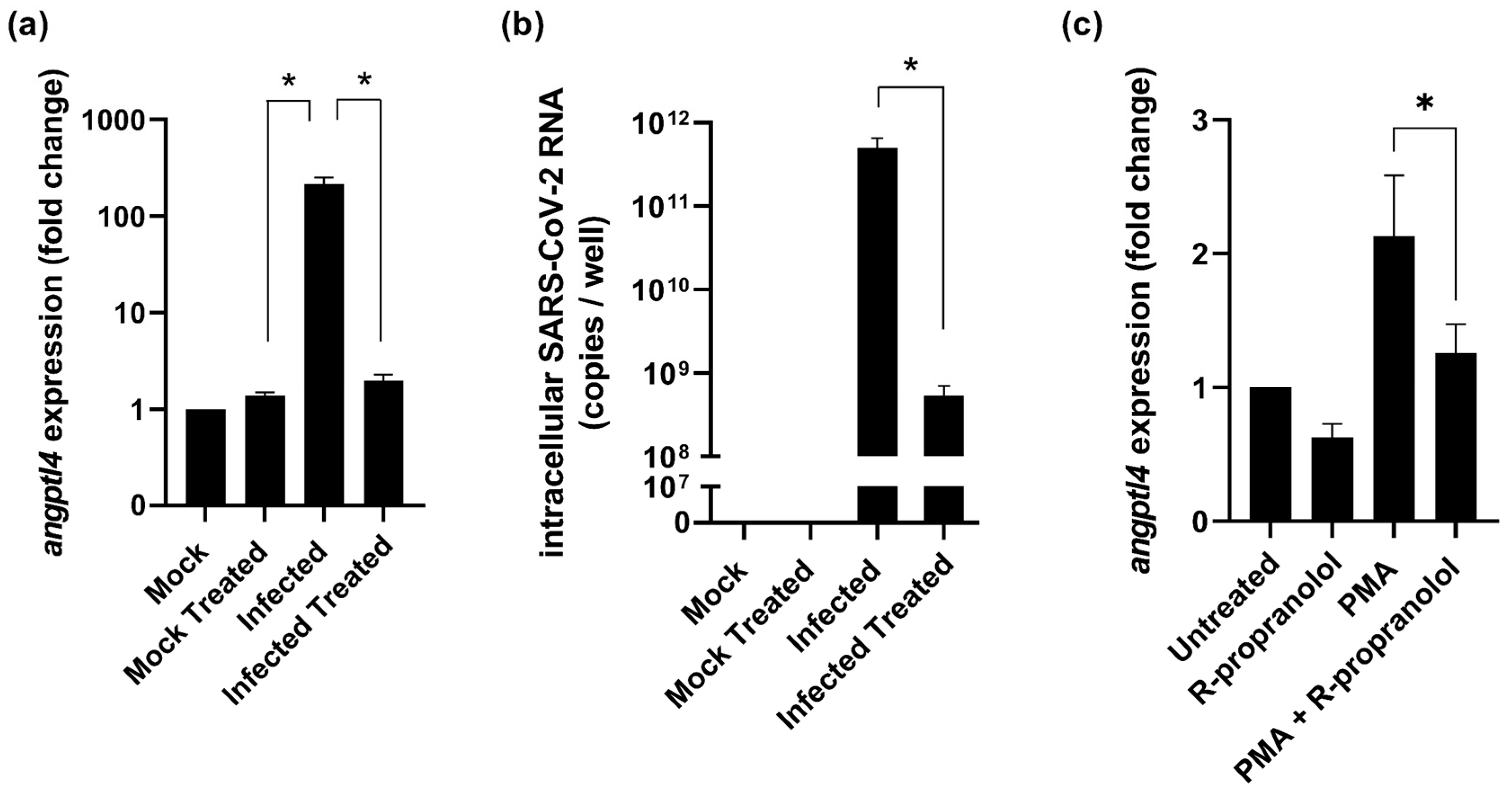
2.2. R-Propranolol Downregulates Expression of the Angiogenic Factor angptl4
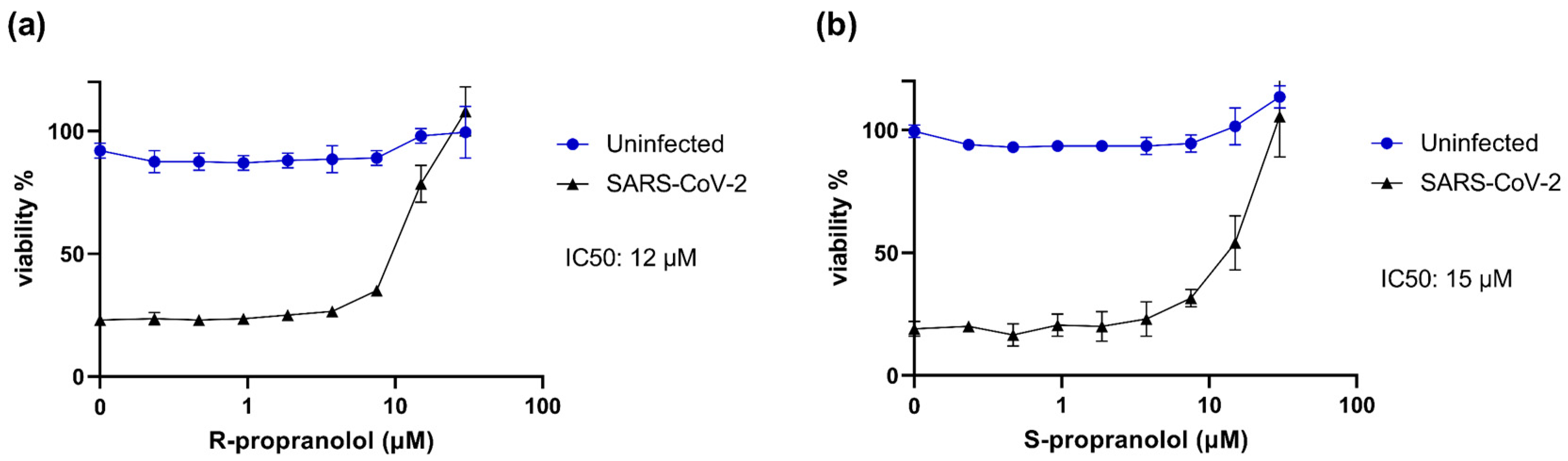
2.3. R- and S-Propranolol Inhibit SARS-CoV-2 Replication in Cell Culture
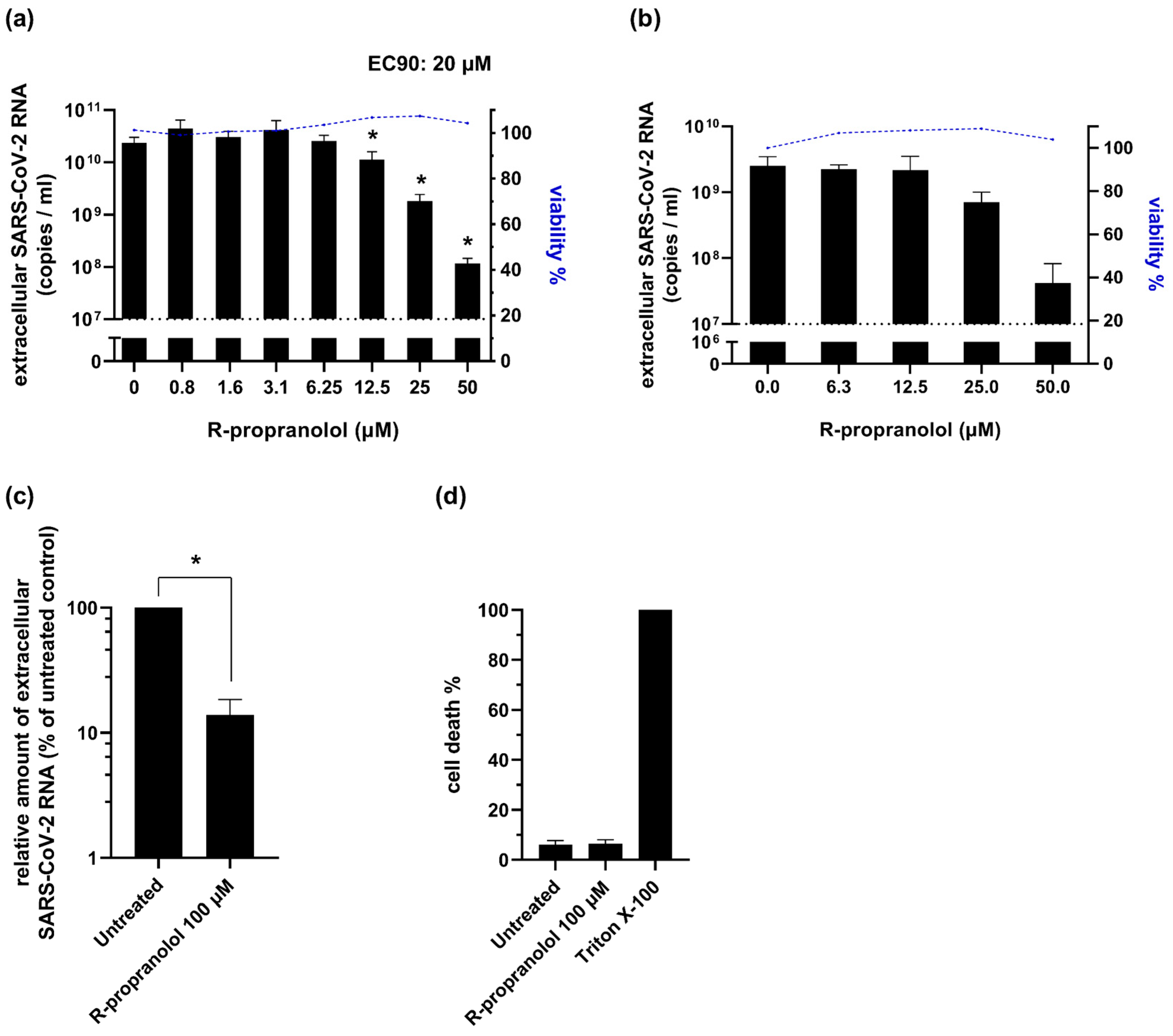
2.4. R-Propranolol Inhibits SARS-CoV-2 Replication in Various Cell Lines and Air–Liquid Interface-Cultured Primary Human Airway Epithelial Cells
2.5. R-Propranolol Inhibits SARS-CoV-2 at a Post-Entry Step of the Replication Cycle
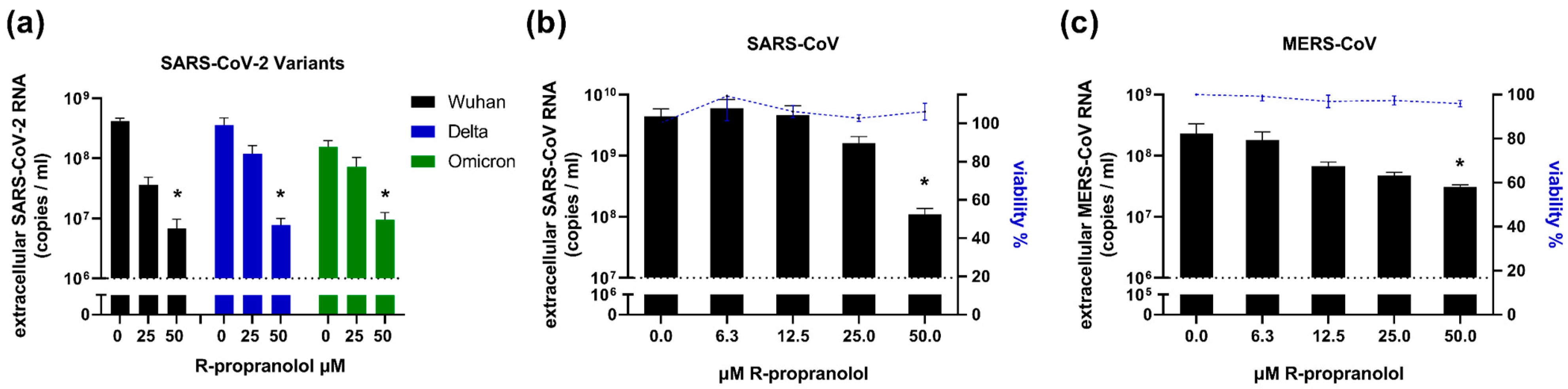
2.6. R-Propranolol Has Broad-Spectrum Antiviral Activity against Different Coronaviruses
3. Discussion
4. Materials and Methods
4.1. Compounds and Cell Culture
4.2. Virus Stocks
4.3. Endothelial Cell Infection
4.4. Endothelial Cell (Drug) Treatments
4.5. Cytopathic Effect (CPE) Reduction Assay
4.6. Viral Load Reduction Assays
4.7. Time-of-Addition Assay
4.8. Virucidal Effect Assay
4.9. Viral Infection of ALI-PBEC
4.10. RNA Isolation and RT-qPCR
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bösmüller, H.; Matter, M.; Fend, F.; Tzankov, A. The pulmonary pathology of COVID-19. Virchows Arch. Int. J. Pathol. 2021, 478, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Luo, R.; Zhang, M.; Wang, Y.; Song, T.; Tao, T.; Li, Z.; Jin, L.; Zheng, H.; Chen, W.; et al. A cross-talk between epithelium and endothelium mediates human alveolar–capillary injury during SARS-CoV-2 infection. Cell Death Dis. 2020, 11, 1042. [Google Scholar] [CrossRef] [PubMed]
- Bordoni, V.; Mariotti, D.; Matusali, G.; Colavita, F.; Cimini, E.; Ippolito, G.; Agrati, C. SARS-CoV-2 Infection of Airway Epithelium Triggers Pulmonary Endothelial Cell Activation and Senescence Associated with Type I IFN Production. Cells 2022, 11, 2912. [Google Scholar] [CrossRef] [PubMed]
- Solimando, A.G.; Marziliano, D.; Ribatti, D. SARS-CoV-2 and Endothelial Cells: Vascular Changes, Intussusceptive Microvascular Growth and Novel Therapeutic Windows. Biomedicines 2022, 10, 2242. [Google Scholar] [CrossRef]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in COVID-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef]
- Hassan, M.; Selimovic, D.; El-Khattouti, A.; Soell, M.; Ghozlan, H.; Haikel, Y.; Abdelkader, O.; Megahed, M. Hepatitis C virus-mediated angiogenesis: Molecular mechanisms and therapeutic strategies. World J. Gastroenterol. 2014, 20, 15467–15475. [Google Scholar] [CrossRef]
- Caposio, P.; Orloff, S.L.; Streblow, D.N. The role of cytomegalovirus in angiogenesis. Virus Res. 2011, 157, 204–211. [Google Scholar] [CrossRef] [Green Version]
- Bhatraju, P.K.; Morrell, E.D.; Stanaway, I.B.; Sathe, N.A.; Srivastava, A.; Postelnicu, R.; Green, R.; Andrews, A.; Gonzalez, M.; Kratochvil, C.J.; et al. Angiopoietin-Like4 Is a Novel Marker of COVID-19 Severity. Crit. Care Explor. 2023, 5, e0827. [Google Scholar] [CrossRef]
- Sasaki, M.; North, P.E.; Elsey, J.; Bubley, J.; Rao, S.; Jung, Y.; Wu, S.; Zou, M.-H.; Pollack, B.P.; Kumar, J.; et al. Propranolol exhibits activity against hemangiomas independent of beta blockade. NPJ Precis. Oncol. 2019, 3, 27. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Mai, H.M.; Zheng, J.; Zheng, J.W.; Wang, Y.A.; Qin, Z.P.; Li, K.L. Propranolol inhibits angiogenesis via down-regulating the expression of vascular endothelial growth factor in hemangioma derived stem cell. Int. J. Clin. Exp. Pathol. 2014, 7, 48–55. [Google Scholar]
- Varone, F.; Sgalla, G.; Iovene, B.; Bruni, T.; Richeldi, L. Nintedanib for the treatment of idiopathic pulmonary fibrosis. Expert Opin. Pharmacother. 2018, 19, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Xu, F.; Aondio, G.; Li, Y.; Fumagalli, A.; Lu, M.; Valmadre, G.; Wei, J.; Bian, Y.; Canesi, M.; et al. Efficacy and tolerability of bevacizumab in patients with severe COVID-19. Nat. Commun. 2021, 12, 814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, P.; Luo, R.; Wang, Y.; Li, Z.; Guo, Y.; Yao, Y.; Li, M.; Tao, T.; Chen, W.; et al. Biomimetic Human Disease Model of SARS-CoV-2 Induced Lung Injury and Immune Responses on Organ Chip System. Adv. Sci. 2020, 8, 2002928. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chong, H.C.; Ng, S.Y.; Kwok, K.W.; Teo, Z.; Tan, E.H.P.; Choo, C.C.; Seet, J.E.; Choi, H.W.; Buist, M.L.; et al. Angiopoietin-like 4 Increases Pulmonary Tissue Leakiness and Damage during Influenza Pneumonia. Cell Rep. 2015, 10, 654–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peuschel, K.E. Some clinical evidence of the hypothesis of an indirect antiviral effect of propranolol through immunoactivation. Med. Hypotheses 2011, 76, 689–691. [Google Scholar] [CrossRef]
- Fang, H.; Wang, Y.; Liu, L.; Cheng, K.; Li, P.; Tan, Y.; Hao, X.; Mei, M.; Xu, X.; Yao, Y.; et al. A Host-Harbored Metabolic Susceptibility of Coronavirus Enables Broad-Spectrum Targeting. bioRxiv 2022. [Google Scholar] [CrossRef]
- Stiles, J.; Amaya, C.; Pham, R.; Rowntree, R.K.; Lacaze, M.; Mulne, A.; Bischoff, J.; Kokta, V.; Boucheron, L.E.; Mitchell, D.C.; et al. Propranolol treatment of infantile hemangioma endothelial cells: A molecular analysis. Exp. Ther. Med. 2012, 4, 594–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Q.; Liao, P.; Li, W.; Hu, J.; Chen, C.; Zhang, Y.; Wang, Y.; Chen, L.; Song, K.; Liu, J.; et al. Clinical Use of Propranolol Reduces Biomarkers of Proliferation in Gastric Cancer. Front. Oncol. 2021, 11, 628613. [Google Scholar] [CrossRef]
- Zhou, C.; Chen, X.; Zeng, W.; Peng, C.; Huang, G.; Li, X.; Ouyang, Z.; Luo, Y.; Xu, X.; Xu, B.; et al. Propranolol induced G0/G1/S phase arrest and apoptosis in melanoma cells via AKT/MAPK pathway. Oncotarget 2016, 7, 68314–68327. [Google Scholar] [CrossRef]
- Klann, K.; Bojkova, D.; Tascher, G.; Ciesek, S.; Münch, C.; Cinatl, J. Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication. Mol. Cell 2020, 80, 164–174. [Google Scholar] [CrossRef]
- Salgado-Benvindo, C.; Leijs, A.A.; Thaler, M.; Tas, A.; Arbiser, J.L.; Snijder, E.J.; van Hemert, M.J. Honokiol inhibits SARS-CoV-2 replication in cell culture. bioRxiv 2022. [Google Scholar] [CrossRef]
- Wang, Y.; Thaler, M.; Ninaber, D.K.; van der Does, A.M.; Ogando, N.S.; Beckert, H.; Taube, C.; Salgado-Benvindo, C.; Snijder, E.J.; Bredenbeek, P.J.; et al. Impact of human airway epithelial cellular composition on SARS-CoV-2 infection biology. bioRxiv 2021. [Google Scholar] [CrossRef]
- Drosten, C.; Günther, S.; Preiser, W.; Van Der Werf, S.; Brodt, H.-R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Kovacikova, K.; Morren, B.M.; Tas, A.; Albulescu, I.C.; van Rijswijk, R.; Jarhad, D.B.; Shin, Y.S.; Jang, M.H.; Kim, G.; Lee, H.W.; et al. 6′-β-Fluoro-Homoaristeromycin and 6’-Fluoro-Homoneplanocin A Are Potent Inhibitors of Chikungunya Virus Replication through Their Direct Effect on Viral Nonstructural Protein 1. Antimicrob. Agents Chemother. 2020, 64, e02532-19. [Google Scholar] [CrossRef] [Green Version]
- Ogando, N.S.; Dalebout, T.J.; Zevenhoven-Dobbe, J.C.; Limpens, R.; van der Meer, Y.; Caly, L.; Druce, J.; de Vries, J.J.C.; Kikkert, M.; Bárcena, M.; et al. SARS-coronavirus-2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020, 101, 925–940. [Google Scholar] [CrossRef]
- Salgado-Benvindo, C.; Thaler, M.; Tas, A.; Ogando, N.S.; Bredenbeek, P.J.; Ninaber, D.K.; Wang, Y.; Hiemstra, P.S.; Snijder, E.J.; van Hemert, M.J. Suramin Inhibits SARS-CoV-2 Infection in Cell Culture by Interfering with Early Steps of the Replication Cycle. Antimicrob. Agents Chemother. 2020, 64, e00900-20. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brunink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thaler, M.; Salgado-Benvindo, C.; Leijs, A.; Tas, A.; Ninaber, D.K.; Arbiser, J.L.; Snijder, E.J.; van Hemert, M.J. R-Propranolol Has Broad-Spectrum Anti-Coronavirus Activity and Suppresses Factors Involved in Pathogenic Angiogenesis. Int. J. Mol. Sci. 2023, 24, 4588. https://doi.org/10.3390/ijms24054588
Thaler M, Salgado-Benvindo C, Leijs A, Tas A, Ninaber DK, Arbiser JL, Snijder EJ, van Hemert MJ. R-Propranolol Has Broad-Spectrum Anti-Coronavirus Activity and Suppresses Factors Involved in Pathogenic Angiogenesis. International Journal of Molecular Sciences. 2023; 24(5):4588. https://doi.org/10.3390/ijms24054588
Chicago/Turabian StyleThaler, Melissa, Clarisse Salgado-Benvindo, Anouk Leijs, Ali Tas, Dennis K. Ninaber, Jack L. Arbiser, Eric J. Snijder, and Martijn J. van Hemert. 2023. "R-Propranolol Has Broad-Spectrum Anti-Coronavirus Activity and Suppresses Factors Involved in Pathogenic Angiogenesis" International Journal of Molecular Sciences 24, no. 5: 4588. https://doi.org/10.3390/ijms24054588




