Identifications of QTLs and Candidate Genes Associated with Pseudomonas syringae Responses in Cultivated Soybean (Glycine max) and Wild Soybean (Glycine soja)
Abstract
1. Introduction
2. Results
2.1. Suinong14 (Improved Cultivar) and ZYD00006 (Wild Soybean) Exhibit Distinct Phenotypic Responses to Psgneau001
2.2. Identified 10 QTLs for BSD in Soybean CSSL Populations
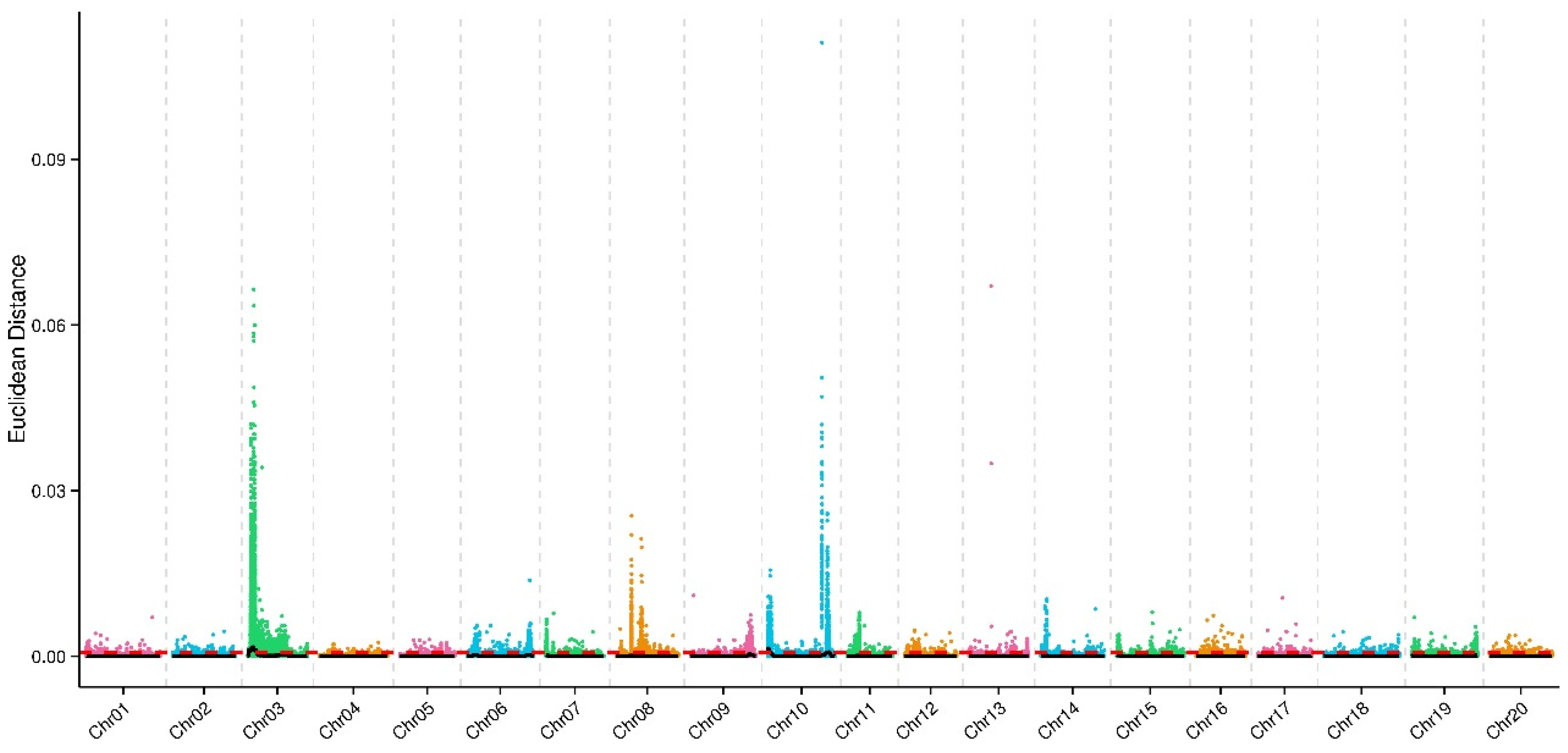
2.3. 3 Intervals Associated with Psgneau001 Resistance Identified via BSA-seq
2.4. Candidate QTL Identification
2.5. Chromosome Fragment Insertion-Based Candidate Interval Verification
2.6. Glyma.10g230200 Identified as the Candidate Gene Response to Psg via Sequencing and Expression Analysis
2.7. Glyma.10g230200 Encodes a WRKY27 and a Gene Differentially Expressed in BSD-Resistant and -Susceptible Soybeans Inoculated with Psgneau001
2.8. Haplotype Analyses Suggest a Link between Glyma.10g230200 and Psgneau001 Resistance
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain Selection
4.2. Plant Materials
4.3. Soybean Planting and Phenotypic Measurement
4.4. QTL Mapping
4.5. BSA-seq and Analysis
4.6. ZYD00006 Chromosome Fragment Insertion Analysis
4.7. Major QTL Gene Predictions and Candidate Gene qPCR Analyses
4.8. Haplotype Analyses of Natural Soybean Varieties
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.H.; Zhou, G.; Ma, J.; Jiang, W.; Jin, L.G.; Zhang, Z.; Guo, Y.; Zhang, J.; Sui, Y.; Zheng, L.; et al. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 2014, 32, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-Genome of Wild and Cultivated Soybeans. Cell 2020, 182, 162–176.e13. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Zhang, Z.; Yu, S.; Kang, Q.; Shi, Y.; Wang, J.; Zhu, R.; Ma, C.; Chen, L.; Wang, J.; et al. Responses of Soybean Genes in the Substituted Segments of Segment Substitution Lines Following a Xanthomonas Infection. Front. Plant Sci. 2020, 11, 972. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Yin, W.; Tang, Q.; Dong, S.; Zheng, X.; Zhang, Z.; Wang, Y. Distribution, Pathotypes, and Metalaxyl Sensitivity of Phytophthora sojae from Heilongjiang and Fujian Provinces in China. Plant Dis. 2010, 94, 881–884. [Google Scholar] [CrossRef]
- Zhou, H.; Lin, J.; Johnson, A.; Morgan, R.L.; Zhong, W.; Ma, W. Pseudomonas syringae type III effector HopZ1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell Host Microbe 2011, 9, 177–186. [Google Scholar] [CrossRef]
- Whitham, S.A.; Qi, M.; Innes, R.W.; Ma, W.; Lopes-Caitar, V.; Hewezi, T. Molecular Soybean-Pathogen Interactions. Annu. Rev. Phytopathol. 2016, 54, 443–468. [Google Scholar] [CrossRef] [PubMed]
- Agbavor, C.; Mirza, B.S.; Wait, A. The Effects of Phyllosphere Bacteria on Plant Physiology and Growth of Soybean Infected with Pseudomonas syringae. Plants 2022, 11, 2634. [Google Scholar] [CrossRef]
- Liu, D.D.; Lan, H.J.; Masoud, H.S.; Ye, M.Y.; Dai, X.Y.; Zhong, C.L.; Tian, S.N.; Liu, J.Z. Silencing GmBIR1 in Soybean Results in Activated Defense Responses. Int. J. Mol. Sci. 2022, 23, 7450. [Google Scholar] [CrossRef]
- Nemchinov, L.G.; Shao, J.; Lee, M.N.; Postnikova, O.A.; Samac, D.A. Resistant and susceptible responses in alfalfa (Medicago sativa) to bacterial stem blight caused by Pseudomonas syringae pv. syringae. PLoS ONE 2017, 12, e0189781. [Google Scholar] [CrossRef]
- Tarakanov, R.I.; Lukianova, A.A.; Evseev, P.V.; Toshchakov, S.V.; Kulikov, E.E.; Ignatov, A.N.; Miroshnikov, K.A.; Dzhalilov, F.S. Bacteriophage Control of Pseudomonas & savastanoi pv. glycinea in Soybean. Plants 2022, 11, 938. [Google Scholar]
- Jia, J.; Wang, X.; Deng, P.; Ma, L.; Baird, S.M.; Li, X.; Lu, S.E. Pseudomonas glycinae sp. nov. isolated from the soybean rhizosphere. MicrobiologyOpen 2020, 9, e1101. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Sakata, N.; Usuki, G.; Ishiga, T.; Hashimoto, Y.; Ishiga, Y. Multiple virulence factors regulated by AlgU contribute to the pathogenicity of Pseudomonas savastanoi pv. glycinea in soybean. PeerJ 2021, 9, e12405. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kim, K.H.; Van, K.; Kim, M.Y.; Lee, S.H. Fine mapping of a resistance gene to bacterial leaf pustule in soybean. Theor. Appl. Genet. 2010, 120, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- Tahir, J.; Hoyte, S.; Bassett, H.; Brendolise, C.; Chatterjee, A.; Templeton, K.; Deng, C.; Crowhurst, R.; Montefiori, M.; Morgan, E.; et al. Multiple quantitative trait loci contribute to resistance to bacterial canker incited by Pseudomonas syringae pv. actinidiae in kiwifruit (Actinidia chinensis). Hortic. Res. 2019, 6, 101. [Google Scholar] [CrossRef] [PubMed]
- González, A.M.; Godoy, L.; Santalla, M. Dissection of Resistance Genes to Pseudomonas syringae pv. phaseolicola in UI3 Common Bean Cultivar. Int. J. Mol. Sci. 2017, 18, 2503. [Google Scholar] [CrossRef]
- Tahir, J.; Brendolise, C.; Hoyte, S.; Lucas, M.; Thomson, S.; Hoeata, K.; McKenzie, C.; Wotton, A.; Funnell, K.; Morgan, E.; et al. QTL Mapping for Resistance to Cankers Induced by Pseudomonas syringae pv. actinidiae (Psa) in a Tetraploid Actinidia chinensis Kiwifruit Population. Pathogens 2020, 9, 967. [Google Scholar] [CrossRef]
- Bisgrove, S.R.; Simonich, M.T.; Smith, N.M.; Sattler, A.; Innes, R.W. A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 1994, 6, 927–933. [Google Scholar]
- Ashfield, T.; Ong, L.E.; Nobuta, K.; Schneider, C.M.; Innes, R.W. Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 2004, 16, 309–318. [Google Scholar] [CrossRef]
- Fu, D.Q.; Ghabrial, S.; Kachroo, A. GmRAR1 and GmSGT1 are required for basal, R gene-mediated and systemic acquired resistance in soybean. Mol. Plant Microbe Interact. 2009, 22, 86–95. [Google Scholar] [CrossRef]
- Wang, J.; Shine, M.B.; Gao, Q.M.; Navarre, D.; Jiang, W.; Liu, C.; Chen, Q.; Hu, G.; Kachroo, A. Enhanced Disease Susceptibility1 Mediates Pathogen Resistance and Virulence Function of a Bacterial Effector in Soybean. Plant Physiol. 2014, 165, 1269–1284. [Google Scholar] [CrossRef] [PubMed]
- Bernacki, M.J.; Czarnocka, W.; Szechyńska-Hebda, M.; Mittler, R.; Karpiński, S. Biotechnological Potential of LSD1, EDS1, and PAD4 in the Improvement of Crops and Industrial Plants. Plants 2019, 8, 290. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Dai, R.; Pike, S.M.; Qiu, W.; Gassmann, W. Functions of EDS1-like and PAD4 genes in grapevine defenses against powdery mildew. Plant Mol. Biol. 2014, 86, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Selote, D.; Shine, M.B.; Robin, G.P.; Kachroo, A. Soybean NDR1-like proteins bind pathogen effectors and regulate resistance signaling. New Phytol. 2014, 202, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Selote, D.; Robin, G.P.; Kachroo, A. GmRIN4 protein family members function nonredundantly in soybean race-specific resistance against Pseudomonas syringae. New Phytol. 2013, 197, 1225–1235. [Google Scholar] [CrossRef]
- Hwang, S.; Lee, T.G. Integration of lodging resistance QTL in soybean. Sci. Rep. 2019, 9, 6540. [Google Scholar] [CrossRef]
- Chen, H.; Pan, X.; Wang, F.; Liu, C.; Wang, X.; Li, Y.; Zhang, Q. Novel QTL and Meta-QTL Mapping for Major Quality Traits in Soybean. Front. Plant Sci. 2021, 12, 774270. [Google Scholar] [CrossRef]
- Lu, X.; Xiong, Q.; Cheng, T.; Li, Q.T.; Liu, X.L.; Bi, Y.D.; Li, W.; Zhang, W.K.; Ma, B.; Lai, Y.C.; et al. A PP2C-1 Allele Underlying a Quantitative Trait Locus Enhances Soybean 100-Seed Weight. Mol. Plant 2017, 10, 670–684. [Google Scholar] [CrossRef]
- Thapa, S.P.; Miyao, E.M.; Michael Davis, R.; Coaker, G. Identification of QTLs controlling resistance to Pseudomonas syringae pv. tomato race 1 strains from the wild tomato, Solanum habrochaites LA1777. Theor. Appl. Genet. 2015, 128, 681–692. [Google Scholar] [CrossRef]
- González, A.M.; Yuste-Lisbona, F.J.; Godoy, L.; Fernández-Lozano, A.; Paula Rodiño, A.; De Ron, A.M.; Rafael Lozano, R.; Santalla, M. Exploring the quantitative resistance to Pseudomonas syringae pv. phaseolicola in common bean (Phaseolus vulgaris L.). Mol. Breed. 2016, 36, 166. [Google Scholar] [CrossRef]
- Lin, F.; Wani, S.H.; Collins, P.J.; Wen, Z.; Li, W.; Zhang, N.; McCoy, A.G.; Bi, Y.; Tan, R.; Zhang, S.; et al. QTL mapping and GWAS for identification of loci conferringpartial resistance to Pythium sylvaticum in soybean(Glycine max (L.) Merr). Mol. Breed. 2020, 40, 54. [Google Scholar] [CrossRef]
- Yang, L.; Tian, Y.; Liu, Y.; Reif, J.C.; Li, Y.; Qiu, L. QTL mapping of qSCN3-1 for resistance to soybean cyst nematode in soybean line Zhongpin 03-5373. Crop J. 2021, 9, 351–359. [Google Scholar] [CrossRef]
- Qi, Z.; Huang, L.; Zhu, R.; Xin, D.; Liu, C.; Han, X.; Jiang, H.; Hong, W.; Hu, G.; Zheng, H.; et al. A high-density genetic map for soybean based on specific length amplified fragment sequencing. PLoS ONE 2014, 9, e104871. [Google Scholar] [CrossRef]
- Wang, J.; Ma, C.; Ma, S.; Zheng, H.; Tian, H.; Wang, X.; Wang, Y.; Jiang, H.; Wang, J.; Zhang, Z.; et al. Genetic variation in GmCRP contributes to nodulation in soybean (Glycine max Merr.). Crop J. 2022, in press. [Google Scholar] [CrossRef]
- Li, C.; Li, Y.H.; Li, Y.; Lu, H.; Hong, H.; Tian, Y.; Li, H.; Zhao, T.; Zhou, X.; Liu, J.; et al. A Domestication-Associated Gene GmPRR3b Regulates the Circadian Clock and Flowering Time in Soybean. Mol. Plant 2020, 13, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Li, W.; Zhang, C.; Yang, L.; Chang, R.Z.; Gaut, B.S.; Qiu, L.J. Genetic diversity in domesticated soybean (Glycine max) and its wild progenitor (Glycine soja) for simple sequence repeat and single-nucleotide polymorphism loci. New Phytol. 2010, 188, 242–253. [Google Scholar] [CrossRef]
- Li, Y.H.; Li, D.; Jiao, Y.Q.; Schnable, J.C.; Li, Y.F.; Li, H.H.; Chen, H.Z.; Hong, H.L.; Zhang, T.; Liu, B.; et al. Identification of loci controlling adaptation in Chinese soya bean landraces via a combination of conventional and bioclimatic GWAS. Plant Biotechnol. J. 2020, 18, 389–401. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, X.; Hu, Y.; Liu, S.; Nan, H.; Li, X.; Fang, C.; Cao, D.; Shi, X.; Kong, L.; et al. Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 2017, 49, 773–779. [Google Scholar] [CrossRef]
- Lam, H.M.; Xu, X.; Liu, X.; Chen, W.; Yang, G.; Wong, F.L.; Li, M.W.; He, W.; Qin, N.; Wang, B.; et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 2010, 42, 1053–1059. [Google Scholar] [CrossRef]
- Lu, S.; Dong, L.; Fang, C.; Liu, S.; Kong, L.; Cheng, Q.; Chen, L.; Su, T.; Nan, H.; Zhang, D.; et al. Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 2020, 52, 428–436. [Google Scholar] [CrossRef]
- Dong, L.; Cheng, Q.; Fang, C.; Kong, L.; Yang, H.; Hou, Z.; Li, Y.; Nan, H.; Zhang, Y.; Chen, Q.; et al. Parallel selection of distinct Tof5 alleles drove the adaptation of cultivated and wild soybean to high latitudes. Mol. Plant 2022, 15, 308–321. [Google Scholar] [CrossRef]
- Jin, T.; Sun, Y.; Shan, Z.; He, J.; Wang, N.; Gai, J.; Li, Y. Natural variation in the promoter of GsERD15B affects salt tolerance in soybean. Plant Biotechnol. J. 2021, 19, 1155–1169. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Yang, S.; Zhang, K.; He, J.; Wu, C.; Ren, Y.; Gai, J.; Li, Y. Natural variation and selection in GmSWEET39 affect soybean seed oil content. New Phytol. 2020, 225, 1651–1666. [Google Scholar] [CrossRef]
- Jiang, H.; Li, Y.; Qin, H.; Li, Y.; Qi, H.; Li, C.; Wang, N.; Li, R.; Zhao, Y.; Huang, S.; et al. Identification of Major QTLs Associated with First Pod Height and Candidate Gene Mining in Soybean. Front. Plant Sci. 2018, 9, 1280. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, Z.; Wen, Y.; Yu, G.; Zou, J.; Huang, S.; Wang, J.; Zhu, J.; Wang, J.; Chen, L.; et al. RNA Sequencing-Associated Study Identifies GmDRR1 as Positively Regulating the Establishment of Symbiosis in Soybean. Mol. Plant Microbe Interact. 2020, 33, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Zhang, Z.; Wang, Z.; Yu, J.; Qin, H.; Mao, X.; Jiang, H.; Xin, D.; Yin, Z.; Zhu, R.; et al. Meta-analysis and transcriptome profiling reveal hub genes for soybean seed storage composition during seed development. Plant Cell Environ. 2018, 41, 2109–2127. [Google Scholar] [CrossRef]
- Li, R.; Jiang, H.; Zhang, Z.; Zhao, Y.; Xie, J.; Wang, Q.; Zheng, H.; Hou, L.; Xiong, X.; Xin, D.; et al. Combined Linkage Mapping and BSA to Identify QTL and Candidate Genes for Plant Height and the Number of Nodes on the Main Stem in Soybean. Int. J. Mol. Sci. 2019, 21, 42. [Google Scholar] [CrossRef]
- Xin, D.; Qi, Z.; Jiang, H.; Hu, Z.; Zhu, R.; Hu, J.; Han, H.; Hu, G.; Liu, C.; Chen, Q. QTL Location and Epistatic Effect Analysis of 100-Seed Weight Using Wild Soybean (Glycine soja Sieb. & Zucc.) Chromosome Segment Substitution Lines. PLoS ONE 2016, 11, e0149380. [Google Scholar]
- Wang, Y.; Gao, H.; He, L.; Zhu, W.; Yan, L.; Chen, Q.; He, C. The PHOSPHATE1 genes participate in salt and Pi signaling pathways and play adaptive roles during soybean evolution. BMC Plant Biol. 2019, 19, 353. [Google Scholar] [CrossRef]
- Gu, Y.; Li, W.; Jiang, H.; Wang, Y.; Gao, H.; Liu, M.; Chen, Q.; Lai, Y.; He, C. Differential expression of a WRKY gene between wild and cultivated soybeans correlates to seed size. J. Exp. Bot. 2017, 68, 2717–2729. [Google Scholar] [CrossRef]
- Zheng, H.; Hou, L.; Xie, J.; Cao, F.; Wei, R.; Yang, M.; Qi, Z.; Zhu, R.; Zhang, Z.; Xin, D.; et al. Construction of Chromosome Segment Substitution Lines and Inheritance of Seed-Pod Characteristics in Wild Soybean. Front. Plant Sci. 2022, 13, 869455. [Google Scholar] [CrossRef]
- Qi, X.; Li, M.W.; Xie, M.; Liu, X.; Ni, M.; Shao, G.; Song, C.; Kay-Yuen Yim, A.; Tao, Y.; Wong, F.L.; et al. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 2014, 5, 4340. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, Y.; Chi, Y.; Fan, B.; Chen, Z. Characterization of Soybean WRKY Gene Family and Identification of Soybean WRKY Genes that Promote Resistance to Soybean Cyst Nematode. Sci. Rep. 2017, 7, 17804. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Dong, H.; Tan, J.; Li, M.; Yu, Y.; Jia, S.; Zhang, C.; Wu, Y.; Liu, Y. Transcriptome analysis of soybean WRKY TFs in response to Peronospora manshurica infection. Genomics 2019, 111, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, H.; Liu, J.; Han, S.; Lin, M.; Guo, Z.; Chen, X. OsWRKY62 and OsWRKY76 Interact with Importin α1s for Negative Regulation of Defensive Responses in Rice Nucleus. Rice 2022, 15, 12. [Google Scholar] [CrossRef]
- Liu, J.; Chen, X.; Liang, X.; Zhou, X.; Yang, F.; Liu, J.; He, S.Y.; Guo, Z. Alternative Splicing of Rice WRKY62 and WRKY76 Transcription Factor Genes in Pathogen Defense. Plant Physiol. 2016, 171, 1427–1442. [Google Scholar] [CrossRef]
- Wang, H.; Bi, Y.; Gao, Y.; Yan, Y.; Yuan, X.; Xiong, X.; Wang, J.; Liang, J.; Li, D.; Song, F. A Pathogen-Inducible Rice NAC Transcription Factor ONAC096 Contributes to Immunity against Magnaprothe oryzae and Xanthomonas oryzae pv. oryzae by Direct Binding to the Promoters of OsRap2.6, OsWRKY62, and OsPAL1. Front. Plant Sci. 2021, 12, 802758. [Google Scholar] [CrossRef]
- Ifnan Khan, M.; Zhang, Y.; Liu, Z.; Hu, J.; Liu, C.; Yang, S.; Hussain, A.; Furqan Ashraf, M.; Noman, A.; Shen, L.; et al. CaWRKY40b in Pepper Acts as a Negative Regulator in Response to Ralstonia solanacearum by Directly Modulating Defense Genes Including CaWRKY40. Int. J. Mol. Sci. 2018, 19, 1403. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, N.; Hu, R.; Xiang, F. Genome-wide identification of soybean WRKY transcription factors in response to salt stress. SpringerPlus 2016, 5, 920. [Google Scholar] [CrossRef]
- Fan, S.; Dong, L.; Han, D.; Zhang, F.; Wu, J.; Jiang, L.; Cheng, Q.; Li, R.; Lu, W.; Meng, F.; et al. GmWRKY31 and GmHDL56 Enhances Resistance to Phytophthora sojae by Regulating Defense-Related Gene Expression in Soybean. Front. Plant Sci. 2017, 8, 781. [Google Scholar] [CrossRef]
- Cui, X.; Yan, Q.; Gan, S.; Xue, D.; Wang, H.; Xing, H.; Zhao, J.; Guo, N. GmWRKY40, a member of the WRKY transcription factor genes identified from Glycine max L., enhanced the resistance to Phytophthora sojae. BMC Plant Biol. 2019, 19, 598. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Dun, H.; Lian, C.; Zhang, X.; Yin, W.; Xia, X. The role of peu-miR164 and its target PeNAC genes in response to abiotic stress in Populus euphratica. Plant Physiol. Biochem. 2017, 115, 418–438. [Google Scholar] [CrossRef] [PubMed]
- Arora, K.; Panda, K.K.; Mittal, S.; Mallikarjuna, M.G.; Thirunavukkarasu, N. In Silico Characterization and Functional Validation of Cell Wall Modification Genes Imparting Waterlogging Tolerance in Maize. Bioinform. Biol. Insights 2017, 11, 1177932217747277. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Cheng, L.; Wang, J.; Liu, J.; Cheng, J.; Yang, Z.; Cao, R.; Han, Y.; Li, H.; Zhang, B. Carotenoid Cleavage Dioxygenase 1 Catalyzes Lutein Degradation to Influence Carotenoid Accumulation and Color Development in Foxtail Millet Grains. J. Agric. Food Chem. 2022, 70, 9283–9294. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.; Somssich, I.E. A DNA-based real-time PCR assay for robust growth quantification of the bacterial pathogen Pseudomonas syringae on Arabidopsis thaliana. Plant Methods 2016, 12, 48. [Google Scholar] [CrossRef]
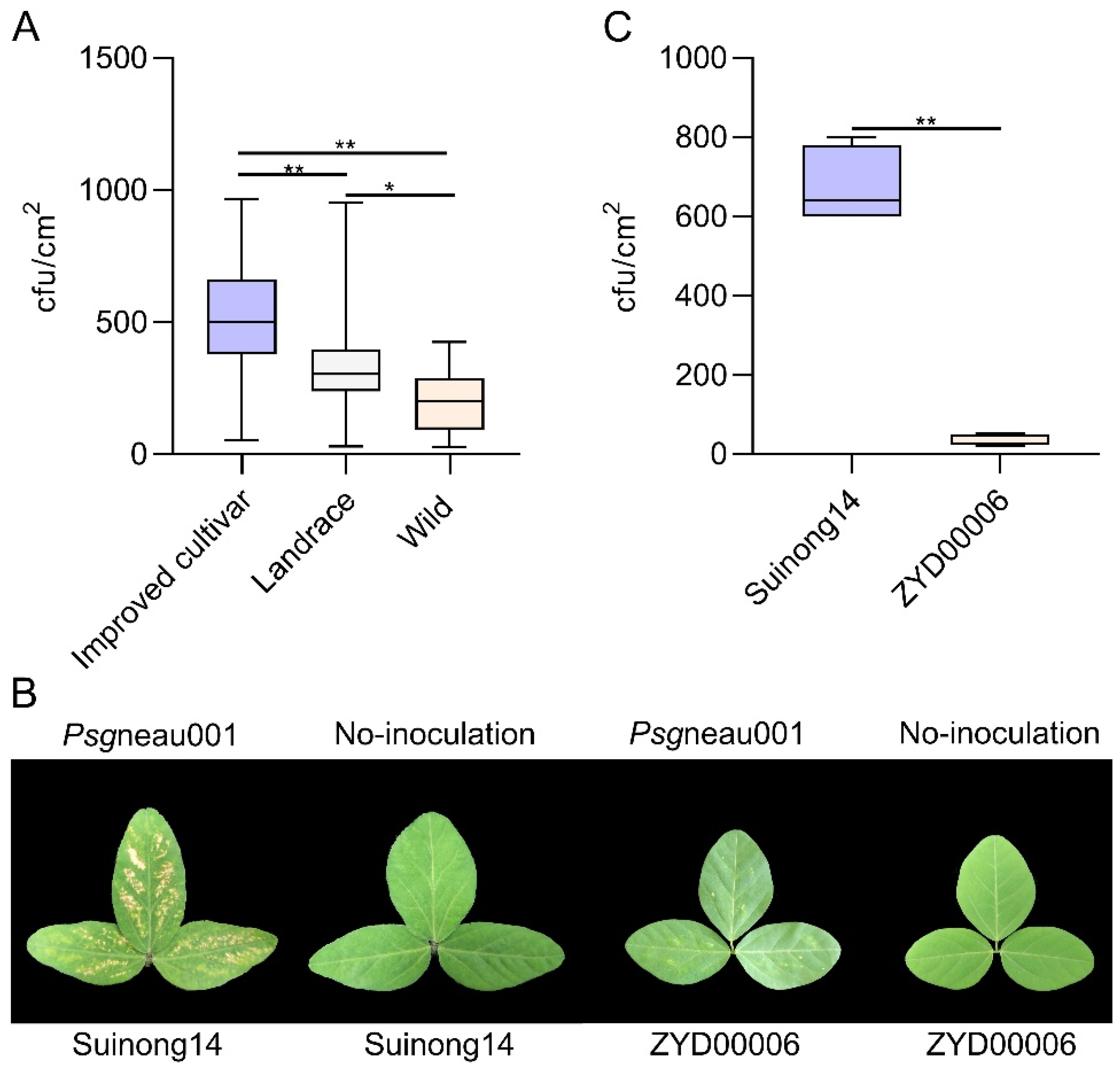




| Trait | Parents | CSSL Population (n = 170) | ||||
|---|---|---|---|---|---|---|
| ZYD00006 | Suinong14 | Mean | SD | Kurtosis | Skewness | |
| Colony-forming units | 34 ** | 680 | 468.35 | 173.087 | −0.140 | −0.360 |
| No. | Chr/LG | QTL | Position (Mb) | LOD | R2 (%) | ADD | Putative Causal Genes or QTLs Identified in Previous Studies |
|---|---|---|---|---|---|---|---|
| 1 | Chr02/D1b | qbsd-02-1 | 1.3 | 2.92 | 5 | −0.48 | |
| 2 | Chr08/A2 | qbsd-08-1 | 7.8 | 2.54 | 2 | 0.7 | |
| 3 | Chr08/A2 | qbsd-08-2 | 9.3 | 3.89 | 3 | 0.9 | |
| 4 | Chr08/A2 | qbsd-08-3 | 11.1 | 2.95 | 1 | 0.24 | |
| 5 | Chr09/K | qbsd-09-1 | 16.4 | 3.42 | 1 | 0.27 | |
| 6 | Chr10/O | qbsd-10-1 | 33.3 | 3.06 | 7 | 1.29 | qXav-10 [4] |
| 7 | Chr10/O | qbsd-10-2 | 46.7 | 4.4 | 8 | 0.75 | q10.1 [31] |
| 8 | Chr11/B1 | qbsd-11-1 | 32.5 | 2.98 | 6 | 0.48 | qSCN3-11 [32] |
| 9 | Chr11/B1 | qbsd-11-2 | 32.7 | 3.43 | 6 | 0.42 | qSCN3-11 [32] |
| 10 | Chr16/J | qbsd-16-1 | 33.2 | 2.89 | 2 | 0.31 | qXav-16 [4] |
| No. | QTL | Chromosome ID | Start Position (bp) | End Position (bp) | Size (Mb) |
|---|---|---|---|---|---|
| 1 | qbsd-bsa-1 | 3 | 380000 | 7390000 | 7.01 |
| 2 | qbsd-bsa-2 | 10 | 0 | 3150000 | 3.15 |
| 3 | qbsd-bsa-3 | 10 | 43910000 | 48050000 | 4.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Feng, H.; Jia, X.; Ma, S.; Ma, C.; Wang, Y.; Pan, S.; Chen, Q.; Xin, D.; Liu, C. Identifications of QTLs and Candidate Genes Associated with Pseudomonas syringae Responses in Cultivated Soybean (Glycine max) and Wild Soybean (Glycine soja). Int. J. Mol. Sci. 2023, 24, 4618. https://doi.org/10.3390/ijms24054618
Wang J, Feng H, Jia X, Ma S, Ma C, Wang Y, Pan S, Chen Q, Xin D, Liu C. Identifications of QTLs and Candidate Genes Associated with Pseudomonas syringae Responses in Cultivated Soybean (Glycine max) and Wild Soybean (Glycine soja). International Journal of Molecular Sciences. 2023; 24(5):4618. https://doi.org/10.3390/ijms24054618
Chicago/Turabian StyleWang, Jinhui, Haojie Feng, Xiaoke Jia, Shengnan Ma, Chao Ma, Yue Wang, Siyang Pan, Qingshan Chen, Dawei Xin, and Chunyan Liu. 2023. "Identifications of QTLs and Candidate Genes Associated with Pseudomonas syringae Responses in Cultivated Soybean (Glycine max) and Wild Soybean (Glycine soja)" International Journal of Molecular Sciences 24, no. 5: 4618. https://doi.org/10.3390/ijms24054618
APA StyleWang, J., Feng, H., Jia, X., Ma, S., Ma, C., Wang, Y., Pan, S., Chen, Q., Xin, D., & Liu, C. (2023). Identifications of QTLs and Candidate Genes Associated with Pseudomonas syringae Responses in Cultivated Soybean (Glycine max) and Wild Soybean (Glycine soja). International Journal of Molecular Sciences, 24(5), 4618. https://doi.org/10.3390/ijms24054618








