Pseudomonas aeruginosa Dps (PA0962) Functions in H2O2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity
Abstract
1. Introduction
2. Results and Discussion
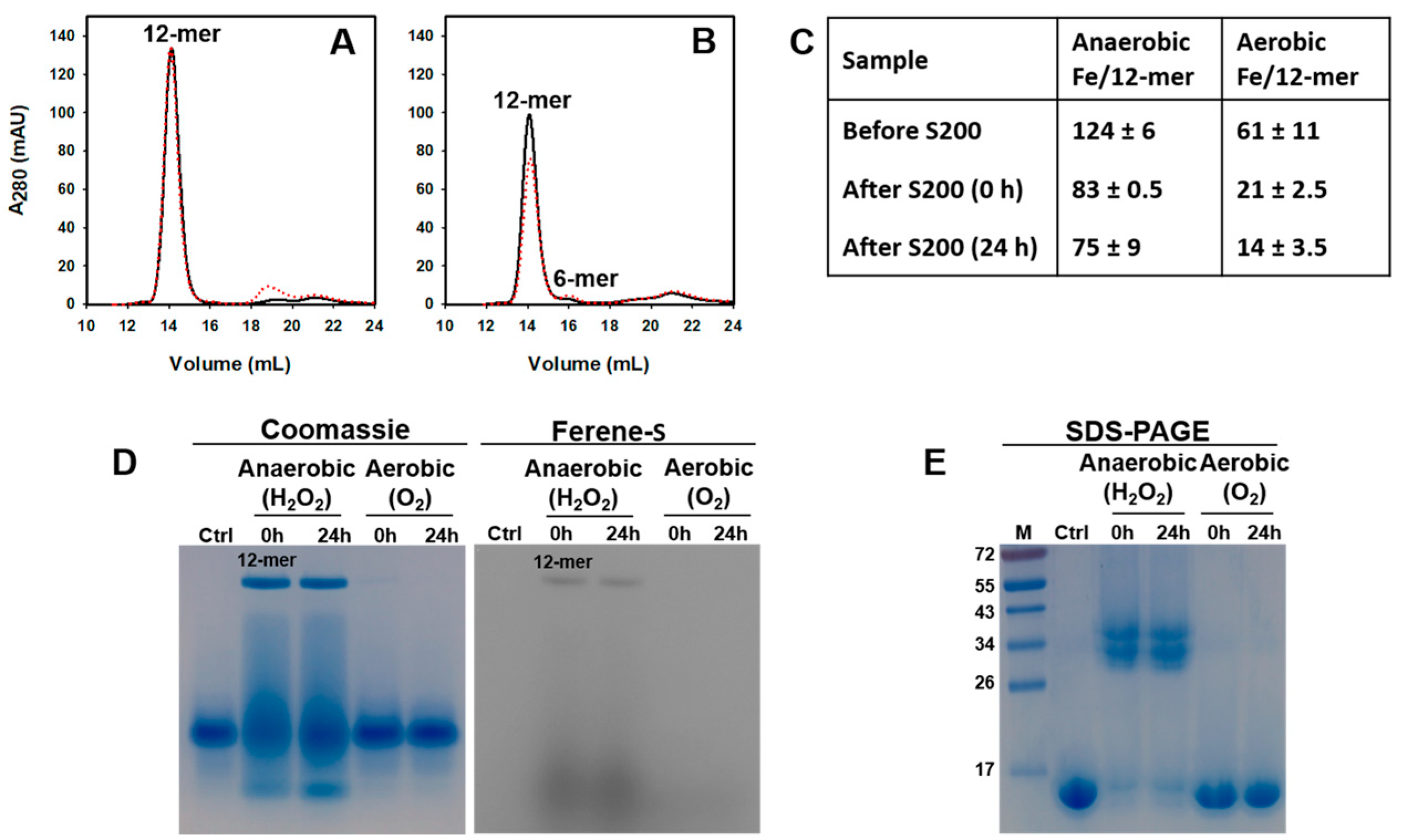
2.1. Pa Dps Oligomerization in Solution
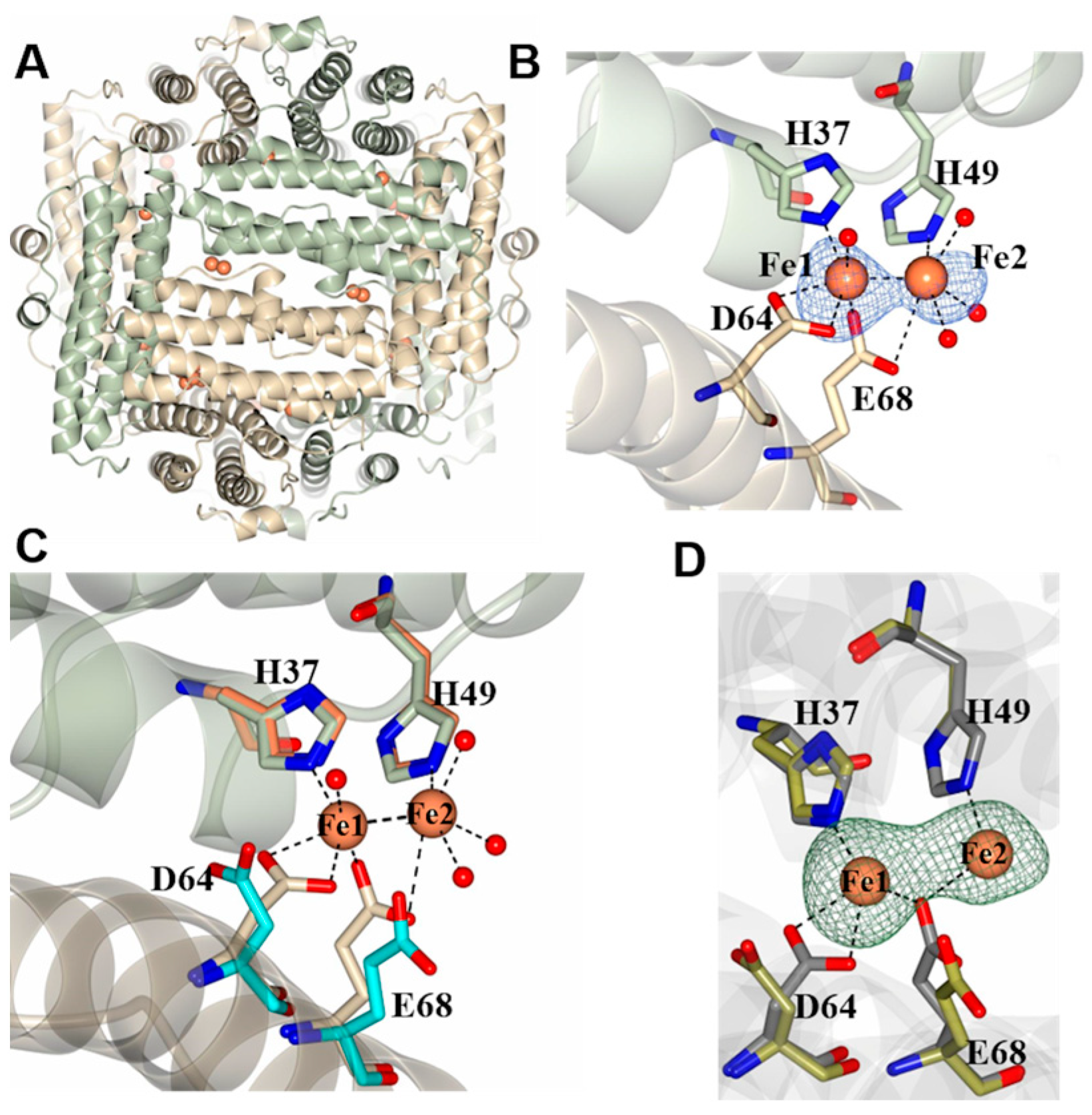
2.2. Pa Dps X-ray Crystal Structure
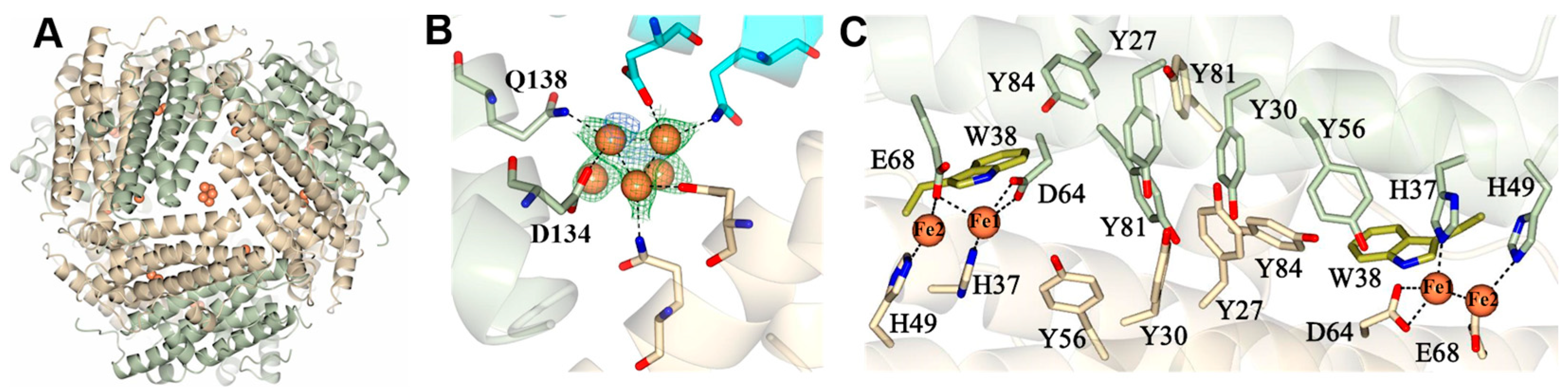
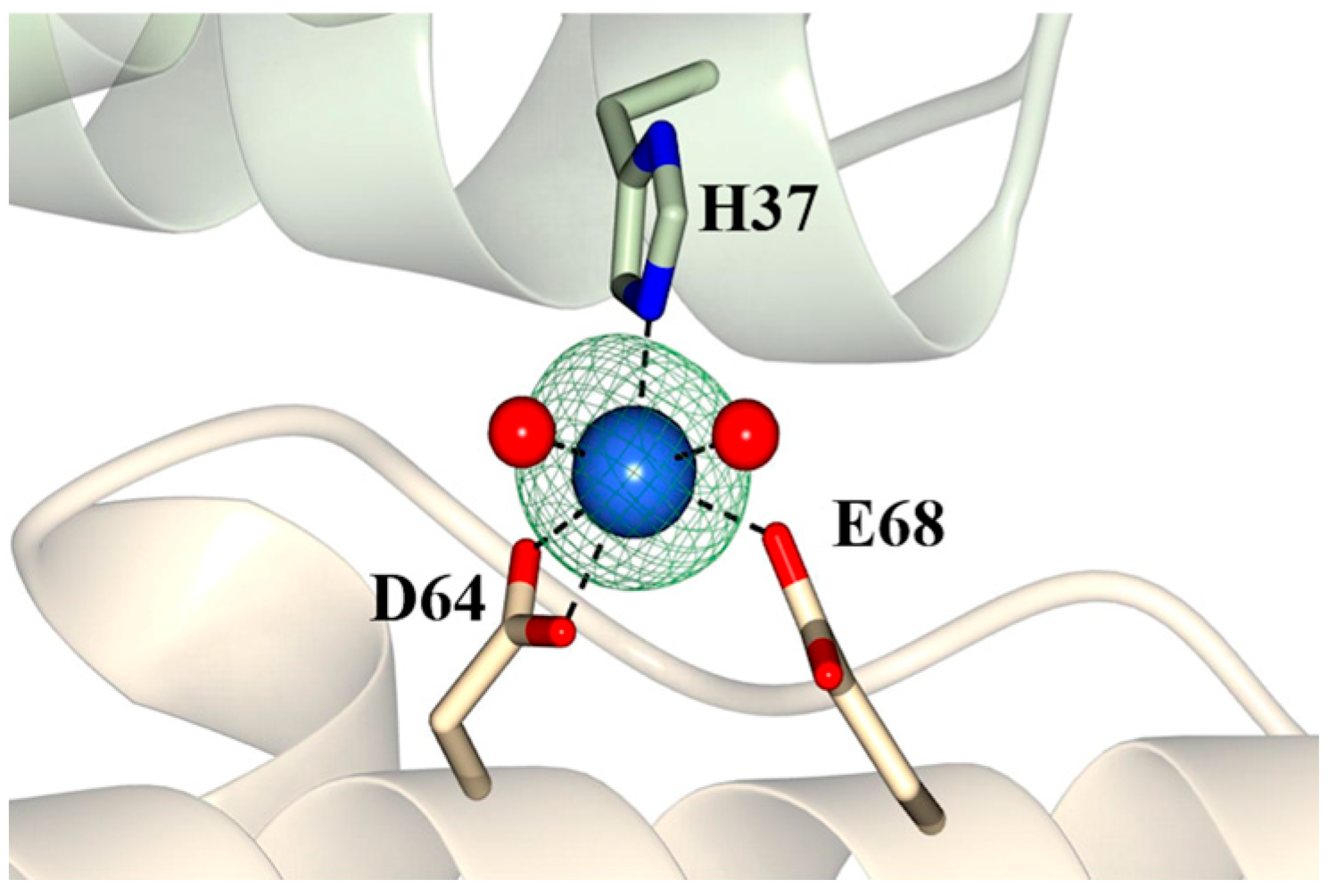
2.3. Pa Dps in Complex with Mn2+
2.4. Pa Catalyzes the Oxidation of Fe2+ When the Oxidant Is H2O2
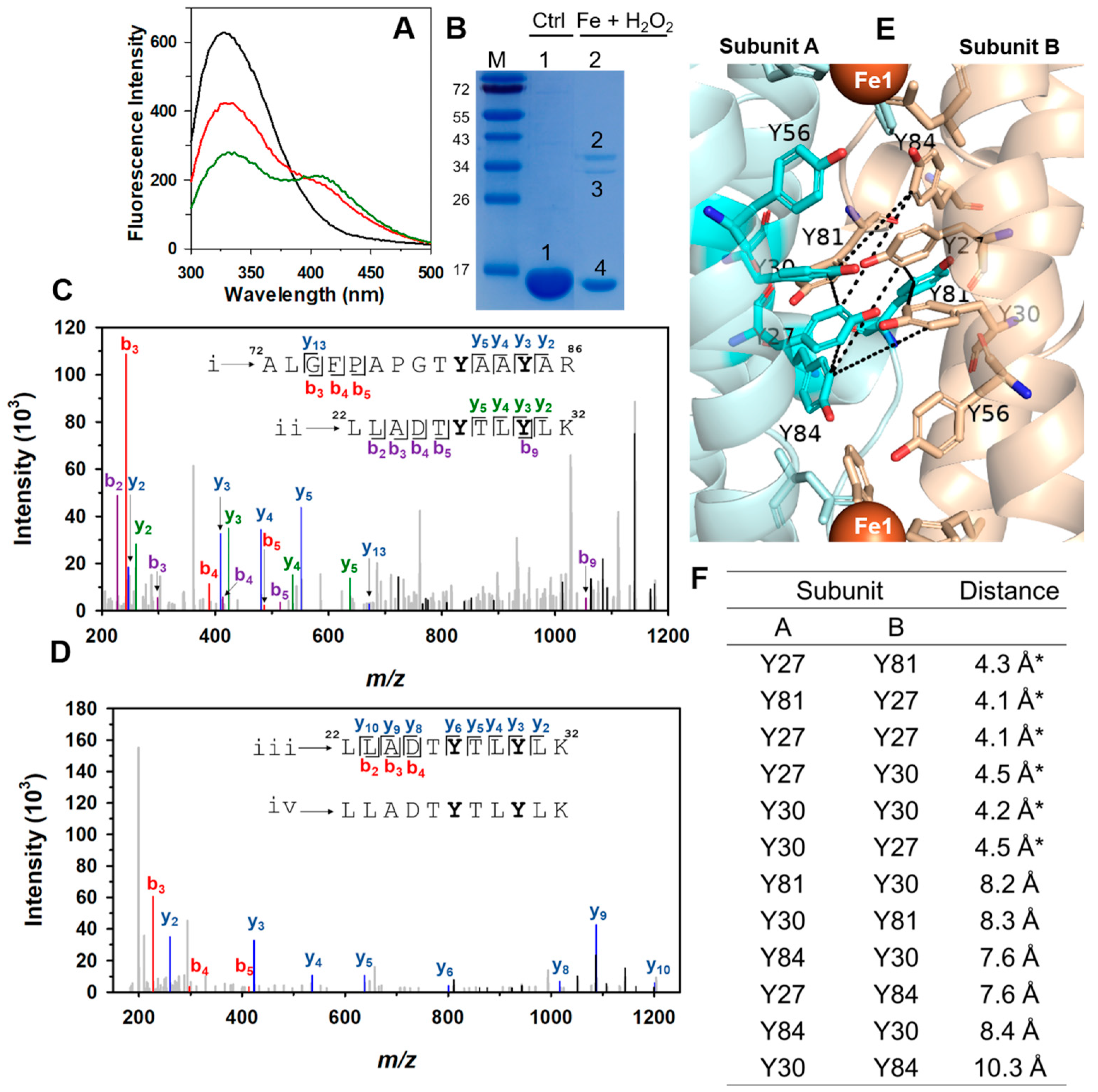
2.5. A Network of Tyr Residues Traps Hydroxyl Radical within the Protein Shell
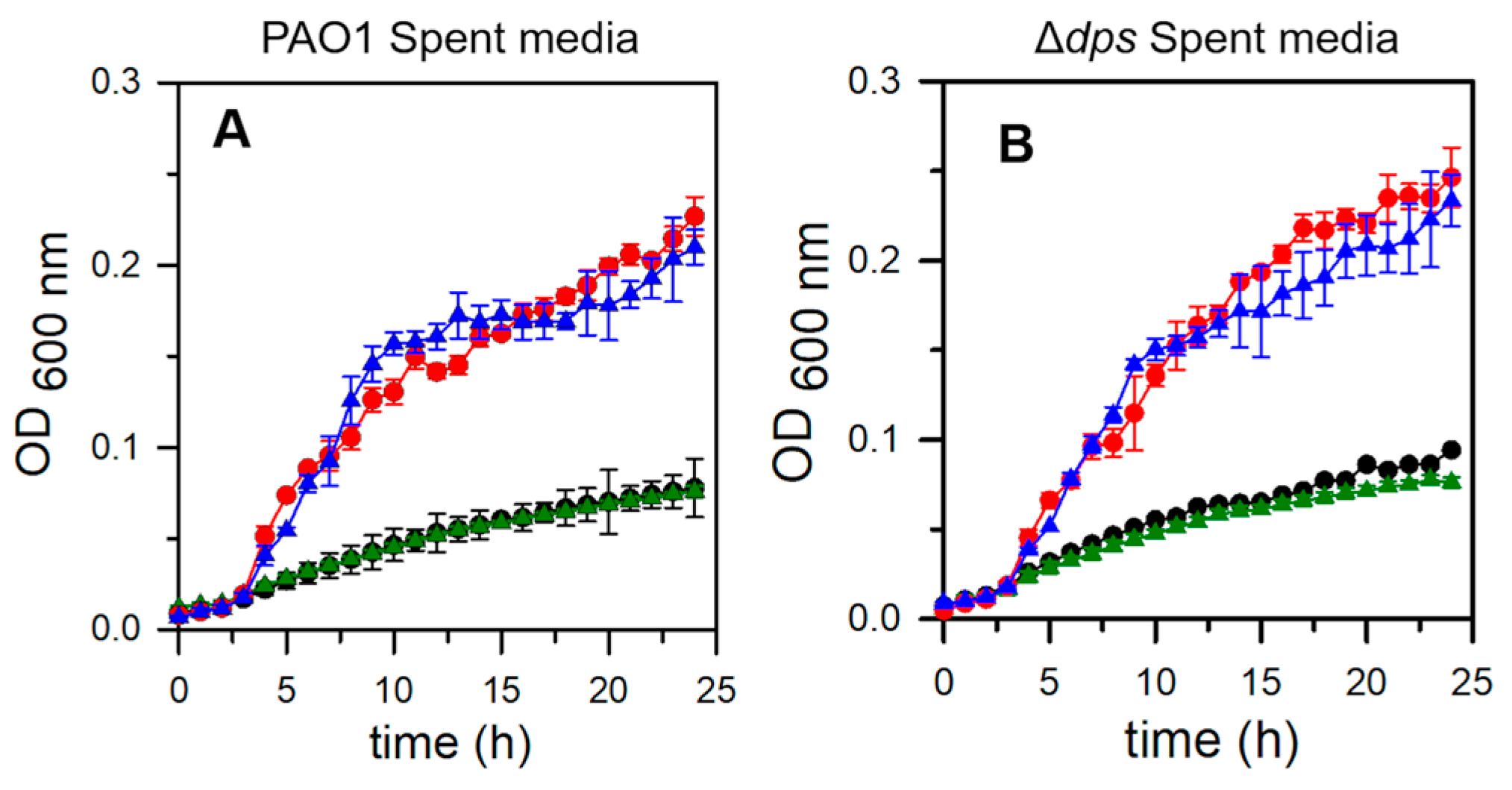
2.6. Pa Dps Protects P. aeruginosa Cells from H2O2-Mediated Oxidative Stress
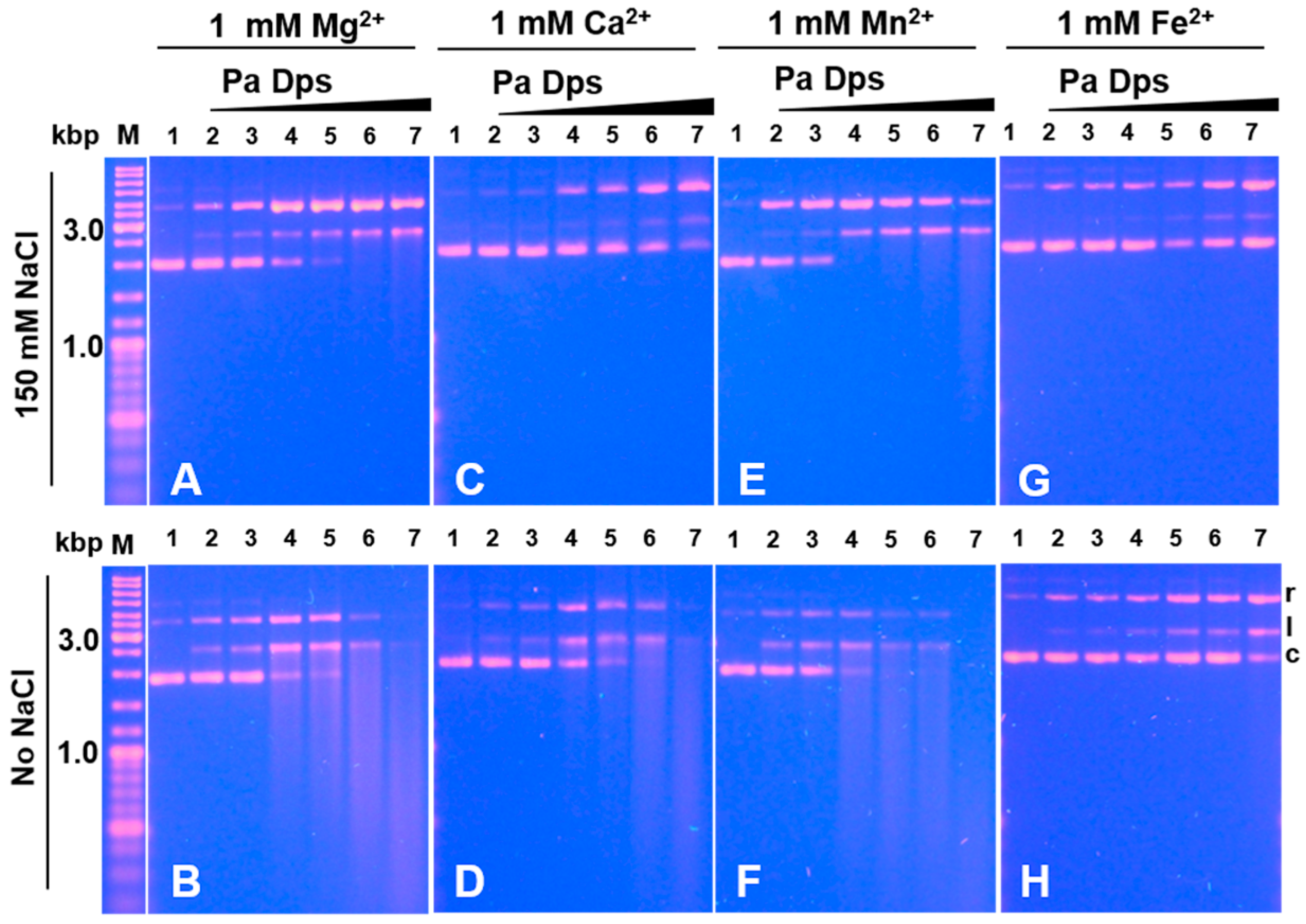
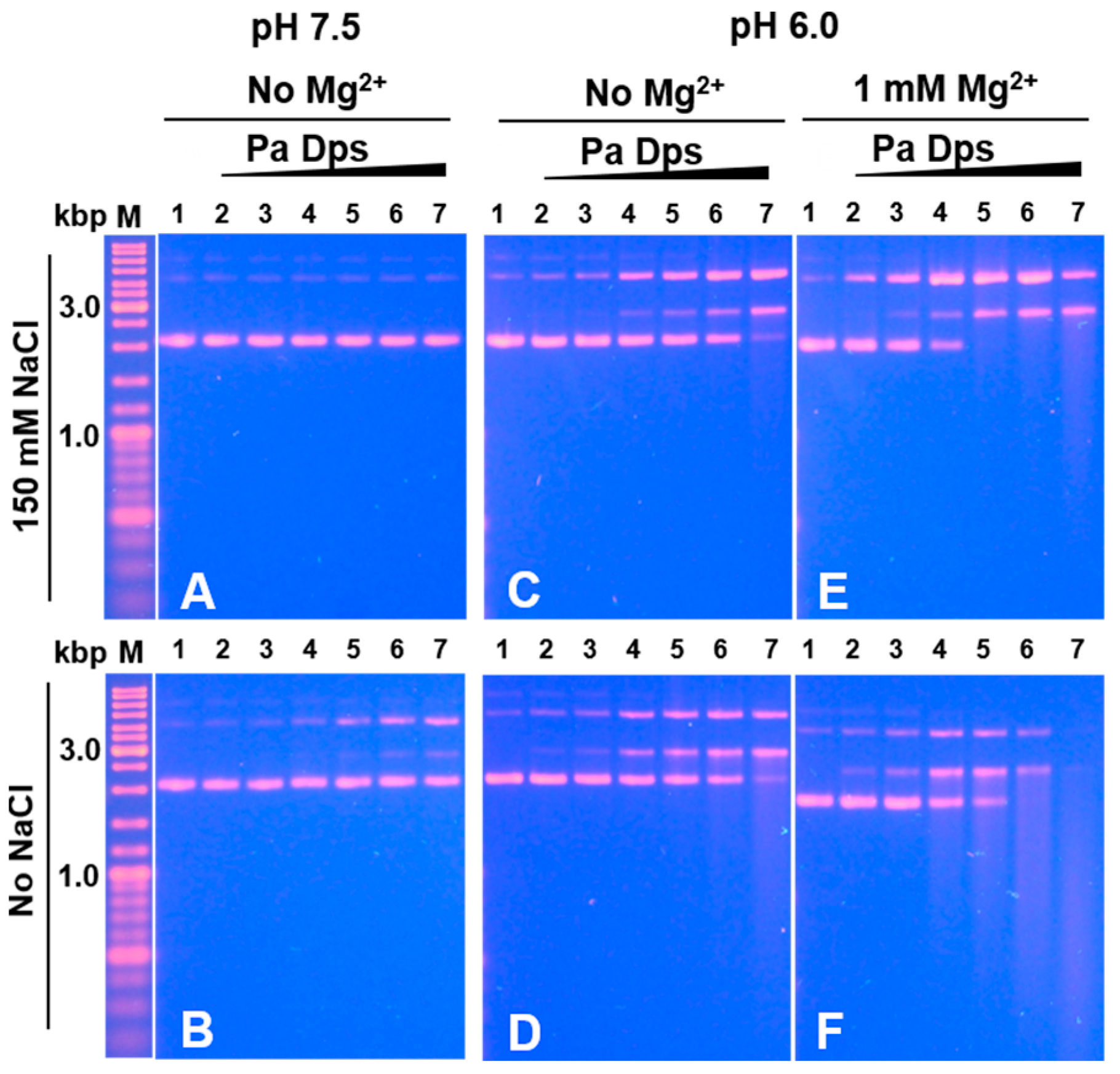
2.7. Pa Dps Cleaves DNA In Vitro
2.8. Pa Dps Does Not Contribute to the Utilization of DNA as a Nutrient Source
3. Materials and Methods
3.1. Chemicals, Bacterial Strains, and Media
3.2. Expression and Purification of Pa Dps
3.3. Titration of Pa Dps with Fe2+ under Aerobic and Anaerobic Conditions
3.4. Proteomics Analysis of Di-Tyrosine Crosslinked Pa Dps
3.5. Crystallization and Data Collection
3.6. Structure Solution and Refinement
3.7. Tolerance of P. aeruginosa PAO1 and Δdps Strains to H2O2
3.8. Utilization of DNA as a Nutrient Source
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nielsen, F.H. Evolutionary events culminating in specific minerals becoming essential for life. Eur. J. Nutr. 2000, 39, 62–66. [Google Scholar] [CrossRef]
- Kehrer, J.P. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology 2000, 149, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Crone, S.; Vives-Florez, M.; Kvich, L.; Saunders, A.M.; Malone, M.; Nicolaisen, M.H.; Martinez-Garcia, E.; Rojas-Acosta, C.; Catalina Gomez-Puerto, M.; Calum, H.; et al. The environmental occurrence of Pseudomonas aeruginosa. APMIS 2020, 128, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Burrows, L.L. The Therapeutic Pipeline for Pseudomonas aeruginosa Infections. ACS Infect. Dis. 2018, 4, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorli, L.; Luque, S.; Gomez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016, 44, D646–D653. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Jepkorir, G.; Lovell, S.; Nama, P.V.; Weeratunga, S.K.; Battaille, K.P.; Rivera, M. Two Disctinct Ferritin-Like Molecules in P. aeruginosa: The Product of the bfrA Gene is a Bacterial Ferritin (FtnA) not a bacterioferritin (Bfr). Biochemistry 2011, 50, 5236–5248. [Google Scholar] [CrossRef]
- Weeratunga, S.; Gee, C.E.; Lovell, S.; Zeng, Y.; Woodin, C.L.; Rivera, M. Binding of Pseudomonas aeruginosa Apobacterioferritin-Associated Ferredoxin to Bacterioferritin B Promotes Heme Mediation of Electron Delivery and Mobilization of Core Mineral Iron. Biochemistry 2009, 48, 7420–7431. [Google Scholar] [CrossRef]
- Weeratunga, S.; Lovell, S.; Yao, H.; Battaile, K.P.; Fischer, C.J.; Gee, C.E.; Rivera, M. Structural Studies of Bacterioferritin B (BfrB) from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center. Biochemistry 2010, 49, 1160–1175. [Google Scholar] [CrossRef]
- Rivera, M. Bacterioferritin: Structure, Dynamics and Protein-Protein Interactions at Play in Iron Storage and Mobilization. Acc Chem. Res. 2017, 50, 331–340. [Google Scholar] [CrossRef]
- Rivera, M. Bacterioferritin: Structure Function and Protein-Protein Interactions. In Handbook of Porphyrin Science; Kadish, K.K., Smith, K.M., Guilard, R., Eds.; World Scientific Publishing Co.: Hackensack, NJ, USA, 2014; Volume 30, pp. 136–179. [Google Scholar]
- Yao, H.; Soldano, A.; Fontenot, L.; Donnarumma, F.; Lovell, S.; Chandler, J.R.; Rivera, M. Pseudomonas aeruginosa Bacterioferritin Is Assembled from FtnA and BfrB Subunits with the Relative Proportions Dependent on the Environmental Oxygen Availability. Biomolecules 2022, 12, 366. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Wang, Y.; Lovell, S.; Kumar, R.; Ruvinsky, A.M.; Battaile, K.P.; Vakser, I.A.; Rivera, M. The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin. J. Am. Chem. Soc. 2012, 134, 13470–13481. [Google Scholar] [CrossRef]
- Eshelman, K.; Yao, H.; Punchi Hewage, A.N.D.; Deay, J.J.; Chandler, J.R.; Rivera, M. Inhibiting the BfrB:Bfd Interaction in Pseudomonas aeruginosa Causes Irreversible Iron Accumulation in Bacterioferritin and Iron Deficiency in the Bacterial Cell. Metallomics 2017, 9, 646–659. [Google Scholar] [CrossRef] [PubMed]
- Punchi Hewage, A.N.D.; Fontenot, L.; Guidry, J.; Weldeghiorghis, T.; Mehta, A.K.; Donnarumma, F.; Rivera, M. Mobilization of Iron Stored in Bacterioferritin Is Required for Metabolic Homeostasis in Pseudomonas aeruginosa. Pathogens 2020, 9, 980. [Google Scholar] [CrossRef]
- Soldano, A.; Yao, H.; Chandler, J.R.; Rivera, M. Inhibiting Iron Mobilization from Bacterioferritin in Pseudomonas aeruginosa Impairs Biofilm Formation Irrespective of Environmental Iron Availability. ACS Infect. Dis. 2020, 6, 447–458. [Google Scholar] [CrossRef]
- Soldano, A.; Yao, H.; Punchi Hewage, A.N.D.; Meraz, K.; Annor-Gyamfi, J.K.; Bunce, R.A.; Battaile, K.P.; Lovell, S.; Rivera, M. Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells. ACS Infect. Dis. 2021, 7, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Punchi Hewage, A.N.D.; Yao, H.; Nammalwar, B.; Gnanasekaran, K.K.; Lovell, S.; Bunce, R.A.; Eshelman, K.; Phaniraj, S.M.; Lee, M.M.; Peterson, B.R.; et al. Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity. J. Am. Chem. Soc. 2019, 141, 8171–8184. [Google Scholar] [CrossRef]
- Almiron, M.; Link, A.J.; Furlong, D.; Kolter, R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992, 6, 2646–2654. [Google Scholar] [CrossRef]
- Martinez, A.; Kolter, R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 1997, 179, 5188–5194. [Google Scholar] [CrossRef]
- Nair, S.; Finkel, S.E. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 2004, 186, 4192–4198. [Google Scholar] [CrossRef]
- Honarmand Ebrahimi, K.; Hagedoorn, P.L.; Hagen, W.R. Unity in the biochemistry of the iron-storage proteins ferritin and bacterioferritin. Chem. Rev. 2015, 115, 295–326. [Google Scholar] [CrossRef]
- Grant, R.A.; Filman, D.J.; Finkel, S.E.; Kolter, R.; Hogle, J.M. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol. 1998, 5, 294–303. [Google Scholar] [CrossRef]
- Haikarainen, T.; Papageorgiou, A.C. Dps-like proteins: Structural and functional insights into a versatile protein family. Cell Mol. Life Sci. 2010, 67, 341–351. [Google Scholar] [CrossRef]
- Ren, B.; Tibbelin, G.; Kajino, T.; Asami, O.; Ladenstein, R. The multi-layered structure of Dps with a novel di-nuclear ferroxidase center. J. Mol. Biol. 2003, 329, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Bellapadrona, G.; Stefanini, S.; Zamparelli, C.; Theil, E.C.; Chiancone, E. Iron translocation into and out of Listeria innocua Dps and size distribution of the protein-enclosed nanomineral are modulated by the electrostatic gradient at the 3-fold "ferritin-like" pores. J. Biol. Chem. 2009, 284, 19101–19109. [Google Scholar] [CrossRef]
- Yao, H.; Rui, H.; Kumar, R.; Eshelman, K.; Lovell, S.; Battaile, K.P.; Im, W.; Rivera, M. Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic. Biochemistry 2015, 54, 1611–1627. [Google Scholar] [CrossRef] [PubMed]
- Bellapadrona, G.; Ardini, M.; Ceci, P.; Stefanini, S.; Chiancone, E. Dps proteins prevent Fenton-mediated oxidative damage by trapping hydroxyl radicals within the protein shell. Free Radic. Biol. Med. 2010, 48, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Ilari, A.; Stefanini, S.; Chiancone, E.; Tsernoglou, D. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat. Struct Biol. 2000, 7, 38–43. [Google Scholar] [CrossRef]
- Zanotti, G.; Papinutto, E.; Dundon, W.; Battistutta, R.; Seveso, M.; Giudice, G.; Rappuoli, R.; Montecucco, C. Structure of the neutrophil-activating protein from Helicobacter pylori. J. Mol. Biol. 2002, 323, 125–130. [Google Scholar] [CrossRef]
- Ceci, P.; Ilari, A.; Falvo, E.; Chiancone, E. The Dps protein of Agrobacterium tumefaciens does not bind to DNA but protects it toward oxidative cleavage: X-ray crystal structure, iron binding, and hydroxyl-radical scavenging properties. J. Biol. Chem. 2003, 278, 20319–20326. [Google Scholar] [CrossRef]
- Malencik, D.A.; Anderson, S.R. Dityrosine as a product of oxidative stress and fluorescent probe. Amino Acids 2003, 25, 233–247. [Google Scholar] [CrossRef]
- Correia, M.; Neves-Petersen, M.T.; Jeppesen, P.B.; Gregersen, S.; Petersen, S.B. UV-light exposure of insulin: Pharmaceutical implications upon covalent insulin dityrosine dimerization and disulphide bond photolysis. PLoS ONE 2012, 7, e50733. [Google Scholar] [CrossRef] [PubMed]
- Biemann, K. Appendix 5. Nomenclature for peptide fragment ions (positive ions). Methods Enzymol. 1990, 193, 886–887. [Google Scholar] [CrossRef]
- Panchaud, A.; Singh, P.; Shaffer, S.A.; Goodlett, D.R. xComb: A cross-linked peptide database approach to protein-protein interaction analysis. J. Proteome Res. 2010, 9, 2508–2515. [Google Scholar] [CrossRef] [PubMed]
- Maura, D.; Hazan, R.; Kitao, T.; Ballok, A.E.; Rahme, L.G. Evidence for Direct Control of Virulence and Defense Gene Circuits by the Pseudomonas aeruginosa Quorum Sensing Regulator, MvfR. Sci. Rep. 2016, 6, 34083. [Google Scholar] [CrossRef]
- Lee, S.Y.; Lim, C.J.; Droge, P.; Yan, J. Regulation of Bacterial DNA Packaging in Early Stationary Phase by Competitive DNA Binding of Dps and IHF. Sci. Rep. 2015, 5, 18146. [Google Scholar] [CrossRef] [PubMed]
- Guerra, J.P.L.; Jacinto, J.P.; Tavares, P. Miniferritins: Small multifunctional protein cages. Coordin. Chem. Rev. 2021, 449, 214187. [Google Scholar] [CrossRef]
- Davey, C.A.; Richmond, T.J. DNA-dependent divalent cation binding in the nucleosome core particle. Proc. Natl. Acad. Sci. USA 2002, 99, 11169–11174. [Google Scholar] [CrossRef] [PubMed]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.L.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete Genome Sequence of Pseudomonas aeruginosa PA01, an Opportunistic Pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Hmelo, L.R.; Borlee, B.R.; Almblad, H.; Love, M.E.; Randall, T.E.; Tseng, B.S.; Lin, C.; Irie, Y.; Storek, K.M.; Yang, J.J.; et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 2015, 10, 1820–1841. [Google Scholar] [CrossRef]
- Choi, K.H.; Schweizer, H.P. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat. Protoc. 2006, 1, 153–161. [Google Scholar] [CrossRef]
- Ikemura, T. Codon Usage and tRNA Content in Unicellular and Multicellular Organisms. Mol. Biol. Evol. 1985, 2, 13–34. [Google Scholar] [PubMed]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Kapp, E.A.; Lothian, A.; Roberts, A.M.; Vasil'ev, Y.V.; Boughton, B.A.; Barnham, K.J.; Kok, W.M.; Hutton, C.A.; Masters, C.L.; et al. Characterization and Identification of Dityrosine Cross-Linked Peptides Using Tandem Mass Spectrometry. Anal. Chem. 2017, 89, 6136–6145. [Google Scholar] [CrossRef]
- Kabsch, W. Automatic Indexing of Rotation Diffraction Patterns. J. Appl. Cryst. 1988, 21, 67–72. [Google Scholar] [CrossRef]
- Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.R. An Introduction to Data Reduction: Space-Group Determination, scaling and intentisy statistics. Acta Cryst. 2011, D67, 282–292. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Cryst. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowan, K. Features and Development of Coot. Acta Cryst. 2010, D66, 486–501. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.r.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-Atom Structure Validation for Macromolecular Crystallography. Acta Cryst. D 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Potterton, L.; McNicholas, S.; Krissinel, E.; Grubber, J.; Cowtan, K.; Emsley, P.; Murshudov, G.N.; Cohen, S.; Perrakis, A.; Noble, M. Developments in the CCP4 Molecular-Graphics Project. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2288–2294. [Google Scholar] [CrossRef]
- Krissinel, E. Enhanced fold recognition using efficient short fragment clustering. J. Mol. Biochem. 2012, 1, 76–85. [Google Scholar]
- Zhao, G.; Ceci, P.; Ilari, A.; Giangiacomo, L.; Laue, T.M.; Chiancone, E.; Chasteen, N.D. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J Biol. Chem. 2002, 277, 27689–27696. [Google Scholar] [CrossRef]
- Su, M.; Cavallo, S.; Stefanini, S.; Chiancone, E.; Chasteen, N.D. The so-called Listeria innocua ferritin is a Dps protein. Iron incorporation, detoxification, and DNA protection properties. Biochemistry 2005, 44, 5572–5578. [Google Scholar] [CrossRef]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 72–82. [Google Scholar] [CrossRef]
- Diederichs, K.; Karplus, P.A. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 1997, 4, 269–275. [Google Scholar] [CrossRef]
- Weiss, M.S. Global indicators of X-ray data quality. J. Appl. Crystallogr. 2001, 34, 130–135. [Google Scholar] [CrossRef]
- Karplus, P.A.; Diederichs, K. Linking crystallographic model and data quality. Science 2012, 336, 1030–1033. [Google Scholar] [CrossRef]
- Evans, P. Resolving some old problems in protein crystallography. Science 2012, 336, 986–987. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajapaksha, N.; Soldano, A.; Yao, H.; Donnarumma, F.; Kashipathy, M.M.; Seibold, S.; Battaile, K.P.; Lovell, S.; Rivera, M. Pseudomonas aeruginosa Dps (PA0962) Functions in H2O2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity. Int. J. Mol. Sci. 2023, 24, 4669. https://doi.org/10.3390/ijms24054669
Rajapaksha N, Soldano A, Yao H, Donnarumma F, Kashipathy MM, Seibold S, Battaile KP, Lovell S, Rivera M. Pseudomonas aeruginosa Dps (PA0962) Functions in H2O2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity. International Journal of Molecular Sciences. 2023; 24(5):4669. https://doi.org/10.3390/ijms24054669
Chicago/Turabian StyleRajapaksha, Nimesha, Anabel Soldano, Huili Yao, Fabrizio Donnarumma, Maithri M. Kashipathy, Steve Seibold, Kevin P. Battaile, Scott Lovell, and Mario Rivera. 2023. "Pseudomonas aeruginosa Dps (PA0962) Functions in H2O2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity" International Journal of Molecular Sciences 24, no. 5: 4669. https://doi.org/10.3390/ijms24054669
APA StyleRajapaksha, N., Soldano, A., Yao, H., Donnarumma, F., Kashipathy, M. M., Seibold, S., Battaile, K. P., Lovell, S., & Rivera, M. (2023). Pseudomonas aeruginosa Dps (PA0962) Functions in H2O2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity. International Journal of Molecular Sciences, 24(5), 4669. https://doi.org/10.3390/ijms24054669






