Abstract
Cytochromes P450 are ancient enzymes diffused in organisms belonging to all kingdoms of life, including viruses, with the largest number of P450 genes found in plants. The functional characterization of cytochromes P450 has been extensively investigated in mammals, where these enzymes are involved in the metabolism of drugs and in the detoxification of pollutants and toxic chemicals. The aim of this work is to present an overview of the often disregarded role of the cytochrome P450 enzymes in mediating the interaction between plants and microorganisms. Quite recently, several research groups have started to investigate the role of P450 enzymes in the interactions between plants and (micro)organisms, focusing on the holobiont Vitis vinifera. Grapevines live in close association with large numbers of microorganisms and interact with each other, regulating several vine physiological functions, from biotic and abiotic stress tolerance to fruit quality at harvest.
1. Cytochromes P450 Enzymes: General Features
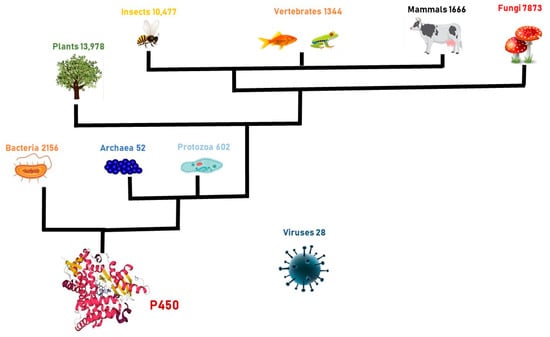
Cytochromes P450 (P450s or CYPs) are heme–thiolate terminal monooxygenases that transfer one atom from molecular oxygen to X–H bonds, where X may be the -C, -N, or S of a substrate, with a concomitant reduction in the remaining oxygen atom to water [1]. The CYP-mediated molecular oxygen activation gives rise to ketones, alcohols, aldehydes, epoxides, and carboxylic acids. Radicals or reactive oxygenated intermediates generated by CYPs can also lead to the formation of oxidative C–C bond cleavage [2], dealkylation [3], desaturation [4], dehydration [5], the oxidative rearrangement of carbon skeletons [6], and decarboxylation [7]. CYPs are a large superfamily of enzymes that show an absorbance peak at 450 nm when their heme is reduced and complexed with carbon monoxide. These enzymes were discovered in 1958, while studying the spectrophotometric properties of pigments in a microsomal fraction of rat livers [8]. A few years later, it was demonstrated that this pigment is a heme-containing protein, and it was called “P-450” (P for pigment) [9]. The heme group plays essential roles in catalysis, providing the P450s’ carbon monoxide-binding ability; moreover, the signature sequence FxxGxxxCxG in the heme-binding domain functions as the fifth ligand to the heme iron. CYPs are ancient enzymes that are extensively diffused in the organisms of all kingdoms of life (Figure 1), including viruses [10], with the largest number of P450 genes being present in plants [11]. Plant P450s are classified into the A-type group and are involved in plant-specific biochemical pathways [12]. The non-A-type P450 enzymes [13] form distinct clades and are more closely related to non-plant P450 enzymes than to the A-type. CYPs are classified into families on the basis of the sequence identity of their amino acid sequences, and the symbol CYP is the root followed by an Arabic numeral representing family therefore, the subfamily is indicated by a letter and the gene is represented by a number [14]. The features and functions of P450 enzymes have been studied from bacteria to mammals (see http://drnelson.uthsc.edu/CytochromeP450.html, accessed on 15 November 2022) [14]. Human P450 enzymes metabolize drugs and synthesize endogenous compounds essential for human physiology. Often, alterations in specific P450s affect the biological processes that they mediate, leading to a disease [15].
2. Cytochrome P450 Reactivity
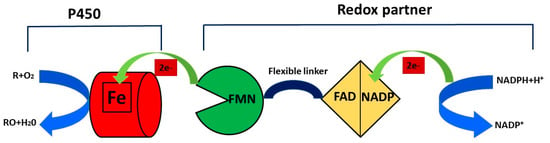
CYPs, through the activation of molecular oxygen, are able to monooxygenate the substrate, followed by the insertion of a single atom of oxygen into an organic substrate and a reduction in the another oxygen atom to water according to the following scheme:
RH + O2 + NAD(P)H + H+ → ROH + H2O + NAD(P)+
NAD(P)H supplies electron equivalents through an electron transfer chain that involves different redox partners, the nature of which is utilized to classify cytochromes P450 into 10 classes [16]. The organic substrate (R) binds to the heme group of the enzyme and this binding allows the transfer of an electron from NADPH through cytochrome P450 reductase (CPR) to the hem domain that reduces the iron (Fe) from the ferric state (Fe3+) to the ferrous state (Fe2+). The molecular oxygen binds to ferrous P450, forming a ferrous CYP–dioxygen complex, and the second electron is transferred from CPR to the ferrous CYP–dioxygen complex, forming the peroxo complex, and this complex is rapidly protonated twice to form one molecule of water and an iron–oxo complex. The oxidized reaction product (RO) is formed during the last step of the catalytic cycle when the atom of oxygen in the iron–oxo complex binds to the organic substrate (R) [17] (Figure 2).
3. The Role of Cytochromes P450 in Plants
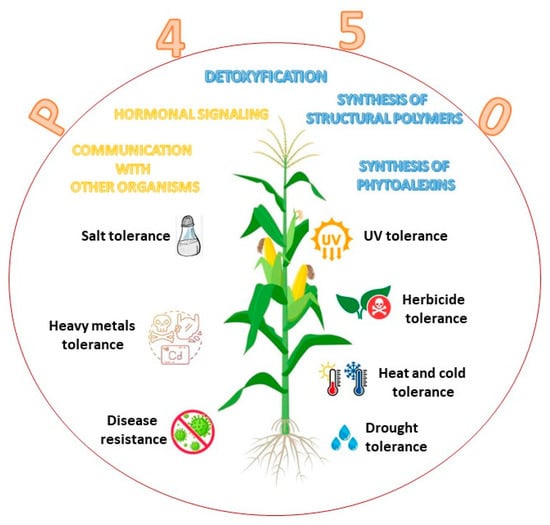
In plant cytochromes, P450 enzymes are present in several organelles and organs, namely, the endoplasmic reticulum, mitochondria, and chloroplast [18]. They are also found in shoot, bulbs, hypocotyl, roots, and the endosperm of germinating seeds. In both plants and animals, CYPs have an absorbance range from 447 to 452 nm and constitute the largest family of enzymes in the metabolism of a plant, where they are grouped into ten clans [19,20]. The most studied family is CYP51, with a pivotal role in regulating the synthesis of sterols [21] and triterpenes [22]. CYP71 impacts shikimate products and intermediates [23,24]. The CYP72 clan has a role in the catabolism of isoprenoid hormones [25], and the CYP74 synthetizes oxylipin derivatives and allene oxide in the octadecanoid and jasmonate pathways [26,27]. CYP85 participates in the metabolism of cyclic terpenes and sterols in the brassinosteroid and in abscisic acid and gibberellin pathways [28]. The CYP86 hydroxylates fatty acids [29], the CYP97 hydroxylates carotenoids [30], and CYP710 control the synthesis of sterol C-22 desaturase [5]. The CYP450 family in plants has a molecular mass between 45 and 62 kDa [4,31,32]. In plants, P450 enzymes are involved in the biosynthesis of structural polymers, defense against pathogen infection, communication with other organisms, hormonal signaling, herbicide resistance, and stress tolerance (Figure 3).
Plant hormone metabolism is also regulated by cytochrome P450 enzymes that control cell division and cell expansion, vascular differentiation, fruit growth, root development, and flower formation. Cytochromes P450 protect plants from dehydration [3], UV stress [33], and drought [34]. Moreover, P450s play a role in physiological process, such as detoxification, adaptation responses to heavy metals, salts, and chemicals (e.g., herbicides) [35,36]. In addition, many P450s participate in the biosynthesis of cell wall components [37]. CYP97 engages in the biosynthetic pathway of xanthophylls, hydroxylating the beta and gamma rings of carotenoids [38] (Table 1).
In Solanum lycopersicum, the CYP78 subfamily modulates the number and length of the lateral shoots and fruit ripening time [39]. In Solanum tuberosum, CYP72 is involved in the biosynthesis of two steroidal glycoalkaloids that catalyze the 26- and 22-hydroxylation steps. The knockdown plants of CYP72 are sterile, and tubers do not sprout during storage.
Tuber sprouting and the biosynthesis of glycoalkaloid in potatoes, two traits that significantly impact potato breeding and are important for the industry, can be controlled by tools provided by the functional analyses of the CYP72 family [40]. The cuticle of anther and the pollen exine are protective envelopes of the male gametophyte and the pollen grain. The fatty acid hydroxylase CYP703 is essential for male fertility in rice because the CYP703A3-2 mutant shows that pollen exine is defective and that the anthers exhibit a reduced level of cutin monomers and wax components [41]. Cucumber and melon contain cucurbitacins, highly oxygenated triterpenoids responsible for the bitter taste [47]. Cucurbitacins derive from the tetracyclic triterpenoid cucurbitadienol that is further oxidized to cucurbitacin at different positions by CYP81 and CYP87 [42,43,48] and CYP72 and CYP71 [22,44]. In Olea europaea, CYP72 catalyzes the oxidative C–C bond cleavage in the biosynthesis of secoxy-iridoids, a series of phenolic compounds that are essential for the flavor and quality of olive oil [45]. CYP716 is involved in the biosynthesis of the triterpenic acids present in apple fruit [46], and CYP97 is involved in the biosynthetic pathway of lutein in Sorghum [32].
4. Cytochrome P450 Enzyme in Plant–Microorganism Interaction
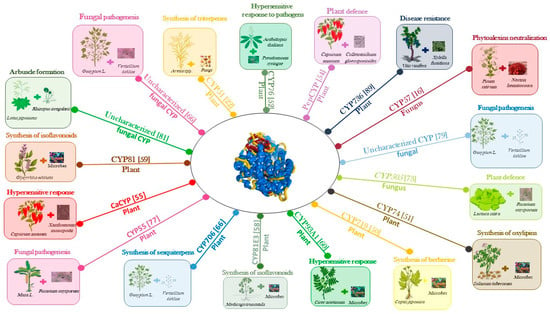
The main focus of this review is an overview of the role of the cytochrome P450 enzyme in plants, including crop plants, focusing on their involvement in the interaction between plants and (micro)organisms (Figure 4). In this context, biotic stresses were considered as a consequence of the plant interaction with the organism, causing the stress (e.g., pathogens). CYPs contribute to the synthesis of terpenoids, phytoalexins, alkaloids, and cyanogenic glucosides, which play important roles in the response of plant to biotic stresses.
4.1. CYPs in Plant–Bacteria Interaction
In Coptis japonica, the biosynthesis of berberine, an antimicrobial benzylisoquinoline, is mediated by the methylenedioxy bridge-forming enzyme CYP719 [49]. In potatoes, the antimicrobial compound oxylipin is synthetized by the pathogen-inducible divinyl ether synthase CYP74 [50]. When the bacterial pathogen Pseudomonas syringae infects A. thaliana, it causes a hypersensitive response that induces the expression of the CYP76C2 gene [51,52]. CYP76C2 gene expression is associated with leaf senescence, the aging of cell cultures, and wounding [51,53]. The pepCYP gene product plays a role in the defense mechanism when the pathogenic fungus Colletotrichum gloeosporioides invades and colonizes the fruits of pepper [54]. CYP89 is another cytochrome P450 in pepper that plays a pivotal role in plant defense response against pathogen infections [55]. Isoflavonoids are phenylpropanoid metabolites that act as antimicrobial compounds [56] that are present in legumes [57]. CYP81 and CYP93 are involved in isoflavonoid biosynthesis in Medicago truncatula [58], Glycyrrhiza echinata [59], and Cicer arietnum [60]. In response to bacterial and fungal pathogen attack, many plants rapidly induce the biosynthesis of phytoalexins, which are low molecular weight antimicrobial compounds showing great structural diversity. CYP71 and CYP79 are responsible for the biosynthesis of the phytalexin camalexin in A. thaliana [13].
4.2. Cytochrome P450 Enzyme in Plant–Fungi Interaction
In fungi, cytochrome P450 enzymes regulate a plethora of different physiological mechanisms including fertility and fitness. In filamentous fungi, P450 enzymes play a housekeeping role, being involved in sterol synthesis [61], and CYP51 has been identified as a good target for the development of antifungal drugs [62]. The diversification and expansion of the P450 families has been related to fungal pathogenicity [63,64]. P450 enzymes are involved in the responses of the host plants to pathogen attacks, neutralizing the production of antibiotic phytoalexins [65]. CYP57 is a pisatin demethylase of the fungus Nectria haematococca, a pathoghen of peas. This P450 enzyme inactivates the isoflavonoid pisatin that is produced by peas when the fungus invades the plant [16]. In Nicotiana benthamiana, CYP51 participates in the synthesis of antimicrobial triterpenes and is involved in antifungal activity [22]. In cotton, CYP70 participates in the synthesis of gossypol and related sesquiterpene induced by Verticillium dahlia infection [66]. The fungus Fusarium oxysporum causes severe vascular wilt disease in several crop plants [67]. Antagonistic F. oxysporum strains protect plants from pathogenic fungi, and they have been successfully used as biological control agents in agriculture [68]. The CYP505 family belonging to class VIII [16] includes a flavocytochrome wherein an N-terminal heme domain is fused to the C terminal FAD/FMN reductase domain [69]. CYP505 members hydroxylate fatty acids in the subterminal omega position, a step that is fundamental for these molecules to be used as an energy source [70]. Oxidized fatty acids are endogenous signal molecules with an important role in the activation of plant defense mechanisms [71] during interactions between plants and fungi [72]. Pathogenic formae specialis of F. oxysporum have a CYP505 enzyme that is differentially expressed when the fungus behaves as a pathogen or as an antagonist. In a particular case, using lettuce plants, it was found that F. oxysporum CYP505A1 is expressed in the host plant during the early phases of the interaction, both in pathogenesis and in antagonism, while its expression is silenced only in the late phase when the fungus behaves as a pathogen [73].
CYP505A1 can mono-hydroxylate lauric, palmitic, and stearic acids present in the cortical cell membranes of the plant, and these hydroxylated compounds might activate the plant defense system [73]. There is increasing evidence that P450 enzymes have a pivotal role in plant defense from pathogenic fungi, and the mechanism can be summarized as the activation of the synthesis of secondary metabolites [74]. Other examples of fungal P450 upregulation are found during the invasion of Heterobasidion annosums [75], Moniliophthora perniciosa [64], and Botrytis cinerea [76], specifically of their host plants. F. oxysporum f. sp. cubense, which is pathogenic to banana, upregulates the expression of its nitric oxide reductase CYP55 when it invades the plant [77]. CYP55 is involved in the nitrogen response pathway that is essential for fungal pathogenicity [78]. CYP55 is expressed in F. oxysporum f.sp. vasinfectum during the invasion of the roots of the cotton plant [78]. A cytochrome P450 that is involved in the metabolism of sulfacetamide, a secondary metabolite that is important for fungal pathogenesis, is upregulated in Verticillium dahlia during the first days of infection in cotton [79]. Fusarium graminearum induces the expression of a benzoate 4-monooxygenase cytochrome P450 gene when it invades wheat coleoptiles [79].
Arbuscular mycorrhizas (AMs) are symbiotic associations between the roots of land plants and fungi belonging the phylum Glomeromycota [80]. The fungal hyphopodium invades the root tissue forming the arbuscule, the site where nutrients between the plant and the fungus are exchanged. It has been shown that, when the AM symbiosis is forming between Lotus japonicus and Rhizopus irregularis, there is a high level of expression of cyp genes in the intraradical hyphae, which due to their involvment in the synthesis of sterols for membrane biogenesis during arbuscule formation [81].
5. Cytochrome P450 Enzyme in Vitis vinifera
This section is focused on Vitis vinifera and the role of cytochromes P450 in grapevine metabolism and during the interaction between grapevines and (micro) organisms. In this context, the interaction of grapevines both above and below ground with several organisms, such as arthropods (spiders, mites, insects) and microorganisms (fungi, oomycetes, bacteria, viruses), nematodes, birds, and mammals, as well as other plants is pivotal in understanding the complex productive system of a vineyard.
The grapevine is a perennial plant of extremely high economic importance worldwide. In vineyards, it lives in association with many microorganisms. Grapevines, together with several fruit crops, are highly valuable but most of the premium cultivars used for winemaking, especially the extensively used European Vitis vinifera cultivars, are highly susceptible to several pathogenic microorganisms including fungi, bacteria, phytoplasma, and viruses. In fact, grapevines interact both above and below ground with arthropods (e.g., insects) microorganisms (e.g., fungi, bacteria, viruses), nematodes, animals (e.g., birds and mammals), and other plants. Many interactions with different organisms are beneficial, providing ecological “services” to the vineyards. The classical example is related to earthworms, which improves soil fertility by transforming organic matter into humus, creating soil porosity where the roots can penetrate and actively up- take nutrients in symbiosis with soil microorganisms. Bacteria, fungi, and viruses are organized in communities and everyone can be beneficial, neutral, or harmful to the grapevines. In this context, microorganisms interact with each other and impact several plant functions. Therefore, the grapevine associated with its microbial communities constitutes a supra-organism, called holobiont, and the mechanism of functioning is related to plant–microorganism interactions. In this sense, even the overall health of the grapevine is influenced by the diversity and structure of the microbial communities. The microbiome is a pivotal component of the grapevine, and it is also extremely responsive to biotic and abiotic changes [82]. The close interaction of the grapevines with many microorganisms modulates vine physiology throughout the entire growing season and during all the phenological stages, impacting grapevine holobiont health and above all the fruit chemical makeup at harvest, which consequently affects wine quality. The omics sciences have delivered a new era in plant biology, starting with the precise description of the taxonomic composition of the microbial communities of the grapevines in each single organ and during their annual growth and development. Following those discoveries, scientists are approaching the complex understanding of grapevine microorganisms and the specific functional contributions of microbes to the grapevine holobiont. To date, few studies have addressed the functional characteristics of the microbiota through metabolomic and transcriptomic approaches in the grapevine holobiont. However, the understanding of the complexity of the grapevine holobiont is a pivotal issue for the future of the wine industry worldwide. The potential understanding and consequent manipulation of the microbiota in a grapevine holobiont can lead to better vineyard management focused on urgent reduction of pesticide and chemical fertilizer inputs through biocontrol, which are key factors for the development of the sustainable viticulture of the future.
5.1. Role of Cytochrome P450 in Vitis vinifera
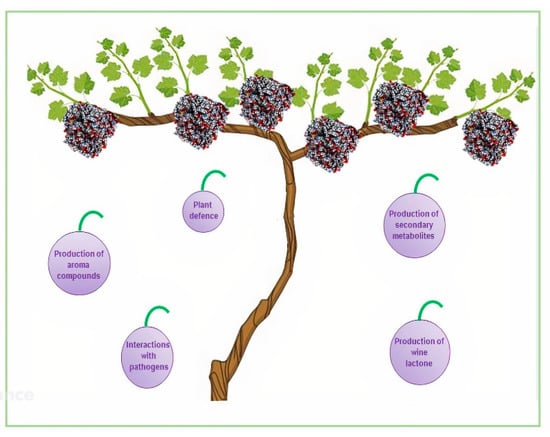
Vitis vinifera has 579 cytochrome P450 sequences belonging to 48 families, a number that is similar to their number in A. thaliana (242), S. lycopersicum (272), and Oryza sativa (309) [83]. Most of the CYP sequences in the grapevine genome are organized in clusters originating from tandem or segmental duplications. Some grapewine P450 genes are induced upon biotic stress, while others are specifically activated during grape berry ripening and might have a role in the production of specific volatiles in berries such as aroma compounds (Figure 5). CYP71, CYP72, CYP75, CYP76, CYP81, CYP82, and CYP89 genes are enlarged in the grapevine genome and are involved in secondary metabolism and are differentially expressed during four different stages of berry development [83,84,85].
The aroma of wine consists of a hundred different volatile compounds at concentrations spanning several orders of magnitude [86]. A characteristic wine aroma depends on trace components with very strong odors [86]. Among monoterpenes, which are important constituents of the aromas in wines, wine lactone is a bicyclic monoterpene lactone that has the most potent odor [87]. (E)-8-carboxylinalool is present both in grapes and wines as a glucose ester with a sugar moiety attached to the carboxyl functional group [88]. CYP76 is highly expressed in maturing V. vinifera berries and oxidizes linalool to (E)-8-carboxylinalool, which acts as a precursor to wine lactone with a nonenzymatic step during wine maturation and aging [83] (Figure 4).
5.2. Cytochromes P450 in Vitis vinifera Interaction with Microorganisms
V. vinifera is highly susceptible to many pests and pathogens, causing great economic loss each year. Currently, we are at the beginning of discovering the involvement of cytochrome P450 enzymes in the interaction between grapevine and its pathogens. Some of the most dangerous pathogens include the bacterium Xylella fastidiosa, the phytopathogenic bacterium Candidatus, phytoplasma solani, and the phytoplasma Flavescence dorée. The overexpression of plant gene coding for cytochrome P450 enzymes after the invasion of the pathogen has been proved for the three microorganisms mentioned above. In detail, CYP736 was involved in the host response to X. fastidiosa infection [89]; moreover, the gene coding for a cytochrome P450 enzyme belonging to CYP2C family was found to be overexpressed after the infection of Candidatus phytoplasma solani [90], and the invasion of Flavescence dorée induced the over-expression of CYP86 [91] (Figure 4).
6. Future Directions and Concluding Remarks
The plant microbiome is a key determinant of plant health and productivity, as well as all tissues of a plant host’s microbial community [92]. The manipulation of the plant microbiome has the potential to reduce the incidence of plant disease [93], increase agricultural production [94], reduce chemical inputs [95], and reduce emissions of greenhouse gasses [96], resulting in more sustainable agriculture. Due to the importance of microbiomes and the cypome complement of both plants and microbes, it is important to decipher the cypome composition of the plant microbiomes and its contribution to the metabolism of the plant as well as to understand the influence of the plant on the expression of the cypome of its microbiome. In recent years, our understanding of the molecular aspects of the grapevine microbiome has greatly increased. However, a lot of research remains to be carried out to precisely decipher and finely characterize the different aspects of microorganisms’detection by the grapevines and the subsequent activation and establishment of physiological plant reactions. This applies, for example, to the precise characterization of the role of P450 in comparative studies of the genetic diversity of resistance genes and other defense-related genes in various cultivated and wild grapevines. The enormous impact of the obtained knowledge from grapevine interaction with the microbiome has already contributed, as evidenced from its integration in breeding experiments, either through genetic transformation or through marker-assisted selection, and this research approach will surely become more common in the future.
Author Contributions
D.M. Conceptualization, writing-original draft preparation, review and editing; S.S. Review; P.S. Review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Garfinkel, D. Studies on Pig Liver Microsomes. I. Enzymic and Pigment Composition of Different Microsomal Fractions. Arch. Biochem. Biophys. 1958, 77, 493–509. [Google Scholar] [CrossRef]
- Irmler, S.; Schröder, G.; St-Pierre, B.; Crouch, N.P.; Hotze, M.; Schmidt, J.; Strack, D.; Matern, U.; Schröder, J. Indole Alkaloid Biosynthesis in Catharanthus Roseus: New Enzyme Activities and Identification of Cytochrome P450 CYP72A1 as Secologanin Synthase. Plant J. 2000, 24, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Bak, S.; Kahn, R.A.; Olsen, C.E.; Halkier, B.A. Cloning and Expression in Escherichia Coli of the Obtusifoliol 14α-demethylase of Sorghum bicolor (L.) Moench, a Cytochrome P450 Orthologous to the Sterol 14α-demethylases (CYP51) from Fungi and Mammals. Plant J. 1997, 11, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, T.; Mizutani, M.; Aoki, N.; Watanabe, B.; Saga, H.; Saito, S.; Oikawa, A.; Suzuki, H.; Sakurai, N.; Shibata, D. Cytochrome P450 CYP710A Encodes the Sterol C-22 Desaturase in Arabidopsis and Tomato. Plant Cell 2006, 18, 1008–1022. [Google Scholar] [CrossRef]
- Hansen, C.C.; Sørensen, M.; Veiga, T.A.M.; Zibrandtsen, J.F.S.; Heskes, A.M.; Olsen, C.E.; Boughton, B.A.; Møller, B.L.; Neilson, E.H.J. Reconfigured Cyanogenic Glucoside Biosynthesis in Eucalyptus Cladocalyx Involves a Cytochrome P450 CYP706C55. Plant Physiol. 2018, 178, 1081–1095. [Google Scholar] [CrossRef]
- Nasomjai, P.; Reed, D.W.; Tozer, D.J.; Peach, M.J.G.; Slawin, A.M.Z.; Covello, P.S.; O’Hagan, D. Mechanistic Insights into the Cytochrome P450-mediated Oxidation and Rearrangement of Littorine in Tropane Alkaloid Biosynthesis. ChemBioChem 2009, 10, 2382–2393. [Google Scholar] [CrossRef] [PubMed]
- Koch, B.M.; Sibbesen, O.; Halkier, B.A.; Svendsen, I.; Møller, B.L. The Primary Sequence of Cytochrome P450tyr, the MultifunctionalN-Hydroxylase Catalyzing the Conversion OfL-Tyrosine Top-Hydroxyphenylacetaldehyde Oxime in the Biosynthesis of the Cyanogenic Glucoside Dhurrin in Sorghum bicolor (L.) Moench. Arch. Biochem. Biophys. 1995, 323, 177–186. [Google Scholar] [CrossRef]
- Klingenberg, M. Pigments of Rat Liver Microsomes. Arch. Biochem. Biophys. 1958, 75, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Omura, T.; Sato, R. The Carbon Monoxide-Binding Pigment of Liver Microsomes: II. Solubilization, Purification, and Properties. J. Biol. Chem. 1964, 239, 2379–2385. [Google Scholar] [CrossRef]
- Lamb, D.C.; Lei, L.; Warrilow, A.G.S.; Lepesheva, G.I.; Mullins, J.G.L.; Waterman, M.R.; Kelly, S.L. The First Virally Encoded Cytochrome P450. J. Virol. 2009, 83, 8266–8269. [Google Scholar] [CrossRef]
- Paquette, S.M.; Bak, S.; Feyereisen, R. Intron–Exon Organization and Phylogeny in a Large Superfamily, the Paralogous Cytochrome P450 Genes of Arabidopsis Thaliana. DNA Cell Biol. 2000, 19, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Durst, F.; Nelson, D.R. Diversity and Evolution of Plant P450 and P450-Reductases. Drug Metab. Drug Interact. 1995, 12, 189–206. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, K.A. Cytochrome P450s as Genes for Crop Improvement. Curr. Opin. Plant Biol. 2001, 4, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R. The Cytochrome P450 Homepage. Hum. Genom. 2009, 4, 59. [Google Scholar] [CrossRef]
- Pikuleva, I.A.; Waterman, M.R. Cytochromes P450: Roles in Diseases. J. Biol. Chem. 2013, 288, 17091–17098. [Google Scholar] [CrossRef] [PubMed]
- Hannemann, F.; Bichet, A.; Ewen, K.M.; Bernhardt, R. Cytochrome P450 Systems—Biological Variations of Electron Transport Chains. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2007, 1770, 330–344. [Google Scholar] [CrossRef]
- Guengerich, F.P. Mechanisms of Cytochrome P450-Catalyzed Oxidations. ACS Catal. 2018, 8, 10964–10976. [Google Scholar] [CrossRef] [PubMed]
- Morant, M.; Bak, S.; Møller, B.L.; Werck-Reichhart, D. Plant Cytochromes P450: Tools for Pharmacology, Plant Protection and Phytoremediation. Curr. Opin. Biotechnol. 2003, 14, 151–162. [Google Scholar] [CrossRef]
- Laffaru Singpho, N.; Sharma, J.G. Importance of Cytochrome P450 Gene Family from Metabolite Biosynthesis to Stress Tolerance: A Review. In Proceedings of the IOP Conference Series: Earth and Environmental Science, Jakarta, Indonesia, 25–26 September 2021; Volume 775, p. 012012. [Google Scholar]
- Nelson, D.; Werck-Reichhart, D. A P450-centric View of Plant Evolution. Plant J. 2011, 66, 194–211. [Google Scholar] [CrossRef]
- Jain, K.S.; Khedkar, V.M.; Arya, N.; Rane, P.V.; Chaskar, P.K.; Coutinho, E.C. Design, Synthesis & Evaluation of Condensed 2H-4-Arylaminopyrimidines as Novel Antifungal Agents. Eur. J. Med. Chem. 2014, 77, 166–175. [Google Scholar]
- Geisler, K.; Hughes, R.K.; Sainsbury, F.; Lomonossoff, G.P.; Rejzek, M.; Fairhurst, S.; Olsen, C.-E.; Motawia, M.S.; Melton, R.E.; Hemmings, A.M. Biochemical Analysis of a Multifunctional Cytochrome P450 (CYP51) Enzyme Required for Synthesis of Antimicrobial Triterpenes in Plants. Proc. Natl. Acad. Sci. USA 2013, 110, E3360–E3367. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, X.T.; Xu, Y.Q.; Zhong, Y.; Li, Y.X.; Chen, J.K.; Li, X.; Nan, P. Global transcriptome analysis profiles metabolic pathways in traditional herb Astragalus membranaceus Bge. var. mongolicus (Bge.) Hsiao. BMC Genom. 2015, 16, S15. [Google Scholar]
- Williams, D.; De Luca, V. Plant cytochrome P450s directing monoterpene indole alkaloid (MIA) and benzylisoquinoline alkaloid (BIA) biosynthesis. Phytochem. Rev. 2022, 1–30. [Google Scholar] [CrossRef]
- Magome, H.; Nomura, T.; Hanada, A.; Takeda-Kamiya, N.; Ohnishi, T.; Shinma, Y.; Katsumata, T.; Kawaide, H.; Kamiya, Y.; Yamaguchi, S. CYP714B1 and CYP714B2 Encode Gibberellin 13-Oxidases That Reduce Gibberellin Activity in Rice. Proc. Natl. Acad. Sci. USA 2013, 110, 1947–1952. [Google Scholar] [CrossRef]
- Hoffmann, I.; Jernerén, F.; Oliw, E.H. Expression of Fusion Proteins of Aspergillus Terreus Reveals a Novel Allene Oxide Synthase. J. Biol. Chem. 2013, 288, 11459–11469. [Google Scholar] [CrossRef]
- Yoeun, S.; Rakwal, R.; Han, O. Dual Positional Substrate Specificity of Rice Allene Oxide Synthase-1: Insight into Mechanism of Inhibition by Type II Ligand Imidazole. BMB Rep. 2013, 46, 151. [Google Scholar] [CrossRef]
- Ohnishi, T.; Watanabe, B.; Sakata, K.; Mizutani, M. CYP724B2 and CYP90B3 Function in the Early C-22 Hydroxylation Steps of Brassinosteroid Biosynthetic Pathway in Tomato. Biosci. Biotechnol. Biochem. 2006, 70, 2071–2080. [Google Scholar] [CrossRef]
- Delventhal, R.; Falter, C.; Strugala, R.; Zellerhoff, N.; Schaffrath, U. Ectoparasitic Growth of Magnaporthe on Barley Triggers Expression of the Putative Barley Wax Biosynthesis Gene CYP96B22 Which Is Involved in Penetration Resistance. BMC Plant Biol. 2014, 14, 26. [Google Scholar] [CrossRef]
- Cui, H.; Yu, X.; Wang, Y.; Cui, Y.; Li, X.; Liu, Z.; Qin, S. Evolutionary Origins, Molecular Cloning and Expression of Carotenoid Hydroxylases in Eukaryotic Photosynthetic Algae. BMC Genom. 2013, 14, 457. [Google Scholar] [CrossRef]
- Vasav, A.P.; Barvkar, V.T. Phylogenomic Analysis of Cytochrome P450 Multigene Family and Their Differential Expression Analysis in Solanum Lycopersicum L. Suggested Tissue Specific Promoters. BMC Genom. 2019, 20, 116. [Google Scholar] [CrossRef]
- Kim, J.-E.; Cheng, K.M.; Craft, N.E.; Hamberger, B.; Douglas, C.J. Over-Expression of Arabidopsis Thaliana Carotenoid Hydroxylases Individually and in Combination with a β-Carotene Ketolase Provides Insight into in Vivo Functions. Phytochemistry 2010, 71, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Mogi, S.; Kaneko, T.; Kojima, H.; Katoh, S.; Sano, A.; Kojima, S. Relationship between Tissue Hydroxyl Radical and Oxidatively Modified Macromolecule Levels. Geriatr. Gerontol. Int. 2014, 14, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Seebeck, T.; Schrenker, D.; Yu, O. CYP709B3, a Cytochrome P450 Monooxygenase Gene Involved in Salt Tolerance in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 169. [Google Scholar] [CrossRef]
- Iquebal, M.A.; Soren, K.R.; Gangwar, P.; Shanmugavadivel, P.S.; Aravind, K.; Singla, D.; Jaiswal, S.; Jasrotia, R.S.; Chaturvedi, S.K.; Singh, N.P. Discovery of Putative Herbicide Resistance Genes and Its Regulatory Network in Chickpea Using Transcriptome Sequencing. Front. Plant Sci. 2017, 8, 958. [Google Scholar] [CrossRef] [PubMed]
- Dimaano, N.G.; Iwakami, S. Cytochrome P450-mediated Herbicide Metabolism in Plants: Current Understanding and Prospects. Pest Manag. Sci. 2021, 77, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Grausem, B.; Widemann, E.; Verdier, G.; Nosbüsch, D.; Aubert, Y.; Beisson, F.; Schreiber, L.; Franke, R.; Pinot, F. CYP 77 A 19 and CYP 77 A 20 Characterized from Solanum Tuberosum Oxidize Fatty Acids in Vitro and Partially Restore the Wild Phenotype in an A Rabidopsis Thaliana Cutin Mutant. Plant Cell Environ. 2014, 37, 2102–2115. [Google Scholar] [CrossRef]
- Fiore, A.; Dall’Osto, L.; Cazzaniga, S.; Diretto, G.; Giuliano, G.; Bassi, R. A Quadruple Mutant of Arabidopsis Reveals a β-Carotene Hydroxylation Activity for LUT1/CYP97C1 and a Regulatory Role of Xanthophylls on Determination of the PSI/PSII Ratio. BMC Plant Biol. 2012, 12, 50. [Google Scholar] [CrossRef]
- Chakrabarti, M.; Zhang, N.A.; Sauvage, C.; Muños, S.; Blanca, J.; Cañizares, J.; Diez, M.J.; Schneider, R.; Mazourek, M.; McClead, J. A Cytochrome P450 Regulates a Domestication Trait in Cultivated Tomato. Proc. Natl. Acad. Sci. USA 2013, 110, 17125–17130. [Google Scholar] [CrossRef]
- Umemoto, N.; Nakayasu, M.; Ohyama, K.; Yotsu-Yamashita, M.; Mizutani, M.; Seki, H.; Saito, K.; Muranaka, T. Two Cytochrome P450 Monooxygenases Catalyze Early Hydroxylation Steps in the Potato Steroid Glycoalkaloid Biosynthetic Pathway. Plant Physiol. 2016, 171, 2458–2467. [Google Scholar] [CrossRef]
- Yang, X.; Wu, D.I.; Shi, J.; He, Y.I.; Pinot, F.; Grausem, B.; Yin, C.; Zhu, L.; Chen, M.; Luo, Z. Rice CYP703A3, a Cytochrome P450 Hydroxylase, Is Essential for Development of Anther Cuticle and Pollen Exine. J. Integr. Plant Biol. 2014, 56, 979–994. [Google Scholar] [CrossRef]
- Shang, Y.; Ma, Y.; Zhou, Y.; Zhang, H.; Duan, L.; Chen, H.; Zeng, J.; Zhou, Q.; Wang, S.; Gu, W. Biosynthesis, Regulation, and Domestication of Bitterness in Cucumber. Science 2014, 346, 1084–1088. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, Y.; Zeng, J.; Duan, L.; Xue, X.; Wang, H.; Lin, T.; Liu, Z.; Zeng, K.; Zhong, Y. Convergence and Divergence of Bitterness Biosynthesis and Regulation in Cucurbitaceae. Nat. Plants 2016, 2, 16183. [Google Scholar] [CrossRef] [PubMed]
- Moses, T.; Pollier, J.; Faizal, A.; Apers, S.; Pieters, L.; Thevelein, J.M.; Geelen, D.; Goossens, A. Unraveling the Triterpenoid Saponin Biosynthesis of the African Shrub Maesa Lanceolata. Mol. Plant 2015, 8, 122–135. [Google Scholar] [CrossRef]
- Rodríguez-López, C.E.; Hong, B.; Paetz, C.; Nakamura, Y.; Koudounas, K.; Passeri, V.; Baldoni, L.; Alagna, F.; Calderini, O.; O’Connor, S.E. Two Bi-functional Cytochrome P450 CYP72 Enzymes from Olive (Olea europaea) Catalyze the Oxidative C-C Bond Cleavage in the Biosynthesis of Secoxy-iridoids–Flavor and Quality Determinants in Olive Oil. New Phytol. 2021, 229, 2288–2301. [Google Scholar] [CrossRef] [PubMed]
- Andre, C.M.; Legay, S.; Deleruelle, A.; Nieuwenhuizen, N.; Punter, M.; Brendolise, C.; Cooney, J.M.; Lateur, M.; Hausman, J.; Larondelle, Y. Multifunctional Oxidosqualene Cyclases and Cytochrome P450 Involved in the Biosynthesis of Apple Fruit Triterpenic Acids. New Phytol. 2016, 211, 1279–1294. [Google Scholar] [CrossRef]
- Takase, S.; Kera, K.; Nagashima, Y.; Mannen, K.; Hosouchi, T.; Shinpo, S.; Kawashima, M.; Kotake, Y.; Yamada, H.; Saga, Y. Allylic Hydroxylation of Triterpenoids by a Plant Cytochrome P450 Triggers Key Chemical Transformations That Produce a Variety of Bitter Compounds. J. Biol. Chem. 2019, 294, 18662–18673. [Google Scholar] [CrossRef]
- Zhang, J.; Dai, L.; Yang, J.; Liu, C.; Men, Y.; Zeng, Y.; Cai, Y.; Zhu, Y.; Sun, Y. Oxidation of Cucurbitadienol Catalyzed by CYP87D18 in the Biosynthesis of Mogrosides from Siraitia Grosvenorii. Plant Cell Physiol. 2016, 57, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Ikezawa, N.; Tanaka, M.; Nagayoshi, M.; Shinkyo, R.; Sakaki, T.; Inouye, K.; Sato, F. Molecular Cloning and Characterization of CYP719, a Methylenedioxy Bridge-Forming Enzyme That Belongs to a Novel P450 Family, from Cultured Coptis Japonica Cells. J. Biol. Chem. 2003, 278, 38557–38565. [Google Scholar] [CrossRef]
- Stumpe, M.; Kandzia, R.; Göbel, C.; Rosahl, S.; Feussner, I. A Pathogen-Inducible Divinyl Ether Synthase (CYP74D) from Elicitor-Treated Potato Suspension Cells. FEBS Lett. 2001, 507, 371–376. [Google Scholar] [CrossRef]
- Godiard, L.; Sauviac, L.; Dalbin, N.; Liaubet, L.; Callard, D.; Czernic, P.; Marco, Y. CYP76C2, an Arabidopsis Thaliana Cytochrome P450 Gene Expressed during Hypersensitive and Developmental Cell Death. FEBS Lett. 1998, 438, 245–249. [Google Scholar] [CrossRef]
- Balint-Kurti, P. The Plant Hypersensitive Response: Concepts, Control and Consequences. Mol. Plant Pathol. 2019, 20, 1163–1178. [Google Scholar] [CrossRef]
- Höfer, R.; Boachon, B.; Renault, H.; Gavira, C.; Miesch, L.; Iglesias, J.; Ginglinger, J.-F.; Allouche, L.; Miesch, M.; Grec, S. Dual Function of the Cytochrome P450 CYP76 Family from Arabidopsis Thaliana in the Metabolism of Monoterpenols and Phenylurea Herbicides. Plant Physiol. 2014, 166, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.-J.; Ko, M.K.; Kim, Y.S.; Kim, K.S.; Kostenyuk, I.; Kee, H.K. A Cytochrome P450 Gene Is Differentially Expressed in Compatible and Incompatible Interactions between Pepper (Capsicum annuum) and the Anthracnose Fungus, Colletotrichum Gloeosporioides. Mol. Plant-Microbe Interact. 1999, 12, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-C.; Kim, S.-Y.; Paek, K.-H.; Choi, D.; Park, J.M. Suppression of CaCYP1, a Novel Cytochrome P450 Gene, Compromises the Basal Pathogen Defense Response of Pepper Plants. Biochem. Biophys. Res. Commun. 2006, 345, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A. Isoflavonoids: Biochemistry, Molecular Biology and Biological Functions. Compr. Nat. Prod. Chem. 1999, 1, 773–823. [Google Scholar]
- Dixon, R.A.; Sumner, L.W. Legume Natural Products: Understanding and Manipulating Complex Pathways for Human and Animal Health. Plant Physiol. 2003, 131, 878–885. [Google Scholar] [CrossRef]
- Liu, C.; Huhman, D.; Sumner, L.W.; Dixon, R.A. Regiospecific Hydroxylation of Isoflavones by Cytochrome P450 81E Enzymes from Medicago Truncatula. Plant J. 2003, 36, 471–484. [Google Scholar] [CrossRef]
- Akashi, T.; Aoki, T.; Ayabe, S. CYP81E1, a Cytochrome P450 CDNA of Licorice (Glycyrrhiza echinata L.), Encodes Isoflavone 2′-Hydroxylase. Biochem. Biophys. Res. Commun. 1998, 251, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Overkamp, S.; Hein, F.; Barz, W. Cloning and Characterization of Eight Cytochrome P450 CDNAs from Chickpea (Cicer arietinum L.) Cell Suspension Cultures. Plant Sci. 2000, 155, 101–108. [Google Scholar] [CrossRef]
- Kelly, D.E.; Kraševec, N.; Mullins, J.; Nelson, D.R. The CYPome (Cytochrome P450 Complement) of Aspergillus Nidulans. Fungal Genet. Biol. 2009, 46, S53–S61. [Google Scholar] [CrossRef]
- Becher, R.; Wirsel, S.G.R. Fungal Cytochrome P450 Sterol 14α-Demethylase (CYP51) and Azole Resistance in Plant and Human Pathogens. Appl. Microbiol. Biotechnol. 2012, 95, 825–840. [Google Scholar] [CrossRef]
- Soanes, D.M.; Alam, I.; Cornell, M.; Wong, H.M.; Hedeler, C.; Paton, N.W.; Rattray, M.; Hubbard, S.J.; Oliver, S.G.; Talbot, N.J. Comparative Genome Analysis of Filamentous Fungi Reveals Gene Family Expansions Associated with Fungal Pathogenesis. PLoS ONE 2008, 3, e2300. [Google Scholar] [CrossRef]
- Leal, G.A.; Gomes, L.H.; Albuquerque, P.S.B.; Tavares, F.C.A.; Figueira, A. Searching for Moniliophthora Perniciosa Pathogenicity Genes. Fungal Biol. 2010, 114, 842–854. [Google Scholar] [CrossRef]
- George, H.L.; VanEtten, H.D. Characterization of Pisatin-Inducible Cytochrome P450s in Fungal Pathogens of Pea That Detoxify the Pea Phytoalexin Pisatin. Fungal Genet. Biol. 2001, 33, 37–48. [Google Scholar] [CrossRef]
- Luo, P.; Wang, Y.; Wang, G.; Essenberg, M.; Chen, X. Molecular Cloning and Functional Identification of (+)-δ-cadinene-8-hydroxylase, a Cytochrome P450 Mono-oxygenase (CYP706B1) of Cotton Sesquiterpene Biosynthesis. Plant J. 2001, 28, 95–104. [Google Scholar] [CrossRef]
- Armstrong, G.M.; Armstrong, J.K. Formae Speciales and Races of Fusarium Oxysporum Causing Wilt Diseases. In Fusarium: Disease, Biology, and Taxonomy; Nelson, P.E., Toussoun, T.A., Cook., R.J., Eds.; Pennsylvania State University, University Park: State College, PA, USA, 1981; pp. 391–399. [Google Scholar]
- Lemanceau, P.; Bakker, P.A.H.M.; de Kogel, W.J.; Alabouvette, C.; Schippers, B. Antagonistic Effect of Nonpathogenic Fusarium Oxysporum Fo47 and Pseudobactin 358 upon Pathogenic Fusarium Oxysporum f. Sp. Dianthi. Appl. Env. Microbiol. 1993, 59, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, N.; Takemae, A.; Shoun, H. Cytochrome P450foxy, a Catalytically Self-Sufficient Fatty Acid Hydroxylase of the Fungus Fusarium Oxysporum. J. Biochem. 1996, 119, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Kitazume, T.; Tanaka, A.; Takaya, N.; Nakamura, A.; Matsuyama, S.; Suzuki, T.; Shoun, H. Kinetic Analysis of Hydroxylation of Saturated Fatty Acids by Recombinant P450foxy Produced by an Escherichia Coli Expression System. Eur. J. Biochem. 2002, 269, 2075–2082. [Google Scholar] [CrossRef]
- Kachroo, A.; Kachroo, P. Fatty Acid-Derived Signals in Plant Defense. Annu. Rev. Phytopathol. 2009, 47, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Fauth, M.; Schweizer, P.; Buchala, A.; Markstädter, C.; Riederer, M.; Kato, T.; Kauss, H. Cutin Monomers and Surface Wax Constituents Elicit H2O2 in Conditioned Cucumber Hypocotyl Segments and Enhance the Activity of Other H2O2Elicitors. Plant Physiol. 1998, 117, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Minerdi, D.; Sadeghi, S.J.; Pautasso, L.; Morra, S.; Aigotti, R.; Medana, C.; Gilardi, G.; Gullino, M.L.; Gilardi, G. Expression and Role of CYP505A1 in Pathogenicity of Fusarium Oxysporum f. Sp. Lactucae. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2020, 1868, 140268. [Google Scholar] [CrossRef] [PubMed]
- Takaoka, S.; Kurata, M.; Harimoto, Y.; Hatta, R.; Yamamoto, M.; Akimitsu, K.; Tsuge, T. Complex Regulation of Secondary Metabolism Controlling Pathogenicity in the Phytopathogenic Fungus Alternaria Alternata. New Phytol. 2014, 202, 1297–1309. [Google Scholar] [CrossRef]
- Karlsson, B.; Tsopelas, P.; Zamponi, L.; Capretti, P.; Soulioti, N.; Swedjemark, G. Susceptibility to Heterobasidion Parviporum in Picea Abies Clones Grown in Different Environments. For. Pathol. 2008, 38, 83–89. [Google Scholar] [CrossRef]
- Siewers, V.; Viaud, M.; Jimenez-Teja, D.; Collado, I.G.; Gronover, C.S.; Pradier, J.-M.; Tudzynsk, B.; Tudzynski, P. Functional Analysis of the Cytochrome P450 Monooxygenase Gene Bcbot1 of Botrytis Cinerea Indicates That Botrydial Is a Strain-Specific Virulence Factor. Mol. Plant-Microbe Interact. 2005, 18, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Myburg, A.A.; van den Berg, N.; Viljoen, A. Pathogenicity Associated Genes in Fusarium oxysporum f. sp. cubense Race 4. S. Afr. J. Sci. 2013, 109, 5. [Google Scholar]
- López-Berges, M.S.; Rispail, N.; Prados-Rosales, R.C.; di Pietro, A. A Nitrogen Response Pathway Regulates Virulence Functions in Fusarium Oxysporum via the Protein Kinase TOR and the BZIP Protein MeaB. Plant Cell 2010, 22, 2459–2475. [Google Scholar] [CrossRef]
- Zhang, X.-W.; Jia, L.-J.; Zhang, Y.; Jiang, G.; Li, X.; Zhang, D.; Tang, W.-H. In Planta Stage-Specific Fungal Gene Profiling Elucidates the Molecular Strategies of Fusarium Graminearum Growing inside Wheat Coleoptiles. Plant Cell 2012, 24, 5159–5176. [Google Scholar] [CrossRef]
- Schubler, A. The Glomeromycota: A Species List with New Families and New Genera. 2010. Available online: http://www.amf-phylogeny.com (accessed on 15 November 2022).
- Handa, Y.; Nishide, H.; Takeda, N.; Suzuki, Y.; Kawaguchi, M.; Saito, K. RNA-Seq Transcriptional Profiling of an Arbuscular Mycorrhiza Provides Insights into Regulated and Coordinated Gene Expression in Lotus Japonicus and Rhizophagus Irregularis. Plant Cell Physiol. 2015, 56, 1490–1511. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial Biogeography of Wine Grapes Is Conditioned by Cultivar, Vintage, and Climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef]
- Ilc, T.; Arista, G.; Tavares, R.; Navrot, N.; Duchene, E.; Velt, A.; Choulet, F.; Paux, E.; Fischer, M.; Nelson, D.R. Annotation, Classification, Genomic Organization and Expression of the Vitis Vinifera CYPome. PLoS ONE 2018, 13, e0199902. [Google Scholar] [CrossRef]
- Martin, D.M.; Aubourg, S.; Schouwey, M.B.; Daviet, L.; Schalk, M.; Toub, O.; Lund, S.T.; Bohlmann, J. Functional Annotation, Genome Organization and Phylogeny of the Grapevine (Vitis vinifera) Terpene Synthase Gene Family Based on Genome Assembly, FLcDNA Cloning, and Enzyme Assays. BMC Plant Biol. 2010, 10, 226. [Google Scholar] [CrossRef] [PubMed]
- Parage, C.; Tavares, R.; Réty, S.; Baltenweck-Guyot, R.; Poutaraud, A.; Renault, L.; Heintz, D.; Lugan, R.; Marais, G.A.B.; Aubourg, S. Structural, Functional, and Evolutionary Analysis of the Unusually Large Stilbene Synthase Gene Family in Grapevine. Plant Physiol. 2012, 160, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Francis, I.L.; Newton, J.L. Determining Wine Aroma from Compositional Data. Aust. J. Grape Wine Res. 2005, 11, 114–126. [Google Scholar] [CrossRef]
- Guth, H. Determination of the Configuration of Wine Lactone. Helv. Chim. Acta 1996, 79, 1559–1571. [Google Scholar] [CrossRef]
- D’Ambrosio, M.; Harghel, P.; Guantieri, V. Isolation of Intact Glycosidic Aroma Precursors from Grape Juice by Hydrophilic Interaction Liquid Chromatography. Aust. J. Grape Wine Res. 2013, 19, 189–192. [Google Scholar] [CrossRef]
- Cheng, D.W.; Lin, H.; Takahashi, Y.; Walker, M.A.; Civerolo, E.L.; Stenger, D.C. Transcriptional Regulation of the Grape Cytochrome P450 Monooxygenase Gene CYP736B Expression in Response to Xylella Fastidiosa Infection. BMC Plant Biol. 2010, 10, 135. [Google Scholar] [CrossRef]
- Škrlj, B.; Novak, M.P.; Brader, G.; Anžič, B.; Ramšak, Ž.; Gruden, K.; Kralj, J.; Kladnik, A.; Lavrač, N.; Roitsch, T. New Cross-Talks between Pathways Involved in Grapevine Infection with ‘Candidatus Phytoplasma Solani’Revealed by Temporal Network Modelling. Plants 2021, 10, 646. [Google Scholar] [CrossRef]
- Bertazzon, N.; Bagnaresi, P.; Forte, V.; Mazzucotelli, E.; Filippin, L.; Guerra, D.; Zechini, A.; Cattivelli, L.; Angelini, E. Grapevine Comparative Early Transcriptomic Profiling Suggests That Flavescence Dorée Phytoplasma Represses Plant Responses Induced by Vector Feeding in Susceptible Varieties. BMC Genom. 2019, 20, 526. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The Rhizosphere Microbiome and Plant Health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Bloemberg, G.V.; Lugtenberg, B.J.J. Molecular Basis of Plant Growth Promotion and Biocontrol by Rhizobacteria. Curr. Opin. Plant Biol. 2001, 4, 343–350. [Google Scholar] [CrossRef]
- Bakker, M.G.; Manter, D.K.; Sheflin, A.M.; Weir, T.L.; Vivanco, J.M. Harnessing the Rhizosphere Microbiome through Plant Breeding and Agricultural Management. Plant Soil 2012, 360, 1–13. [Google Scholar] [CrossRef]
- Adesemoye, A.O.; Torbert, H.A.; Kloepper, J.W. Plant Growth-Promoting Rhizobacteria Allow Reduced Application Rates of Chemical Fertilizers. Microb. Ecol. 2009, 58, 921–929. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, M.; Verma, S.; Choudhary, P.; Chakdar, H. Plant Microbiome: Trends and Prospects for Sustainable Agriculture. In Plant Microbe Symbiosis; Springer: Berlin/Heidelberg, Germany, 2020; pp. 129–151. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).