Peroxisomes Are Highly Abundant and Heterogeneous in Human Parotid Glands
Abstract
:1. Introduction
2. Results
2.1. Peroxisomes Are Highly Abundant in the Human Parotid Gland
2.2. Peroxisomal β-Oxidation Enzymes Are Expressed at High Levels in the Human Parotid Gland
2.3. Plasmalogen Synthesizing Enzymes Are Expressed at Significantly High Levels in Human Parotid Glands
2.4. Cholesterol Synthesizing Enzymes Were Expressed Significantly Higher in the Human Parotid Glands
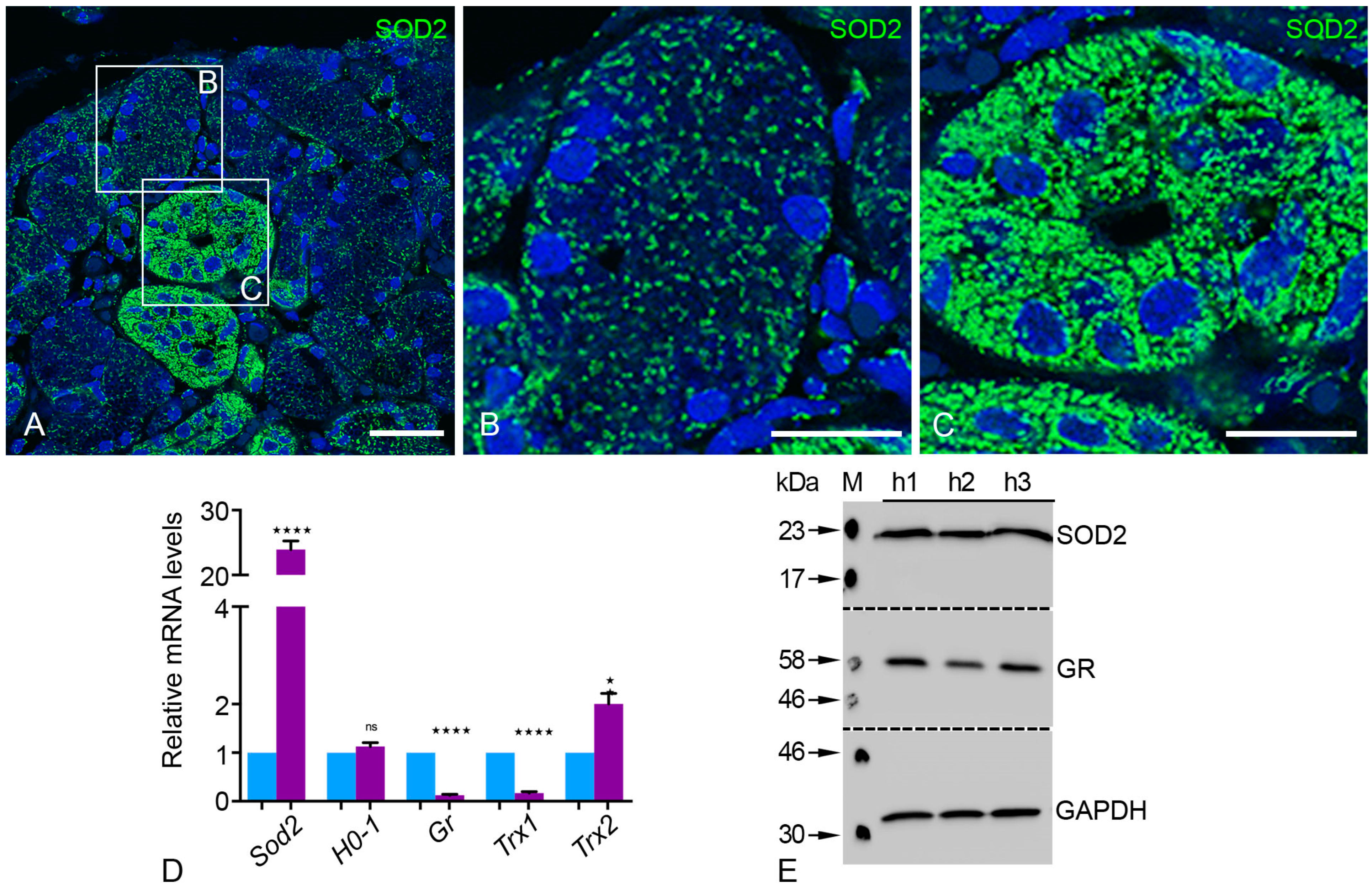
2.5. Peroxisomal Antioxidative Enzymes Were Detected in the Human Parotid Gland
2.6. Antioxidative Enzymes of Different Cell Compartments Were Also Abundant in Human Parotid Glands
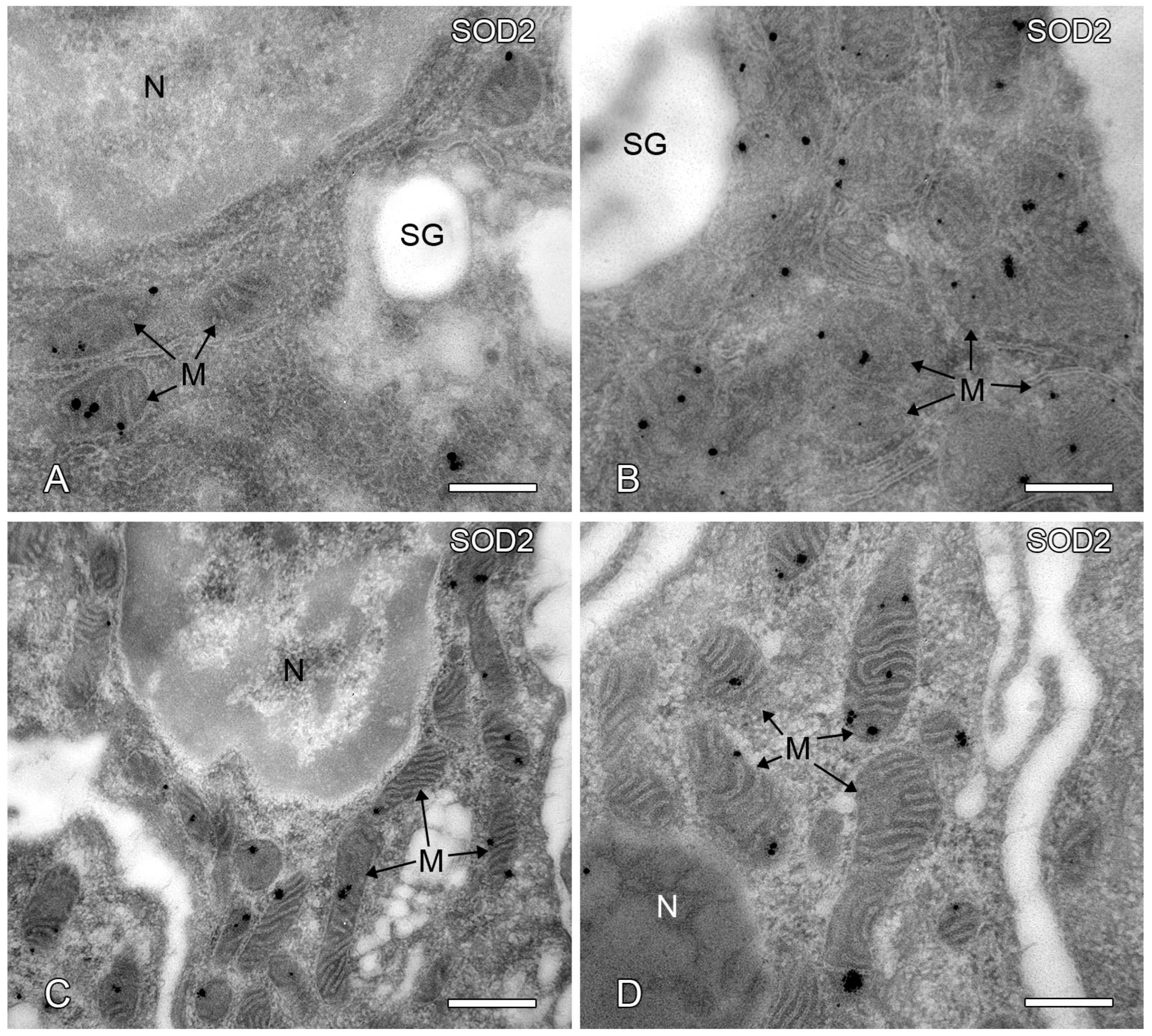
2.7. Post-Embedding Immunoelectron Microscopy of SOD2 Localization
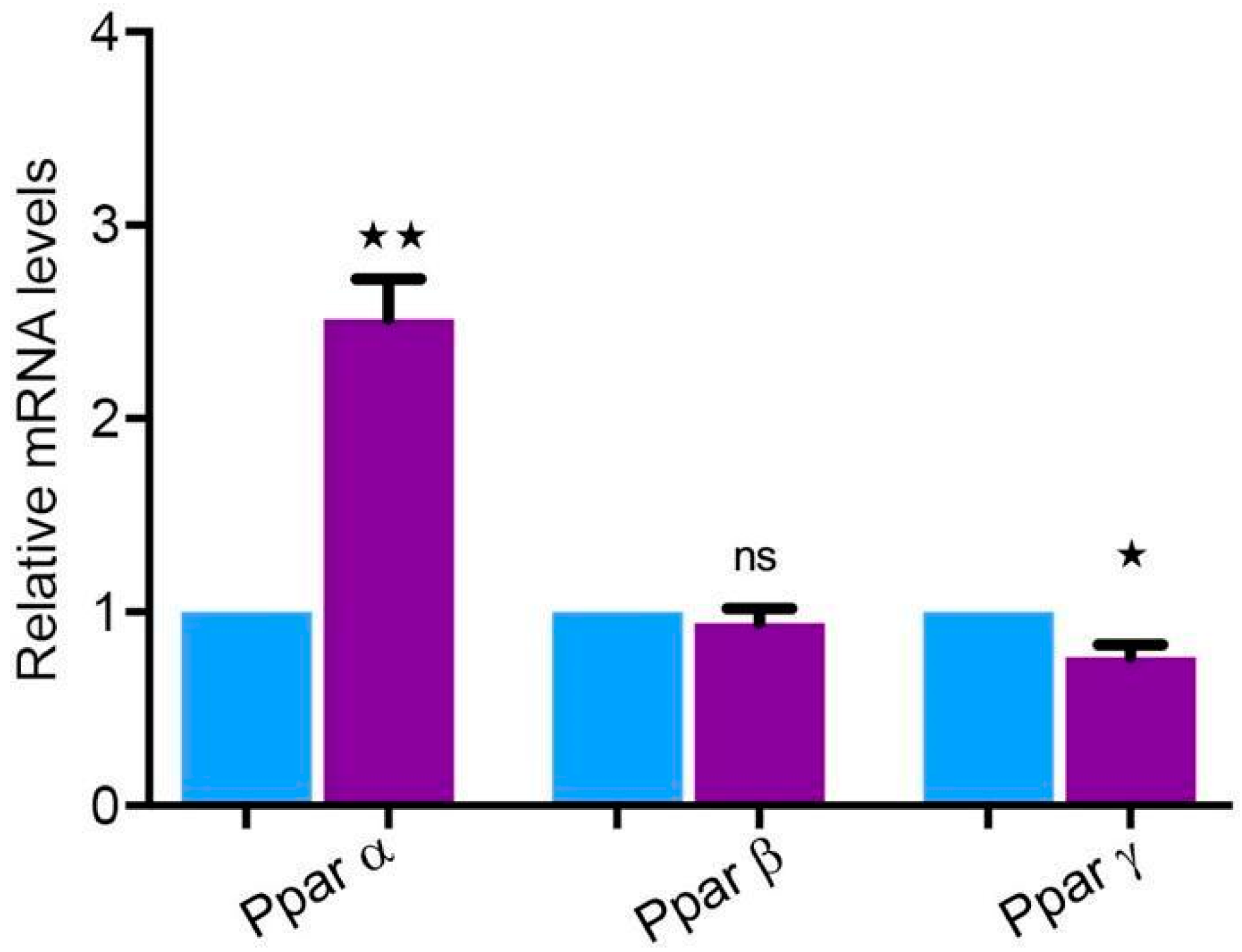
2.8. Peroxisome Proliferator-Activated Receptors (PPARs) Are Highly Expressed in the Parotid Gland
3. Discussion
3.1. Marker Proteins for the Correct Identification of the Parotid Gland
3.2. Peroxisomes in the Parotid Gland
3.3. In the Human Parotid Gland, Peroxisomal ß-Oxidation, Cholesterol Production, and Plasmalogen Synthesis Enzymes Are Highly Expressed
3.4. PPARs Are Expressed at a Significantly High Level in the Human Parotid Gland
4. Materials & Methods
4.1. Surgical Removal and Fixation of the Human Parotid Glands
4.2. Paraffin Embedding, Sectioning, and Immunofluorescence
4.3. Fixation and Embedding for Electron Microscopy
4.4. Cytochemical Localization of Catalase Activity with the Alkaline DAB Method
4.5. Post-Embedding Immunoelectron Microscopy
4.6. Homogenization of Human Parotid Glands to Obtain Tissue Lysates for Western Blotting
4.7. Western Blot Analysis
4.8. RNA Isolation
4.9. cDNA-Synthesis
4.10. Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR)
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Omon, M.; Shiga, Y. Sialographic investigations on the anatomy of mouse parotid glands. Anat. Rec. 1986, 214, 165–167. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, G.N. The Physiology and Biochemistry of the Mouth, 4th ed.; Blackwell: Oxford, UK, 1978; ISBN 0632001380. [Google Scholar]
- Watermann, C.; Valerius, K.P.; Wagner, S.; Wittekindt, C.; Klussmann, J.P.; Baumgart-Vogt, E.; Karnati, S. Step-by-step protocol to perfuse and dissect the mouse parotid gland and isolation of high-quality RNA from murine and human parotid tissue. BioTechniques 2016, 60, 200–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karnati, S.; Baumgart-Vogt, E. Peroxisomes in airway epithelia and future prospects of these organelles for pulmonary cell biology. Histochem. Cell Biol. 2009, 131, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Wanders, R.J.A.; Baes, M.; Ribeiro, D.; Ferdinandusse, S.; Waterham, H.R. The physiological functions of human peroxisomes. Physiol. Rev. 2023, 103, 957–1024. [Google Scholar] [CrossRef]
- Meyer, M.T.; Watermann, C.; Dreyer, T.; Wagner, S.; Wittekindt, C.; Klussmann, J.P.; Ergün, S.; Baumgart-Vogt, E.; Karnati, S. Differential Expression of Peroxisomal Proteins in Distinct Types of Parotid Gland Tumors. Int. J. Mol. Sci. 2021, 22, 7872. [Google Scholar] [CrossRef]
- Riva, A.; Testa-Riva, F.; del Fiacco, M.; Lantini, M.S. Fine structure and cytochemistry of the intralobular ducts of the human parotid gland. J. Anat. 1976, 122, 627–640. [Google Scholar]
- Riva, A.; Puxeddu, P.; del Fiacco, M.; Testa-Riva, F. Ultrastructural localization of endogenous peroxidase in human parotid and submandibular glands. J. Anat. 1978, 127, 181–191. [Google Scholar]
- Hand, A.R. Morphologic and cytochemical identification of peroxisomes in the rat parotid and other exocrine glands. J. Histochem. Cytochem. 1973, 21, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Graham, R.C.; Karnovsky, M.J. The early stages of absorption of injected horseradish peroxidase in the proximal tubules of mouse kidney: Ultrastructural cytochemistry by a new technique. J. Histochem. Cytochem. 1966, 14, 291–302. [Google Scholar] [CrossRef] [Green Version]
- Sato, A.; Miyoshi, S. Ultrastructure of the main excretory duct epithelia of the rat parotid and submandibular glands with a review of the literature. Anat. Rec. 1988, 220, 239–251. [Google Scholar] [CrossRef]
- Hanker, J.S.; Preece, J.W.; Burkes, E.J.; Romanovicz, D.K. Catalase in salivary gland striated and excretory duct cells. I. The distribution of cytoplasmic and particulate catalase and the presence of catalase-positive rods. Histochem. J. 1977, 9, 711–728. [Google Scholar] [CrossRef]
- Tandler, B.; Walter, R.J. Epon-Maraglas Embedment for Electron Microscopy. Stain Technol. 1977, 52, 238–239. [Google Scholar] [CrossRef] [PubMed]
- Nagato, T.; Tandler, B.; Phillips, C.J. Unusual smooth endoplasmic reticulum in submandibular acinar cells of the male round-eared bat, Tonatia sylvicola. J. Ultrastruct. Res. 1984, 87, 275–284. [Google Scholar] [CrossRef]
- Tandler, B.; Nagato, T.; Phillips, C.J. Ultrastructure of the binary parotid glands in the free-tailed bat, Tadarida thersites. II. Accessory parotid gland. Anat. Rec. 1998, 251, 122–135. [Google Scholar] [CrossRef]
- Venkatesh, S.G.; Goyal, D.; Carenbauer, A.L.; Darling, D.S. Parotid secretory protein binds phosphatidylinositol (3,4) bisphosphate. J. Dent. Res. 2011, 90, 1085–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, S.L.; Krisans, S.K. Rat liver peroxisomes catalyze the initial step in cholesterol synthesis. The condensation of acetyl-CoA units into acetoacetyl-CoA. J. Biol. Chem. 1990, 265, 5731–5735. [Google Scholar] [CrossRef]
- Angermüller, S.; Fahimi, H.D. Selective cytochemical localization of peroxidase, cytochrome oxidase and catalase in rat liver with 3,3′-diaminobenzidine. Histochemistry 1981, 71, 33–44. [Google Scholar]
- Karn, R.C.; Chung, A.G.; Laukaitis, C.M. Shared and unique proteins in human, mouse and rat saliva proteomes: Footprints of functional adaptation. Proteomes 2013, 1, 275–289. [Google Scholar] [CrossRef] [Green Version]
- Kuraji, M.; Matsuno, T.; Satoh, T. Astaxanthin affects oxidative stress and hyposalivation in aging mice. J. Clin. Biochem. Nutr. 2016, 59, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Sowa, P.; Misiolek, M.; Pasinski, B.; Bartosz, G.; Soszynski, M.; Adamczyk-Sowa, M.; Sadowska-Bartosz, I. Oxidative Stress Markers Patients with Parotid Gland Tumors: A Pilot Study. BioMed Res. Int. 2018, 2018, 4340871. [Google Scholar] [CrossRef] [Green Version]
- Ginsburg, I.; Kohen, R.; Shalish, M.; Varon, D.; Shai, E.; Koren, E. The oxidant-scavenging abilities in the oral cavity may be regulated by a collaboration among antioxidants in saliva, microorganisms, blood cells and polyphenols: A chemiluminescence-based study. PLoS ONE 2013, 8, e63062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpenter, G.H. The secretion, components, and properties of saliva. Annu. Rev. Food Sci. Technol. 2013, 4, 267–276. [Google Scholar] [CrossRef]
- Ramachandran, P.; Boontheung, P.; Xie, Y.; Sondej, M.; Wong, D.T.; Loo, J.A. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 2006, 5, 1493–1503. [Google Scholar] [CrossRef] [PubMed]
- Denny, P.; Hagen, F.K.; Hardt, M.; Liao, L.; Yan, W.; Arellanno, M.; Bassilian, S.; Bedi, G.S.; Boontheung, P.; Cociorva, D.; et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J. Proteome Res. 2008, 7, 1994–2006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bingle, L.; Barnes, F.A.; Lunn, H.; Musa, M.; Webster, S.; Douglas, C.W.I.; Cross, S.S.; High, A.S.; Bingle, C.D. Characterisation and expression of SPLUNC2, the human orthologue of rodent parotid secretory protein. Histochem. Cell Biol. 2009, 132, 339–349. [Google Scholar] [CrossRef] [Green Version]
- Grant, P.; Ahlemeyer, B.; Karnati, S.; Berg, T.; Stelzig, I.; Nenicu, A.; Kuchelmeister, K.; Crane, D.I.; Baumgart-Vogt, E. The biogenesis protein PEX14 is an optimal marker for the identification and localization of peroxisomes in different cell types, tissues, and species in morphological studies. Histochem. Cell Biol. 2013, 140, 423–442. [Google Scholar] [CrossRef]
- Karnati, S.; Baumgart-Vogt, E. Peroxisomes in mouse and human lung: Their involvement in pulmonary lipid metabolism. Histochem. Cell Biol. 2008, 130, 719–740. [Google Scholar] [CrossRef]
- Karnati, S.; Graulich, T.; Oruqaj, G.; Pfreimer, S.; Seimetz, M.; Stamme, C.; Mariani, T.J.; Weissmann, N.; Mühlfeld, C.; Baumgart-Vogt, E. Postnatal development of the bronchiolar club cells of distal airways in the mouse lung: Stereological and molecular biological studies. Cell Tissue Res. 2016, 364, 543–557. [Google Scholar] [CrossRef]
- Li, X.; Baumgart, E.; Dong, G.-X.; Morrell, J.C.; Jimenez-Sanchez, G.; Valle, K.D.; Smith, K.S.; Gould, S.J. PEX11 Is Required for Peroxisome Proliferation in Response to 4-Phenylbutyrate but Is Dispensable for Peroxisome Proliferator-Activated Receptor Alpha-Mediated Peroxisome Proliferation. Mol. Cell. Biol. 2002, 22, 8226–8240. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Gould, S.J. PEX11 promotes peroxisome division independently of peroxisome metabolism. J. Cell Biol. 2002, 156, 643–651. [Google Scholar] [CrossRef]
- Li, X.; Baumgart, E.; Morrell, J.C.; Jimenez-Sanchez, G.; Valle, K.D.; Gould, S.J. PEX11 beta deficiency is lethal and impairs neuronal migration but does not abrogate peroxisome function. Mol. Cell. Biol. 2002, 22, 4358–4365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Issemann, I.; Green, S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 1990, 347, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, E. Application of in situ hybridization, cytochemical and immunocytochemical techniques for the investigation of peroxisomes. A review including novel data. Robert Feulgen Prize Lecture 1997. Histochem. Cell Biol. 1997, 108, 185–210. [Google Scholar] [CrossRef] [PubMed]
- Di Cara, F.; Andreoletti, P.; Trompier, D.; Vejux, A.; Bülow, M.H.; Sellin, J.; Lizard, G.; Cherkaoui-Malki, M.; Savary, S. Peroxisomes in Immune Response and Inflammation. Int. J. Mol. Sci. 2019, 20, 3877. [Google Scholar] [CrossRef] [Green Version]
- Wanders, R.J.A.; Waterham, H.R. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 2006, 75, 295–332. [Google Scholar] [CrossRef]
- Huyghe, S.; Mannaerts, G.P.; Baes, M.; van Veldhoven, P.P. Peroxisomal multifunctional protein-2: The enzyme, the patients and the knockout mouse model. Biochim. Biophys. Acta 2006, 1761, 973–994. [Google Scholar] [CrossRef]
- Kawaguchi, K.; Morita, M. ABC Transporter Subfamily D: Distinct Differences in Behavior between ABCD1-3 and ABCD4 in Subcellular Localization, Function, and Human Disease. Biomed Res. Int. 2016, 2016, 6786245. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, H.; Takahata, S. Role of peroxisomal fatty acyl-CoA beta-oxidation in phospholipid biosynthesis. Arch. Biochem. Biophys. 1991, 284, 326–331. [Google Scholar] [CrossRef]
- Stables, M.J.; Gilroy, D.W. Old and new generation lipid mediators in acute inflammation and resolution. Prog. Lipid Res. 2011, 50, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.-C.; Pfeiffer, D.R.; Calhoon, E.A.; Madiai, F.; Marcucci, G.; Liu, S.; Jurkowitz, M.S. Purification, identification, and cloning of lysoplasmalogenase, the enzyme that catalyzes hydrolysis of the vinyl ether bond of lysoplasmalogen. J. Biol. Chem. 2011, 286, 24916–24930. [Google Scholar] [CrossRef] [Green Version]
- Faust, P.L.; Kovacs, W.J. Cholesterol biosynthesis and ER stress in peroxisome deficiency. Biochimie 2014, 98, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Hindy, A.M.; Yoshiga, K.; Takada, K.; Okuda, K. Sterol composition and biosynthesis in mouse salivary glands. Arch. Oral Biol. 1986, 31, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Valle, L.; Alonso, C.C.; Villa, N.; Oliveira-Filho, R.M. Enlargement of salivary glands upon the combined effect of isoproterenol and of a cholesterol synthesis inhibitor (SC-12937). Gen. Pharmacol. Vasc. Syst. 1980, 11, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, W.J.; Olivier, L.M.; Krisans, S.K. Central role of peroxisomes in isoprenoid biosynthesis. Prog. Lipid Res. 2002, 41, 369–391. [Google Scholar] [CrossRef]
- Kurahashi, M.; Nakamura, H.; Inomata, K. Cholesterol metabolism in salivary glands of streptozotocin-induced diabetic rats. Jpn. J. Oral Biol. 1987, 29, 102–106. [Google Scholar] [CrossRef] [Green Version]
- Takeda, I.; Kizu, Y.; Yoshitaka, O.; Saito, I.; Yamane, G.-Y. Possible role of nitric oxide in radiation-induced salivary gland dysfunction. Radiat. Res. 2003, 159, 465–470. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Matsuno, T.; Omata, K.; Satoh, T. Relationship between hyposalivation and oxidative stress in aging mice. J. Clin. Biochem. Nutr. 2017, 61, 40–46. [Google Scholar] [CrossRef] [Green Version]
- Indo, H.P.; Yen, H.-C.; Nakanishi, I.; Matsumoto, K.-I.; Tamura, M.; Nagano, Y.; Matsui, H.; Gusev, O.; Cornette, R.; Okuda, T.; et al. A mitochondrial superoxide theory for oxidative stress diseases and aging. J. Clin. Biochem. Nutr. 2015, 56, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Dukan, S.; Farewell, A.; Ballesteros, M.; Taddei, F.; Radman, M.; Nyström, T. Protein oxidation in response to increased transcriptional or translational errors. Proc. Natl. Acad. Sci. USA 2000, 97, 5746–5749. [Google Scholar] [CrossRef] [Green Version]
- Kesarwala, A.H.; Krishna, M.C.; Mitchell, J.B. Oxidative stress in oral diseases. Oral Dis. 2016, 22, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Quiles, M.; Broekema, M.F.; Kalkhoven, E. PPARgamma in Metabolism, Immunity, and Cancer: Unified and Diverse Mechanisms of Action. Front. Endocrinol. 2021, 12, 624112. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Shen, W.-J.; Bittner, S.; Kraemer, F.B.; Azhar, S. PPARs: Regulators of metabolism and as therapeutic targets in cardiovascular disease. Part II: PPAR-β/δ and PPAR-γ. Future Cardiol. 2017, 13, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M.; Takeshita, A.; Tsukamoto, T.; Gonzalez, F.J.; Osumi, T. Tissue-selective, bidirectional regulation of PEX11 alpha and perilipin genes through a common peroxisome proliferator response element. Mol. Cell. Biol. 2004, 24, 1313–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumgart, E.; Vanhooren, J.C.; Fransen, M.; Marynen, P.; Puype, M.; Vandekerckhove, J.; Leunissen, J.A.; Fahimi, H.D.; Mannaerts, G.P.; van Veldhoven, P.P. Molecular characterization of the human peroxisomal branched-chain acyl-CoA oxidase: cDNA cloning, chromosomal assignment, tissue distribution, and evidence for the absence of the protein in Zellweger syndrome. Proc. Natl. Acad. Sci. USA 1996, 93, 13748–13753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Xu, B.; Wang, Y.; Wei, L. Anti-inflammatory effect of peroxisome proliferator-activated receptor-γ (PPAR-γ) on non-obese diabetic mice with Sjogren’s syndrome. Int. J. Clin. Exp. Pathol. 2014, 7, 4886–4894. [Google Scholar]
- Ahlemeyer, B.; Neubert, I.; Kovacs, W.J.; Baumgart-Vogt, E. Differential expression of peroxisomal matrix and membrane proteins during postnatal development of mouse brain. J. Comp. Neurol. 2007, 505, 1–17. [Google Scholar] [CrossRef]
- Nenicu, A.; Lüers, G.H.; Kovacs, W.; Bergmann, M.; Baumgart-Vogt, E. Peroxisomes in Human and Mouse Testis: Differential Expression of Peroxisomal Proteins in Germ Cells and Distinct Somatic Cell Types of the Testis1. Biol. Reprod. 2007, 77, 1060–1072. [Google Scholar] [CrossRef] [Green Version]
- Newman, G.R.; Jasani, B.; Williams, E.D. A simple post-embedding system for the rapid demonstration of tissue antigens under the electron microscope. Histochem. J. 1983, 15, 543–555. [Google Scholar] [CrossRef]
- Slot, J.W.; Geuze, H.J. Sizing of protein A-colloidal gold probes for immunoelectron microscopy. J. Cell Biol. 1981, 90, 533–536. [Google Scholar] [CrossRef]
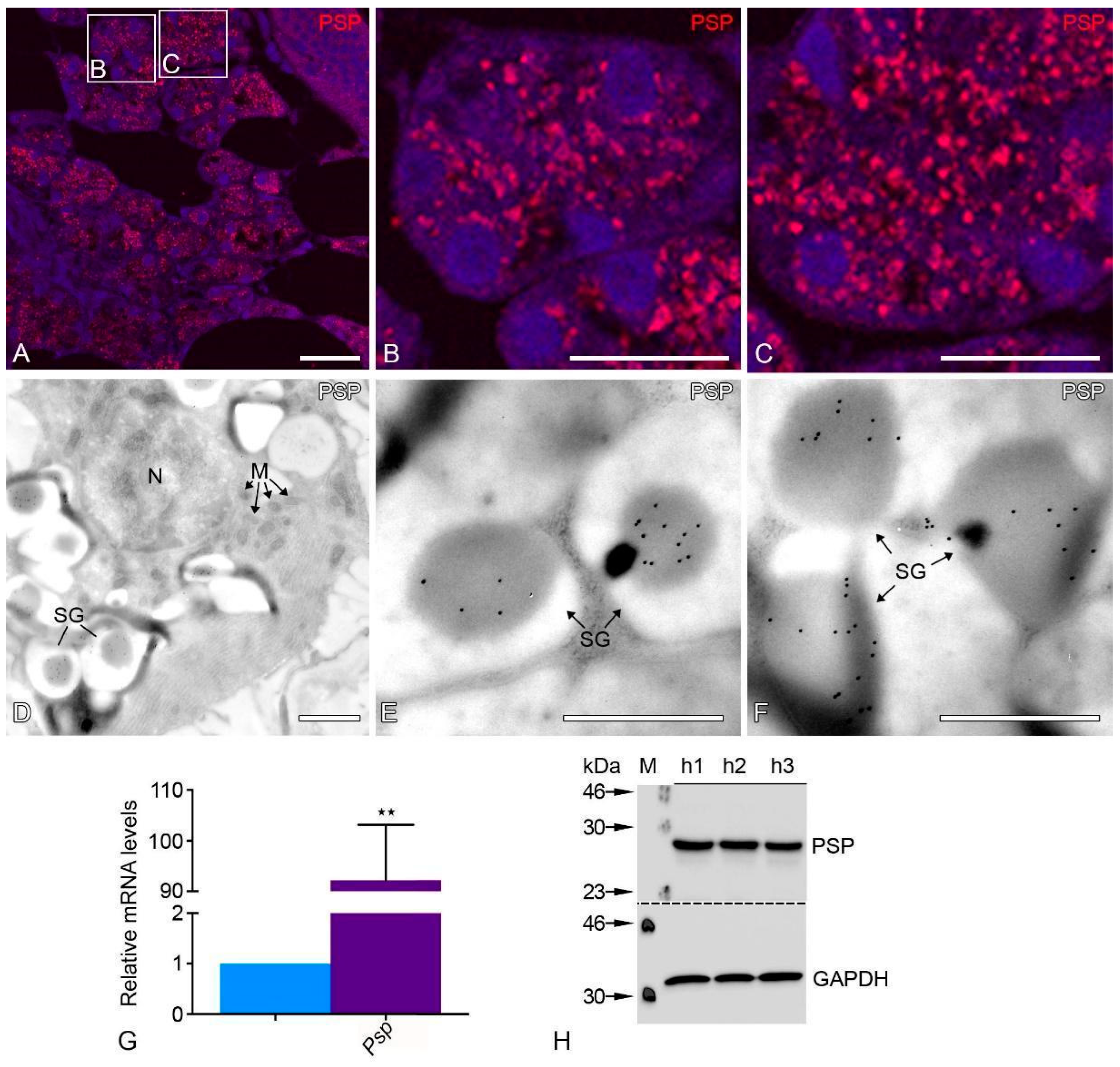

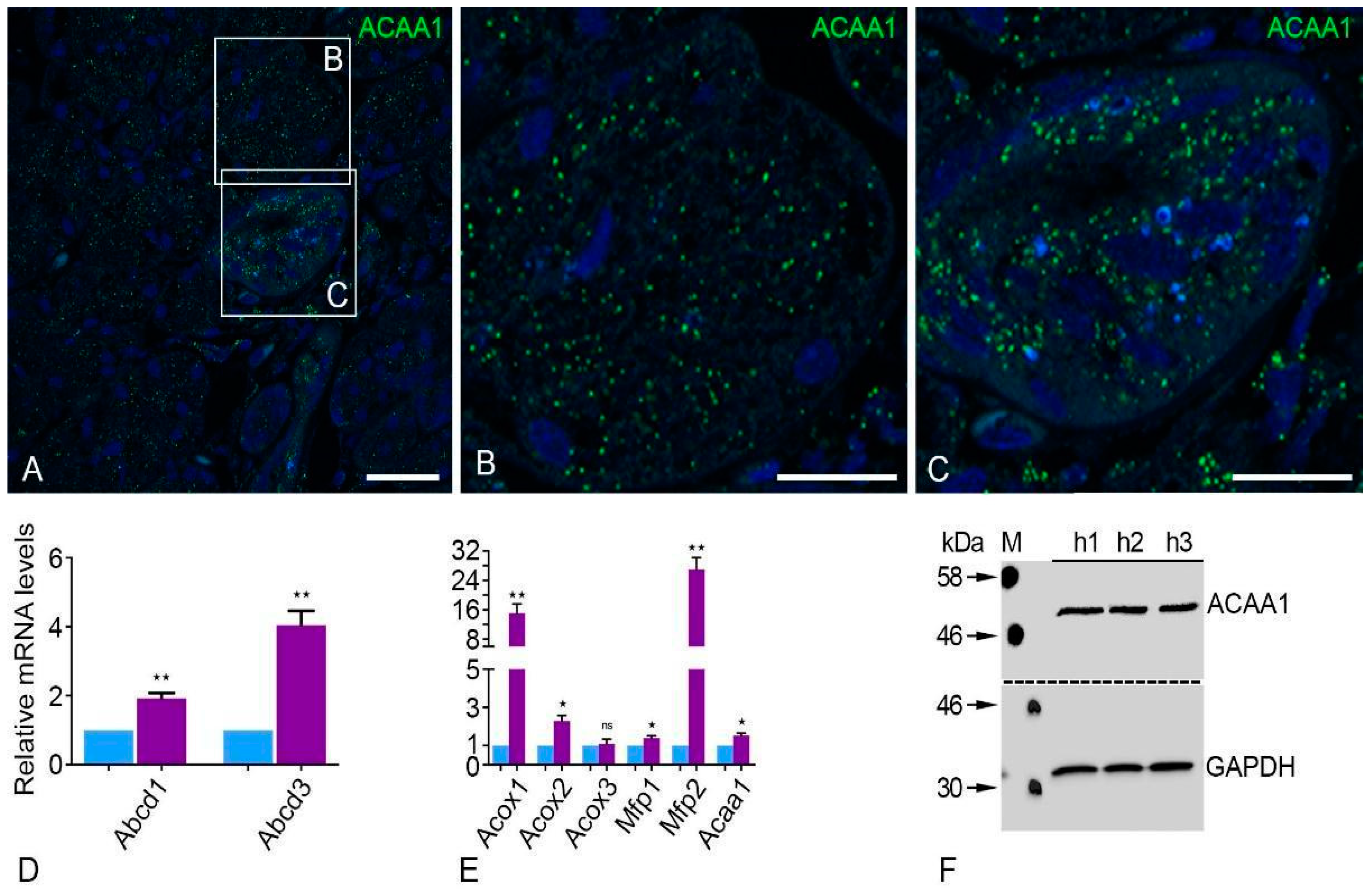
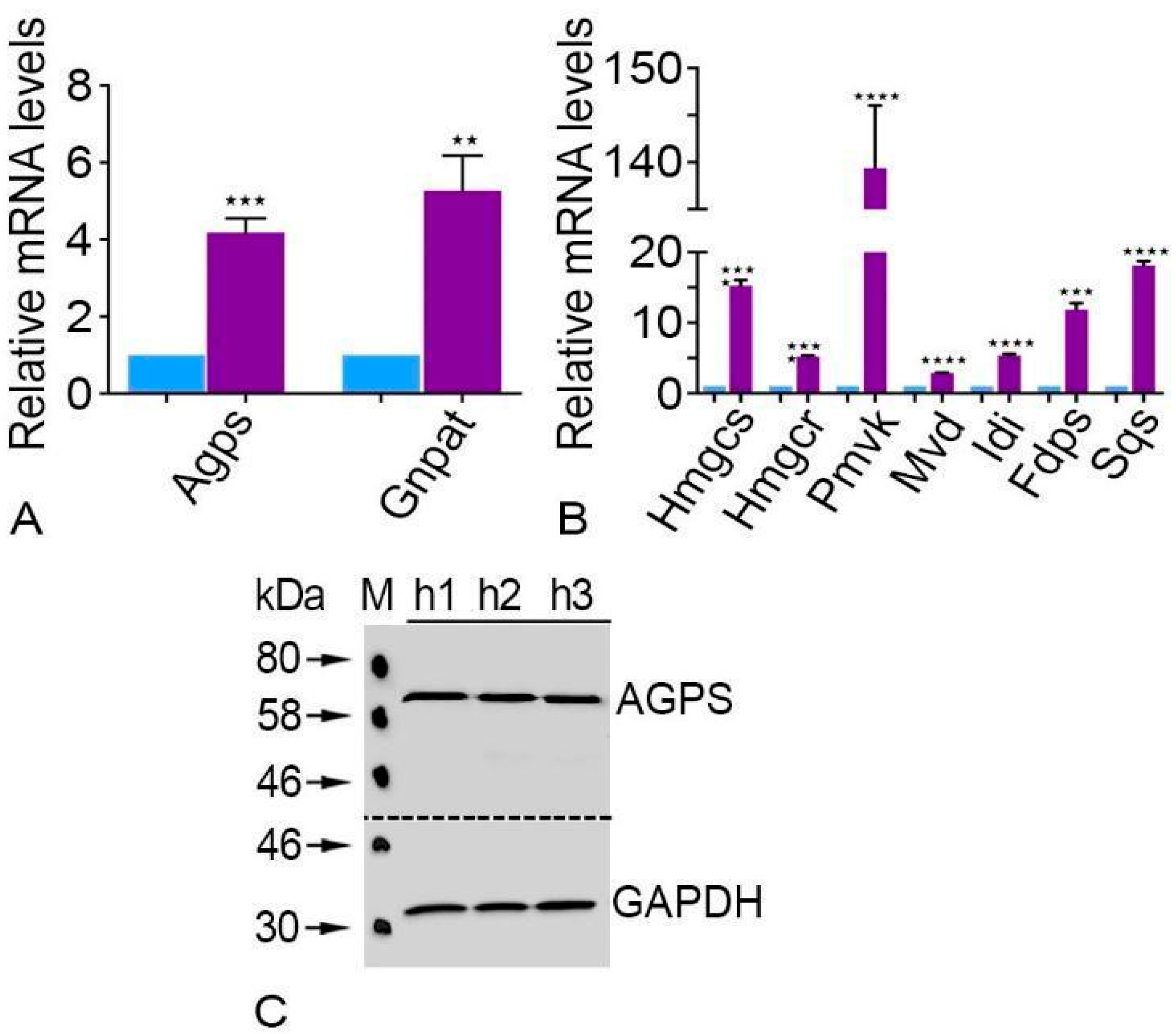
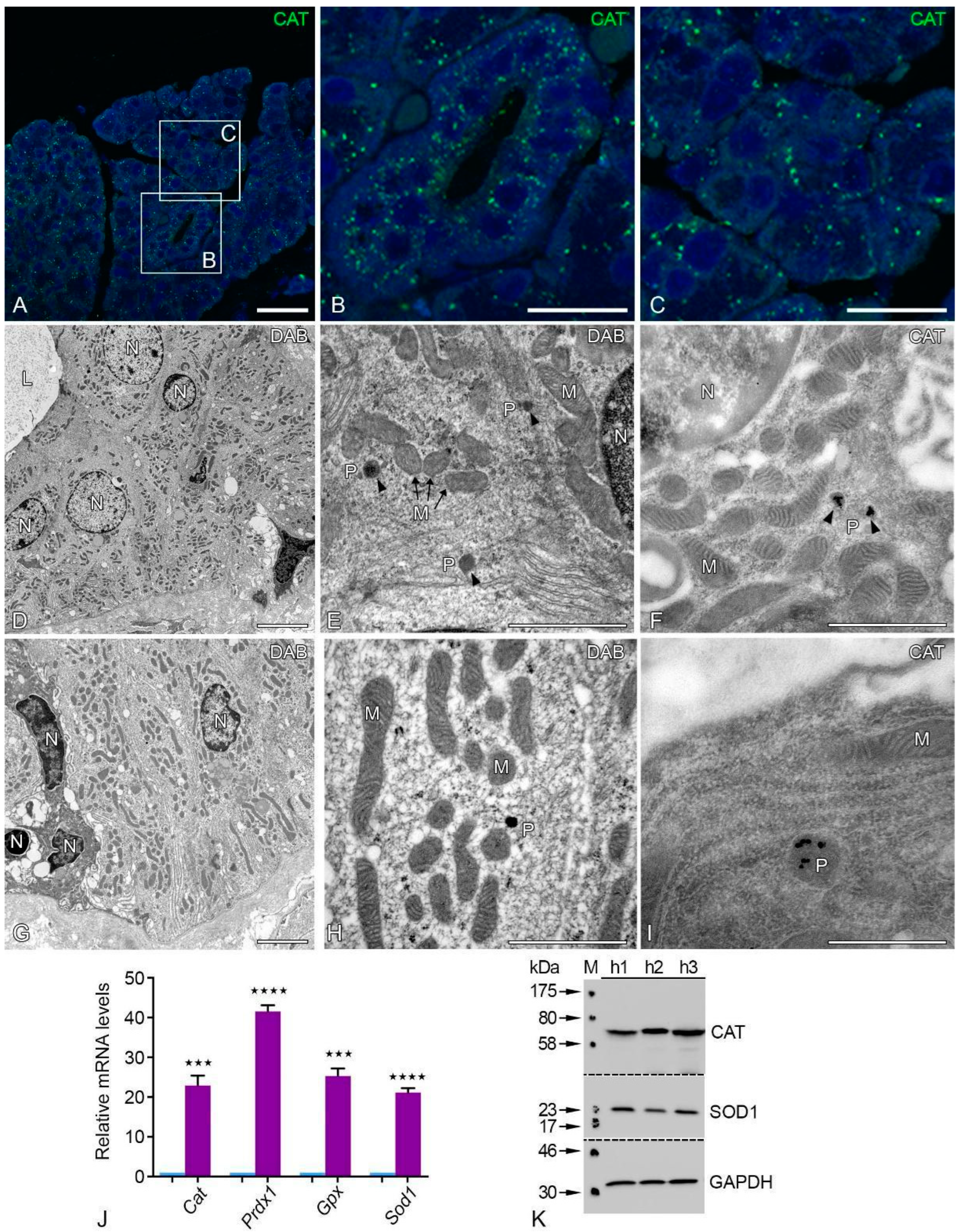
| Primary Antibody | Host | Target Molecular Weight | Dilution (IF) | Dilution (WB) | Supplier |
|---|---|---|---|---|---|
| Cell type specific antigens | |||||
| Parotid secretory Protein, human (SPLUNC2) | Mouse, monoclonal | 25 kDa | 1:1000 | Abbexa, Cambridge, UK, Cat. no.: abx11413 | |
| Peroxisomal biogenesis and metabolic proteins | |||||
| Peroxin 13 (Pex13p), mouse | Rabbit, polyclonal | 44 kDa | 1:1000 | 1:6000 | Gift from Denis I. Crane; School of Biomol. Biophys. Sci., Griffith Univ., Nathan, Brisbane, Australia |
| Peroxin 14 (Pex14p), mouse | Rabbit, polyclonal | 42 kDa | 1:1000 | 1:3000 | Gift from Denis I. Crane |
| Catalase (CAT), mouse | Rabbit, polyclonal | 60 kDa | 1:2000 | 1:5000 | Gift from Denis I. Crane |
| Thiolase | Rabbit, polyclonal | 51 kDa | 1:1000 | 1:5000 | Gift from Nancy E Bravermann; Depts. of Human Genetics and Pediatrics, McGill University-Montreal Montreal, QC, Canada. |
| Alkylglycerone-phosphate synthase (AGPS) | Mouse, monoclonal | 78 kDa | 1:1000 | 1:500 | Santa cruz, Cat no: sc-374201 |
| Antioxidative enzymes from other cell compartments | |||||
| Gluthation reductase | Rabbit, polyclonal | 56 kDa | 1:1000 | Abcam, Cambridge, UK, Cat. no: ab16801 | |
| Superoxide dismutase 1 (SOD-1), | Goat; polyclonal | 17 kDa | 1:5000 | R&D Systems, Minneapolis, MN, USA, Cat. no.; AF3787 | |
| Superoxide dismutase 2 (SOD-2) | Rabbit, polyclonal | 25 kDa | 1:1000 | 1:1000 | Research diagnostics, Inc., NJ, USA, Cat no: RDI-RTSODMabR |
| Other marker proteins of different cell compartments | |||||
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Mouse, monoclonal | 36 kDa | 1:60,000 | Hy Test Ltd., Cat. no.:5G4 | |
| Secondary Antibodies | |||||
| HRP-rabbit | 1:6000 | Cell Signaling Technology, Inc., Danvers, MA, USA | |||
| HRP-mouse | 1:6000 | Cell Signaling Technology, Inc., Danvers, MA, USA | |||
| Bovine anti goat HRP | 1:5000 | Santa cruz, Cat. no: sc-2378 | |||
| Gene Target | Gene Bank Accession No. | Sence Primer (3′-3′) | Antisence Primer (5′-3′) | Annealing Temp °C | PCR Product (bp) |
|---|---|---|---|---|---|
| ABCD1 | NM_000033.3 | GTGGAGGACATGCAAAGGAA | TCACACATAGCCTCCCAACC | 58.1 | 113 |
| ABCD3 | NM_002858.3 | ATGACCCTTGGAACACTTCG | TGCCATCCATATGCAGGTAG | 57.8 | 385 |
| ACOX1 | NM_004035.6 | ATTTCCTTCAGGGGAGCATC | GCCAAGTGTCACATCCTGAA | 57.3 | 137 |
| ACOX2 | NM_003500.3 | CAAATTGTCGGCCTCCTGTA | GAGATCTCTGTGGCGTGGAG | 57.9 | 125 |
| ACOX3 | NM_003501.2 | GGAGTGTGTGGGCTCTTATC | CTCTTGCTCGGTAGGCATC | 57.7 | 107 |
| ACTB | NM_001101.3 | TCCCTGGAGAAGAGCTACGA | AGCACTGTGTTGGCGTACAG | 59.4 | 194 |
| AGPS | NM_003659.3 | AGGGGGATCGTGAGAAGGT | CCAAAGCCAAGTCTCGAATG | 59.6 | 147 |
| CAT | NM_001752.3 | CGTGCTGAATGAGGAACAGA | TTGTCCAGAAGAGCCTGGAT | 57.9 | 150 |
| FDPS | NM_001135821.1 | CAAGGAGGTCCTGGAGTACAA | GGAGACTATCAGCATCCTGTTTC | 58.7 | 113 |
| GAPDH | NM_002046.5 | GTCAACGGATTTGGTCGTATT | TGTAGTTGAGGTCAATGAAGGG | 56.6 | 106 |
| GNPAT | NM_014236.3 | GTGCAGAAAAACGCCTTAGC | GGCTGGTTTTCCTATTGGTG | 58.3 | 150 |
| GPX1 | NM_000581.2 | CAGTTTGGGCATCAGGAGAA | TCGAAGAGCATGAAGTTGGG | 57.8 | 101 |
| GR1/GSR | NM_000637.3 | GTGGCCTCCTATGACTACCT | CATCCAACATTCACGCAAGTG | 57.9 | 137 |
| HMGCR | NM_000859.2 | CGATGCATAGCCATCCTGTAT | GCTGGAATGACAGCTTCACA | 57.7 | 87 |
| HMGCS | NM_001098272.2 | TCTATCCTTCACACAGCTCTTTC | GGCAACAATTCCCACATCTTT | 57.9 | 89 |
| HO-1 | NM_002133.2 | CGGCTTCAAGCTGGTGAT | AGCTCTTCTGGGAAGTAGACA | 57.7 | 114 |
| HPRT | NM_000194.2 | CACTGGCAAAACAATGCAGACT | GTCTGGCTTATATCCAACACTTCGT | 60.2 | 118 |
| IDI | NM_004969.3 | TCTCATTGGGCATGAAGGTC | CATAAAACCTCGGGCTCCTT | 57.6 | 106 |
| MFP1 | NM_001966.3 | ATGGATATGGATGGCCAAGG | GCTCCAGTTGGGGAATATCA | 57.1 | 126 |
| MFP2 | NM_000414.3 | TGTCGTTGCAGGCCTTATT | CCTCCCAAATCATTCACAACAAC | 57.4 | 148 |
| MVD | NM_002461.2 | GGTGGCACCTGTTCTTCTCTCT | CTGATGAGCAGCTGTCTGGAGT | 56.5 | 82 |
| MVK | NM_000431.3 | CTGGACACAAGCTTTCTGGA | AAGCCTGCAACCTCCTTTAG | 57.7 | 83 |
| PEX3 | NM_003630.2 | TTCTTTTGCGGGTCCAGTTA | ACATCTGGGGGAGCAAGAAT | 57.4 | 100 |
| PEX5 | NM_001131023.1 | CTGAGGCAGTGAGTGTTCTT | TCAGCCACCAACTCATCTTC | 57.5 | 100 |
| PEX6 | NM_000287.3 | AACAGTTGGGGAAGCTCCAG | ATGGAACAGGGCTCAGGGTA | 59.9 | 101 |
| PEX7 | NM_000288.3 | CTCAGGAGGTGTATAGTGTTGATT | CAGTTGGATCCCACAATTTGAC | 57.8 | 99 |
| PEX10 | NM_153818.1 | TGGAGTGGAGGAAGGAGGTT | GATGGGTCCACCTGGATGAT | 59.8 | 118 |
| PEX11alpha | NM_003847.2 | GGTAATGAAGCTCAAGAAACTGGAG | TGCTCTGCTCAGTTGCCTGT | 59.9 | 101 |
| PEX11beta | NM_003846.2 | CCAGTCCTGAGTTACAGAAACAGATT | TGACTCAAGGGCATCTGCTG | 60.7 | 101 |
| PEX11gamma | NM_080662.3 | ACCGCCTGATCCGAGTG | CATCAAAGAGTCGCAAGATGGT | 58.7 | 150 |
| PEX12 | NM_000286.2 | AAGCTCTGGAGCACAAACCA | ACACCCCCAACAGCTTTCTT | 59.8 | 103 |
| PEX13 | NM_002618.3 | CCATGTAGTTGCCAGAGCAG | CATCAAGGCTAGCCAGAAGC | 58.3 | 140 |
| PEX14 | NM_004565.2 | CTGCCTTTGGCTTTGATCTC | CGTGGTGTCACGGTAGTCAA | 57.1 | 137 |
| PEX16 | NM_004813.2 | CGAGCTGTCAGAGCTGGTGTACT | ACAGCGACACAGGCAACTTTT | 64.1 | 101 |
| PEX19 | NM_002857.3 | CTCTCAGAGGCTGCAGGGAG | GTGGCATTTTTGGCTAATCCA | 61.7 | 101 |
| PMVK | NM_006556.3 | GCCTTTCGGAAGGACATGAT | GTCACTCACCAGCCAGATG | 58 | 114 |
| PPARalpha | NM_005036.4 | CTGGCCAAGAGAATCTACGAG | ACTGGTTCCATGTTGCCAAG | 57.9 | |
| PPARbeta | NM_177435.2 | AACATGCAAGGCACTGACTG | CTGCCAAAGTGCTGGGATT | 59 | 129 |
| PPARgamma | NM_138712.3 | ATCTTTCAGGGCTGCCAGT | TCGTGGACTCCATATTTGAGG | 58.9 | 131 |
| PRDX6 | NM_004905.2 | TTAGTGCCATGTGCCTTTCA | TAGCAACCCACTGCAAGAAG | 57.7 | 144 |
| PSP/SPLUNC2 | NM_001319164.1 | GAAGTCTGAGGTGGTGTCAAG | TGCCAAGATTGTCAAGAAGAGA | 58.2 | 107 |
| RPL13 | NM_000977. | CGGAATGGCATGGTCTTGA | CCTTACGTCTGCGGATCTTAC | 57.8 | 100 |
| SOD1 | NM_000454.4 | AGGATGAAGAGAGGCATGTTG | ATGGTCTCCTGAGAGTGAGAT | 57.7 | 107 |
| SOD2 | NM_000636.2 | GTTGGCCAAGGGAGATGTTA | CGTTAGGGCTGAGGTTTGT | 57.5 | 110 |
| SQS | NM_001287742.1 | GAAGTCAGTGAGACCAAGAACC | CGCTCTCTGTAGAGCCTTAGA | 58.6 | 76 |
| TBP | NM_003194.4 | TGACCCAGCATCACTGTTTC | GCTGGAACTCGTCTCACTATTC | 58.1 | 118 |
| Thiolase | NM_001607.3 | GATGCCTTCTTACCCCAACA | CCCAACCACTGCATAAGACC | 57.5 | |
| TRX1 | NM_001244938.1 | GGACGCTGCAGGTGATAAA | CACTCTGAAGCAACATCATGAAAG | 57.9 | 102 |
| TRX2 | NM_012473.3 | GTTAGAGAAGATGGTGGCCAAG | GCTGACACCTCATACTCAATGG | 58.7 | 99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watermann, C.; Meyer, M.T.; Wagner, S.; Wittekindt, C.; Klussmann, J.P.; Erguen, S.; Baumgart-Vogt, E.; Karnati, S. Peroxisomes Are Highly Abundant and Heterogeneous in Human Parotid Glands. Int. J. Mol. Sci. 2023, 24, 4783. https://doi.org/10.3390/ijms24054783
Watermann C, Meyer MT, Wagner S, Wittekindt C, Klussmann JP, Erguen S, Baumgart-Vogt E, Karnati S. Peroxisomes Are Highly Abundant and Heterogeneous in Human Parotid Glands. International Journal of Molecular Sciences. 2023; 24(5):4783. https://doi.org/10.3390/ijms24054783
Chicago/Turabian StyleWatermann, Christoph, Malin Tordis Meyer, Steffen Wagner, Claus Wittekindt, Jens Peter Klussmann, Sueleyman Erguen, Eveline Baumgart-Vogt, and Srikanth Karnati. 2023. "Peroxisomes Are Highly Abundant and Heterogeneous in Human Parotid Glands" International Journal of Molecular Sciences 24, no. 5: 4783. https://doi.org/10.3390/ijms24054783
APA StyleWatermann, C., Meyer, M. T., Wagner, S., Wittekindt, C., Klussmann, J. P., Erguen, S., Baumgart-Vogt, E., & Karnati, S. (2023). Peroxisomes Are Highly Abundant and Heterogeneous in Human Parotid Glands. International Journal of Molecular Sciences, 24(5), 4783. https://doi.org/10.3390/ijms24054783







