A Unified Deep Learning Framework for Single-Cell ATAC-Seq Analysis Based on ProdDep Transformer Encoder
Abstract
1. Introduction
2. Results and Discussion
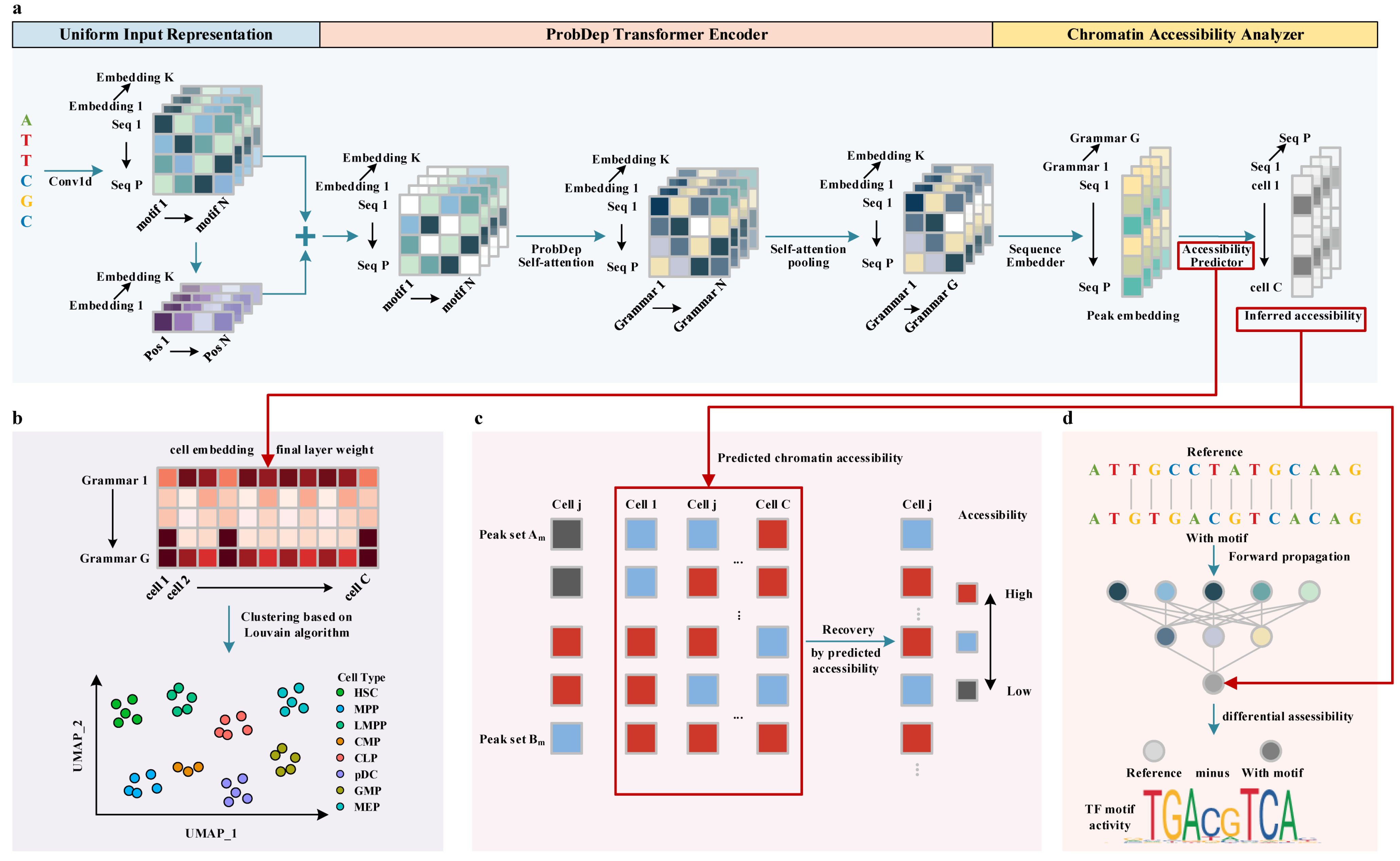
2.1. PROTRAIT Predicts Single-Cell Chromatin Accessibility on Held-Out DNA Sequences
2.2. PROTRAIT Annotates Cell Types by Clustering on Cell Embedding
2.3. PROTRAIT Denoises Single-Cell Chromatin Accessibility Profiles
2.4. PROTRAIT Infers TF Activity at Single-Cell and Single-Nucleotide Resolution
2.5. PROTRAIT Is Scalable to Large Datasets
3. Materials and Methods
3.1. Datasets
3.2. Chromatin Accessibility Modeler Based on ProbDep Transformer Encoder
3.2.1. Uniform Input Representation
3.2.2. ProbDep Transformer Encoder
3.2.3. Chromatin Accessibility Analyzer
3.2.4. Training and Implementation
3.3. Cell Type Annotator Based on Cell Embedding
3.4. scATAC-Seq Data Denoiser Based on Predicted Chromatin Accessibility
3.5. TF Activity Analyzer Based on Differential Accessibility Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baek, S.; Lee, I. Single-cell ATAC sequencing analysis: From data preprocessing to hypothesis generation. Comput. Struct. Biotechnol. J. 2020, 18, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Preissl, S.; Gaulton, K.J.; Ren, B. Characterizing cis-regulatory elements using single-cell epigenomics. Nat. Rev. Genet. 2022, 24, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Zhang, L.; Dollinger, E.; Peng, Q.; Nie, Q.; Xie, X. Predicting transcription factor binding in single cells through deep learning. Sci. Adv. 2020, 6, eaba9031. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, C.; Zhang, X. scDART: Integrating unmatched scRNA-seq and scATAC-seq data and learning cross-modality relationship simultaneously. Genome Biol. 2022, 23, 139. [Google Scholar] [CrossRef]
- Cao, Y.; Fu, L.; Wu, J.; Peng, Q.; Nie, Q.; Zhang, J.; Xie, X. SAILER: Scalable and accurate invariant representation learning for single-cell ATAC-seq processing and integration. Bioinformatics 2021, 37, i317–i326. [Google Scholar] [CrossRef]
- Li, Z.; Kuppe, C.; Ziegler, S.; Cheng, M.; Kabgani, N.; Menzel, S.; Zenke, M.; Kramann, R.; Costa, I.G. Chromatin-accessibility estimation from single-cell ATAC-seq data with scOpen. Nat. Commun. 2021, 12, 6386. [Google Scholar] [CrossRef]
- Fang, R.; Preissl, S.; Li, Y.; Lucero, J.; Wang, X.; Motamedi, A.; Shiau, A.K.; Zhou, X.; Xie, F.; Mukamel, E.A.; et al. Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun. 2021, 12, 1337. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Cieslak, M.C.; Castelfranco, A.M.; Roncalli, V.; Lenz, P.H.; Hartline, D.K. t-Distributed Stochastic Neighbor Embedding (t-SNE): A tool for eco-physiological transcriptomic analysis. Mar. Genom. 2020, 51, 100723. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Saul, N. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Huang, M.; Wang, J.; Torre, E.; Dueck, H.; Shaffer, S.; Bonasio, R.; Murray, J.I.; Raj, A.; Zhang, M.L.; Zhang, N.R. SAVER: Gene expression recovery for single-Cell RNA sequencing. Nat. Methods 2018, 15, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, D.; Sharma, R.; Nainys, J.; Yim, K.; Kathail, P.; Carr, A.J.; Burdziak, C.; Moon, K.R.; Chaffer, C.L.; Pattabiraman, D.; et al. Recovering gene interactions from single-cell data using data diffusion. Cell 2018, 174, 716–729. [Google Scholar] [CrossRef] [PubMed]
- Bravo González-Blas, C.; Minnoye, L.; Papasokrati, D.; Aibar, S.; Hulselmans, G.; Christiaens, V.; Davie, K.; Wouters, J.; Aerts, S. cisTopic: Cis-regulatory topic modeling on single-cell ATAC-seq data. Nat. Methods 2019, 16, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Xu, K.; Tian, K.; Shao, Y.; Tang, L.; Gao, G.; Zhang, M.; Jiang, T.; Zhang, Q.C. SCALE method for single-Cell ATAC-seq analysis via latent feature extraction. Nat. Commun. 2019, 10, 4576. [Google Scholar] [CrossRef] [PubMed]
- de Boer, C.G.; Regev, A. BROCKMAN: Deciphering variance in epigenomic regulators by k-mer factorization. BMC Bioinform. 2018, 19, 253. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Kelley, D.R. scBasset: Sequence-based modeling of single-cell ATAC-seq using convolutional neural networks. Nat. Methods 2022, 19, 1088–1096. [Google Scholar] [CrossRef]
- Kelley, D.R.; Snoek, J.; Rinn, J.L. Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016, 26, 990–999. [Google Scholar] [CrossRef]
- Zhou, J.; Troyanskaya, O.G. Predicting effects of noncoding variants with deep learning–Based sequence model. Nat. Methods 2015, 12, 931–934. [Google Scholar] [CrossRef]
- Kelley, D.R.; Reshef, Y.A.; Bileschi, M.; Belanger, D.; McLean, C.Y.; Snoek, J. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Res. 2018, 28, 739–750. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Liu, Y.; Lu, L.; Tan, X.; Zou, Q. By hybrid neural networks for prediction and interpretation of transcription factor binding sites based on multi-omics. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA, 9–12 December 2021; pp. 594–599. [Google Scholar]
- Wang, Z.; Tan, X.; Li, B.; Shao, Q.; Li, Z.; Yang, Y.; Zhang, Y. BindTransNet: A Transferable Transformer-Based Architecture for Cross-Cell Type DNA-Protein Binding Sites Prediction. In Proceedings of the International Symposium on Bioinformatics Research and Applications, Shenzhen, China, 26–28 November 2021; pp. 203–214. [Google Scholar]
- Beltagy, I.; Peters, M.E.; Cohan, A. Longformer: The long-document transformer. arXiv 2020, arXiv:2004.05150. [Google Scholar]
- Jiang, K.; Peng, P.; Lian, Y.; Xu, W. The encoding method of position embeddings in vision transformer. J. Vis. Commun. Image Rep. 2022, 89, 103664. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, R.; Li, T.; Jiang, J.; Ma, J.; Wang, P. MolRoPE-BERT: An enhanced molecular representation with Rotary Position Embedding for molecular property prediction. J. Mol. Graph. Model. 2022, 118, 108344. [Google Scholar] [CrossRef] [PubMed]
- Schep, A.N.; Wu, B.; Buenrostro, J.D.; Greenleaf, W.J. chromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 2017, 14, 975–978. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gong, M.; Liu, Y.; Xiong, S.; Wang, M.; Zhou, J.; Gong, M. Towards a better understanding of TF-DNA binding prediction from genomic features. Comput. Biol. Med. 2022, 149, 105993. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Zeng, Y.; Liu, Y.; Xiong, S.; Wang, M.; Zhou, J.; Zou, Q. A novel convolution attention model for predicting transcription factor binding sites by combination of sequence and shape. Brief. Bioinform. 2022, 23, bbab525. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Wang, Z.; Xiong, S.; Huang, G.; Gong, M. Uncovering the Relationship between Tissue-Specific TF-DNA Binding and Chromatin Features through a Transformer-Based Model. Genes 2022, 13, 1952. [Google Scholar] [CrossRef]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Pérez, N.M.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Wang, H.; Lee, C.H.; Qi, C.; Tailor, P.; Feng, J.; Abbasi, S.; Atsumi, T.; Morse, H.C. IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood J. Am. Soc. Hematol. 2008, 112, 4028–4038. [Google Scholar] [CrossRef]
- Arinobu, Y.; Mizuno, S.i.; Chong, Y.; Shigematsu, H.; Iino, T.; Iwasaki, H.; Graf, T.; Mayfield, R.; Chan, S.; Kastner, P.; et al. Reciprocal activation of GATA-1 and PU. 1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell 2007, 1, 416–427. [Google Scholar] [CrossRef]
- Kato, H.; Igarashi, K. To be red or white: Lineage commitment and maintenance of the hematopoietic system by the “inner myeloid”. Haematologica 2019, 104, 1919. [Google Scholar] [CrossRef]
- Jenal, M. HIC1 and BCL2A1: Novel Factors Involved in Myeloid Differentiation and Survival. Ph.D. Thesis, Universität Tübingen, Tübingen, Germany, 2009. [Google Scholar]
- Smith, B.W.; Rozelle, S.S.; Gadue, P.; Monti, S.; Chui, D.H.K.; Steinberg, M.H.; Frelinger, A.L.; Michelson, A.D.; Theberge, R.; McComb, M.E.; et al. The Aryl Hydrocarbon Receptor (AhR) Regulates the Production of Bipotential Hematopoietic Progenitor Cells. Blood 2012, 120, 766. [Google Scholar] [CrossRef]
- In’t Hout, F.E.; Gerritsen, M.; Bullinger, L.; Van der Reijden, B.A.; Huls, G.; Vellenga, E.; Jansen, J.H. Transcription factor 4 (TCF4) expression predicts clinical outcome in RUNX1 mutated and translocated acute myeloid leukemia. Haematologica 2020, 105, e454. [Google Scholar] [CrossRef] [PubMed]
- Mann-Nüttel, R.; Ali, S.; Petzsch, P.; Köhrer, K.; Alferink, J.; Scheu, S. The transcription factor reservoir and chromatin landscape in activated plasmacytoid dendritic cells. BMC Genom. Data 2021, 22, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Z.; Zeng, Y.; Zhou, J.; Zou, Q. High-resolution transcription factor binding sites prediction improved performance and interpretability by deep learning method. Brief. Bioinform. 2021, 22, bbab273. [Google Scholar] [CrossRef] [PubMed]
- Horak, C.E.; Mahajan, M.C.; Luscombe, N.M.; Gerstein, M.; Weissman, S.M.; Snyder, M. GATA-1 binding sites mapped in the β-globin locus by using mammalian chIp-chip analysis. Proc. Natl. Acad. Sci. USA 2002, 99, 2924–2929. [Google Scholar] [CrossRef]
- Zhang, K.; Hocker, J.D.; Miller, M.; Hou, X.; Chiou, J.; Poirion, O.B.; Qiu, Y.; Li, Y.E.; Gaulton, K.J.; Wang, A.; et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 2021, 184, 5985–6001. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Zhang, Y.; Yu, Y.; Zhang, J.; Liu, Y.; Zou, Q. A Unified Deep Learning Framework for Single-Cell ATAC-Seq Analysis Based on ProdDep Transformer Encoder. Int. J. Mol. Sci. 2023, 24, 4784. https://doi.org/10.3390/ijms24054784
Wang Z, Zhang Y, Yu Y, Zhang J, Liu Y, Zou Q. A Unified Deep Learning Framework for Single-Cell ATAC-Seq Analysis Based on ProdDep Transformer Encoder. International Journal of Molecular Sciences. 2023; 24(5):4784. https://doi.org/10.3390/ijms24054784
Chicago/Turabian StyleWang, Zixuan, Yongqing Zhang, Yun Yu, Junming Zhang, Yuhang Liu, and Quan Zou. 2023. "A Unified Deep Learning Framework for Single-Cell ATAC-Seq Analysis Based on ProdDep Transformer Encoder" International Journal of Molecular Sciences 24, no. 5: 4784. https://doi.org/10.3390/ijms24054784
APA StyleWang, Z., Zhang, Y., Yu, Y., Zhang, J., Liu, Y., & Zou, Q. (2023). A Unified Deep Learning Framework for Single-Cell ATAC-Seq Analysis Based on ProdDep Transformer Encoder. International Journal of Molecular Sciences, 24(5), 4784. https://doi.org/10.3390/ijms24054784






