The Important Role of m6A-Modified circRNAs in the Differentiation of Intramuscular Adipocytes in Goats Based on MeRIP Sequencing Analysis
Abstract
1. Introduction
2. Results
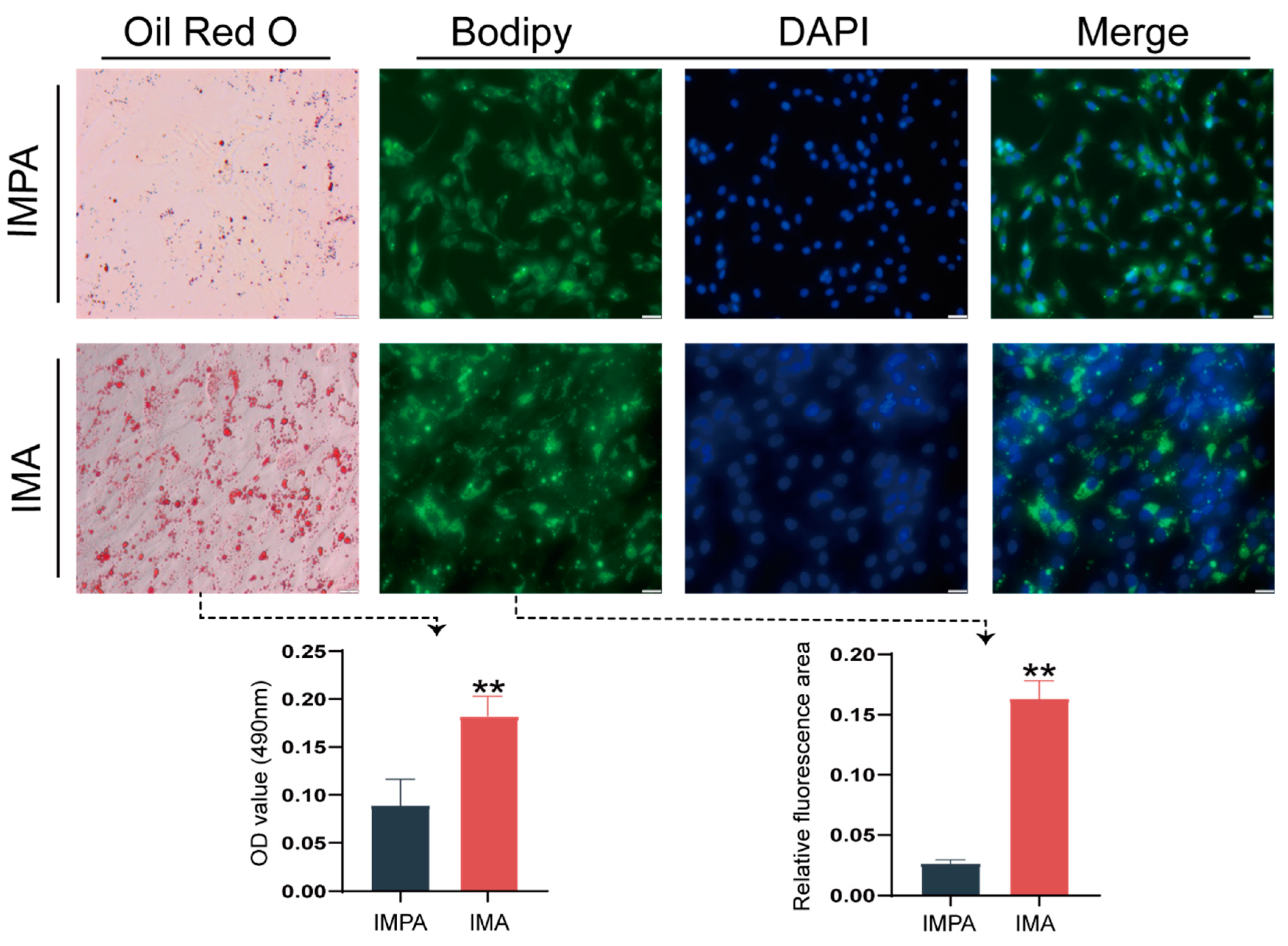
2.1. Identification of an Intramuscular Preadipocyte Differentiation Model in Goats
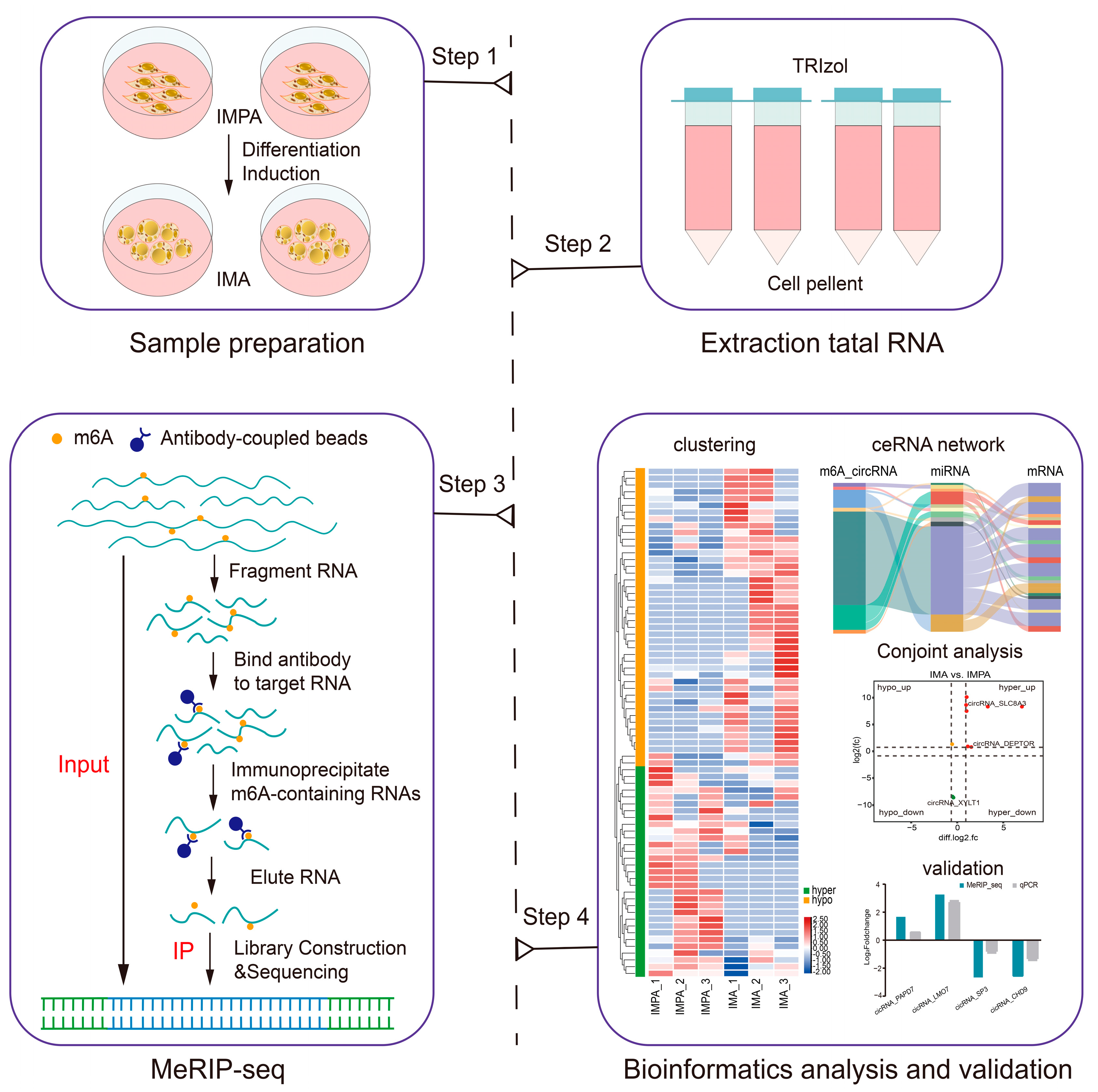
2.2. Overview of the circRNA-seq and MeRIP-seq Data
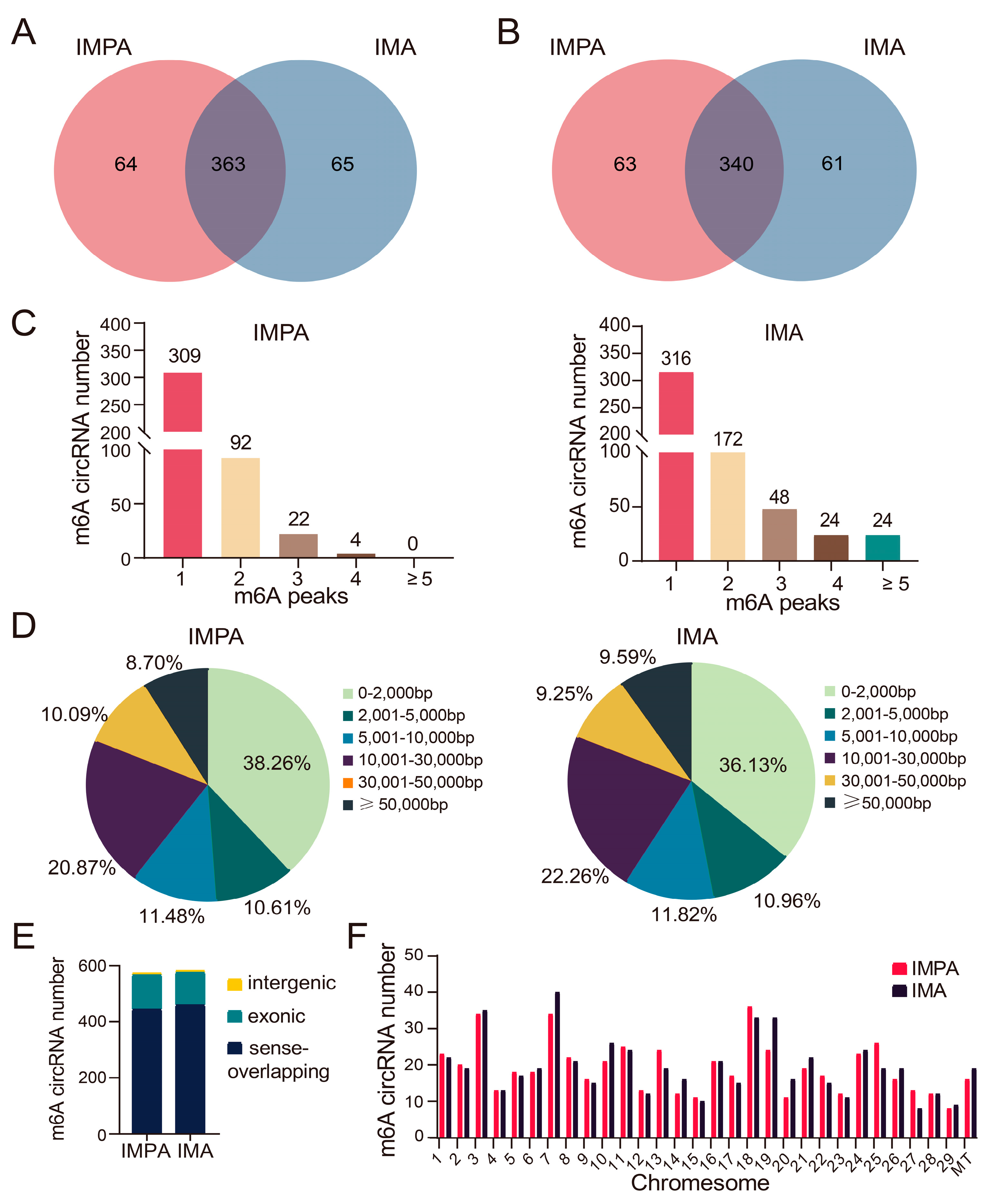
2.3. Characteristics of m6A-Modified circRNAs in the Intramuscular Adipocytes of Goats before and after Differentiation
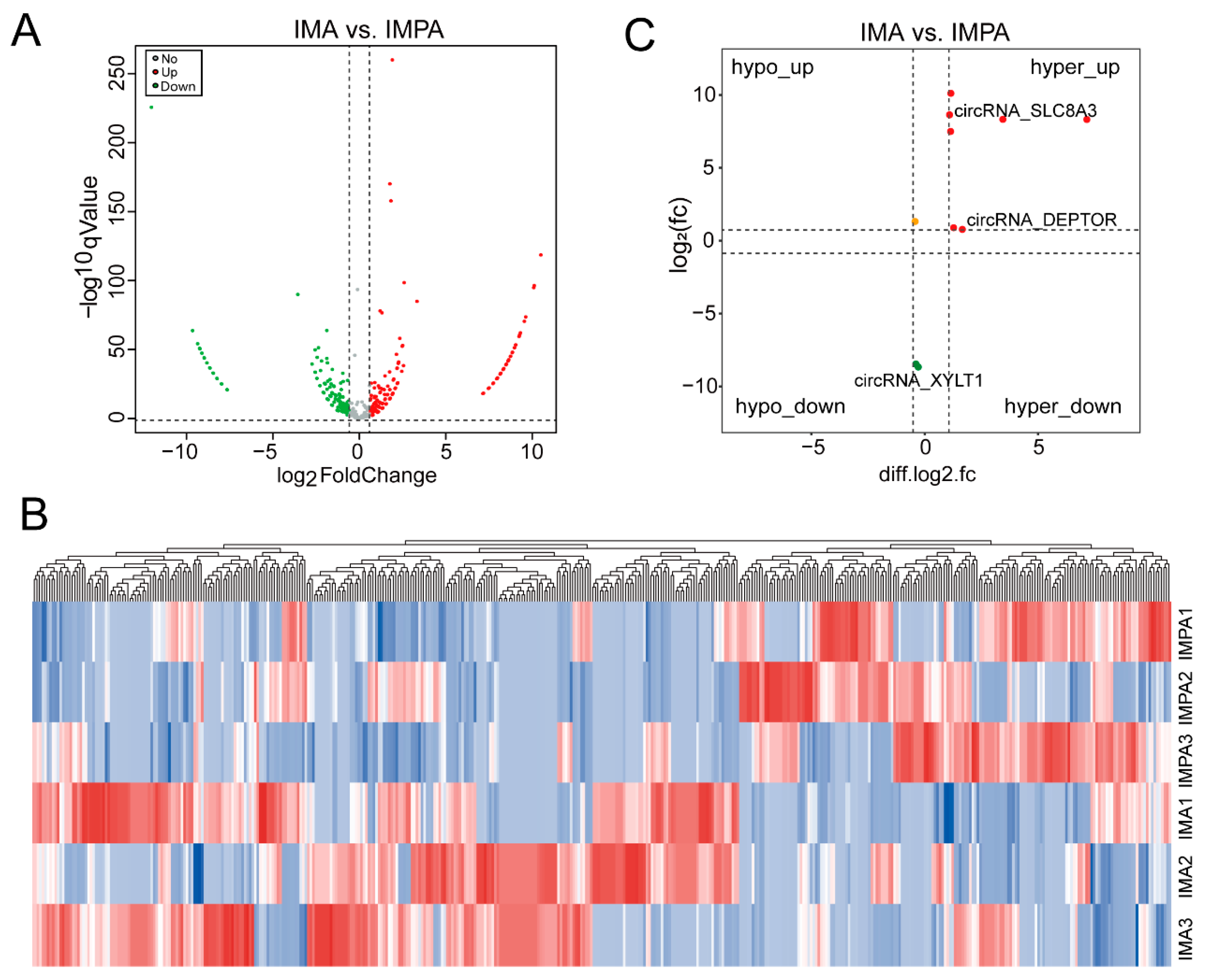
2.4. Differential Expression of m6A-Modified circRNAs in Different Goat Adipocytes
2.5. Regulatory Network of the Differential m6A-circRNAs between IMA and IMPA Groups
2.6. Conjoint Analysis of circRNA-Seq and MeRIP-Seq
2.7. Verification of circRNA Expression Profiles Using qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Isolation and Cell Culture of Goat Intramuscular Preadipocytes
4.2. Preadipocyte Differentiation Induction
4.3. Oil Red O and BODIPY Staining
4.4. RNA Extraction, Library Construction, and Sequencing
4.5. Sequencing Data Analysis
4.6. Bioinformatics Analysis and Statistical Analysis
4.7. Validation of Gene Expression by RT-qPCR Technique
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, K.; Liu, Y.; He, X.; Tao, L.; Jiang, Y.; Lan, R.; Hong, Q.; Chu, M. A Novel SNP in the Promoter Region of IGF1 Associated with Yunshang Black Goat Kidding Number via Promoting Transcription Activity by SP1. Front. Cell Dev. Biol. 2022, 10, 873095. [Google Scholar] [CrossRef]
- Yang, H.; Xu, X.L.; Ma, H.M.; Jiang, J. Integrative analysis of transcriptomics and proteomics of skeletal muscles of the Chinese indigenous Shaziling pig compared with the Yorkshire breed. BMC Genet. 2016, 17, 80. [Google Scholar] [CrossRef]
- Jiang, S.; Wei, H.; Song, T.; Yang, Y.; Peng, J.; Jiang, S. Transcriptome comparison between porcine subcutaneous and intramuscular stromal vascular cells during adipogenic differentiation. PLoS ONE 2013, 8, e77094. [Google Scholar] [CrossRef]
- Xiong, Y.; Xu, Q.; Lin, S.; Wang, Y.; Lin, Y.; Zhu, J. Knockdown of LXRα Inhibits Goat Intramuscular Preadipocyte Differentiation. Int. J. Mol. Sci. 2018, 19, 3037. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Zhang, B.; Zhong, H.; Nie, R.; Ling, Y.; Zhang, H.; Wu, C. Dynamic Expression and Regulatory Network of Circular RNA for Abdominal Preadipocytes Differentiation in Chicken (Gallus gallus). Front. Cell Dev. Biol. 2021, 9, 761638. [Google Scholar] [CrossRef]
- Zhao, R.; Ni, J.; Lu, S.; Jiang, S.; You, L.; Liu, H.; Shou, J.; Zhai, C.; Zhang, W.; Shao, S.; et al. CircUBAP2-mediated competing endogenous RNA network modulates tumorigenesis in pancreatic adenocarcinoma. Aging 2019, 11, 8484–8501. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef]
- Barbagallo, D.; Caponnetto, A.; Brex, D.; Mirabella, F.; Barbagallo, C.; Lauretta, G.; Morrone, A.; Certo, F.; Broggi, G.; Caltabiano, R.; et al. CircSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1. Cancers 2019, 11, 194. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef]
- Wesselhoeft, R.A.; Kowalski, P.S.; Parker-Hale, F.C.; Huang, Y.; Bisaria, N.; Anderson, D.G. RNA Circularization Diminishes Immunogenicity and Can Extend Translation Duration In Vivo. Mol. Cell 2019, 74, 508–520.e4. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, L.; Li, J.; Zhao, B.; Huang, G.; Li, X.; Xie, Z.; Zhou, Z. The Emerging Role of m6A Modification in Regulating the Immune System and Autoimmune Diseases. Front. Cell Dev. Biol. 2021, 9, 755691. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, J.; Wang, X.; Zhou, Z.; Liu, H.; Zhu, D. A Novel YTHDF3-Based Model to Predict Prognosis and Therapeutic Response in Breast Cancer. Front. Mol. Biosci. 2022, 9, 874532. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B.; et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cell 2019, 76, 96–109.e9. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, P.; Wang, J.; Xu, G.; Wang, T.; Feng, J.; Bei, Y.; Xu, J.; Wang, H.; Das, S.; et al. Downregulation of circ-ZNF609 Promotes Heart Repair by Modulating RNA N6-Methyladenosine-Modified Yap Expression. Research 2022, 2022, 9825916. [Google Scholar]
- Hui, T.; Zhu, Y.; Shen, J.; Bai, M.; Fan, Y.; Feng, S.; Wang, Z.; Zhao, J.; Zhang, Q.; Liu, X.; et al. Identification and Molecular Analysis of m6A-circRNAs from Cashmere Goat Reveal Their Integrated Regulatory Network and Putative Functions in Secondary Hair Follicle during Anagen Stage. Animals 2022, 12, 694. [Google Scholar] [CrossRef]
- Lee, E.J.; Lee, H.J.; Kamli, M.R.; Pokharel, S.; Bhat, A.R.; Lee, Y.H.; Choi, B.H.; Chun, T.; Kang, S.W.; Lee, Y.S.; et al. Depot-specific gene expression profiles during differentiation and transdifferentiation of bovine muscle satellite cells, and differentiation of preadipocytes. Genomics 2012, 100, 195–202. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Y.; Bao, X.; Zhu, X.; Kwok, Y.K.; Sun, K.; Chen, X.; Huang, Y.; Jauch, R.; Esteban, M.A.; et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015, 25, 335–350. [Google Scholar] [CrossRef]
- Gong, C.; Li, Z.; Ramanujan, K.; Clay, I.; Zhang, Y.; Lemire-Brachat, S.; Glass, D.J. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev. Cell 2015, 34, 181–191. [Google Scholar] [CrossRef]
- Qiu, X.; Gao, G.; Du, L.; Wang, J.; Wang, Q.; Yang, F.; Zhou, X.; Long, D.; Huang, J.; Liu, Z.; et al. Time-Series Clustering of lncRNA-mRNA Expression during the Adipogenic Transdifferentiation of Porcine Skeletal Muscle Satellite Cells. Curr. Issues Mol. Biol. 2022, 44, 2038–2053. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Lai, L.; Li, X.; Wang, L.; Li, A.; Yang, Q. N6-methyladenosine modification of circular RNA circ-ARL3 facilitates Hepatitis B virus-associated hepatocellular carcinoma via sponging miR-1305. IUBMB life 2021, 73, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Xie, Y.; Yu, T.; Liu, N.; Wang, Z.; Woolsey, R.J.; Tang, Y.; Zhang, X.; Qin, W.; Zhang, Y.; et al. m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 2020, 30, 211–228. [Google Scholar] [CrossRef]
- Guo, C.; Mi, J.; Li, H.; Su, P.; Nie, H. Dysregulated circular RNAs as novel biomarkers in esophageal squamous cell carcinoma: A meta-analysis. Cancer Med. 2021, 10, 7895–7908. [Google Scholar] [CrossRef]
- Ge, P.; Zhang, J.; Zhou, L.; Lv, M.-q.; Li, Y.-x.; Wang, J.; Zhou, D.-x. CircRNA expression profile and functional analysis in testicular tissue of patients with non-obstructive azoospermia. Reprod. Biol. Endocrinol. 2019, 17, 100. [Google Scholar] [CrossRef] [PubMed]
- Ebermann, C.; Schnarr, T.; Müller, S. Recent advances in understanding circular RNAs. F1000Research 2020, 9, F1000 Faculty Rev-655. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Maimaitiyiming, H.; Norman, H.; Zhou, Q.; Wang, S. CD47 deficiency protects mice from diet-induced obesity and improves whole body glucose tolerance and insulin sensitivity. Sci. Rep. 2015, 5, 8846. [Google Scholar] [CrossRef]
- Sun, S.; Luo, J.; Du, H.; Liu, G.; Liu, M.; Wang, J.; Han, S.; Che, H. Widely Targeted Lipidomics and Transcriptomics Analysis Revealed Changes of Lipid Metabolism in Spleen Dendritic Cells in Shrimp Allergy. Foods 2022, 11, 1882. [Google Scholar] [CrossRef]
- Hong, S.M.; Hwang, J.H.; Kim, I.H. Evaluation of the effect of low dietary fermentable carbohydrate content on growth performance, nutrient digestibility, blood characteristics, and meat quality in finishing pigs. Asian-Australas. J. Anim. Sci. 2012, 25, 1294–1299. [Google Scholar] [CrossRef]
- Yang, S.W.; Kim, M.H.; Choi, J.S.; Jin, S.K.; Park, M.J.; Song, Y.M.; Lee, C.Y. Effects of the plane of nutrition during the latter grower and entire finisher phases on grow-finish pig performance in summer. J. Anim. Sci. 2019, 61, 10–17. [Google Scholar] [CrossRef]
- Chen, X.; Yang, T.; Wang, W.; Xi, W.; Zhang, T.; Li, Q.; Yang, A.; Wang, T. Circular RNAs in immune responses and immune diseases. Theranostics 2019, 9, 588–607. [Google Scholar] [CrossRef]
- Feng, H.; Yousuf, S.; Liu, T.; Zhang, X.; Huang, W.; Li, A.; Xie, L.; Miao, X. The comprehensive detection of miRNA and circRNA in the regulation of intramuscular and subcutaneous adipose tissue of Laiwu pig. Sci. Rep. 2022, 12, 16542. [Google Scholar] [CrossRef] [PubMed]
- Wrann, C.D.; White, J.P.; Salogiannnis, J.; Laznik-Bogoslavski, D.; Wu, J.; Ma, D.; Lin, J.D.; Greenberg, M.E.; Spiegelman, B.M. Exercise induces hippocampal BDNF through a PGC-1α/FNDC5 pathway. Cell Metab. 2013, 18, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Nagai, K.; Nagata, Y.; Doronbekov, K.; Nishimura, S.; Yoshioka, S.; Fujita, T.; Shiga, K.; Miyake, T.; Taniguchi, Y.; et al. Exploration of genes showing intramuscular fat deposition-associated expression changes in musculus longissimus muscle. Anim. Genet. 2006, 37, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.C.; Derks, M.F.L.; Lopes, M.S.; Madsen, O.; Harlizius, B.; van Son, M.; Grindflek, E.H.; Gòdia, M.; Gjuvsland, A.B.; Otto, P.I.; et al. Fine Mapping of a Major Backfat QTL Reveals a Causal Regulatory Variant Affecting the CCND2 Gene. Front. Genet. 2022, 13, 871516. [Google Scholar] [CrossRef]
- Duarte, I.N.H.; Bessa, A.F.O.; Rola, L.D.; Genuíno, M.V.H.; Rocha, I.M.; Marcondes, C.R.; Regitano, L.C.A.; Munari, D.P.; Berry, D.P.; Buzanskas, M.E. Cross-population selection signatures in Canchim composite beef cattle. PLoS ONE 2022, 17, e0264279. [Google Scholar] [CrossRef]
- Yi, Y.; Hu, W.; Zhao, C.; Wu, M.; Zeng, H.; Xiong, M.; Lv, W.; Wu, Y.; Zhang, Q. Deciphering the Emerging Roles of Adipocytes and Adipose-Derived Stem Cells in Fat Transplantation. Cell Transplant. 2021, 30, 963689721997799. [Google Scholar] [CrossRef]
- Xu, H.; Lin, C.; Li, T.; Zhu, Y.; Yang, J.; Chen, S.; Chen, J.; Chen, X.; Chen, Y.; Guo, A.; et al. N6-Methyladenosine-Modified circRNA in the Bovine Mammary Epithelial Cells Injured by Staphylococcus aureus and Escherichia coli. Front. Immunol. 2022, 13, 873330. [Google Scholar] [CrossRef]
- Zhang, C.; Cui, H.; Huang, C.; Kong, F.; Yang, Q.; Miao, P.; Cao, Z.; Zhang, W.; Chang, D. Interactions of circRNAs with methylation: An important aspect of circRNA biogenesis and function (Review). Mol. Med. Rep. 2022, 25, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Lin, S.; Wang, Y.; Zhu, J.; Lin, Y. Fibroblast growth factor 10 (FGF10) promotes the adipogenesis of intramuscular preadipocytes in goat. Mol. Biol. Rep. 2018, 45, 1881–1888. [Google Scholar] [CrossRef]
- Li, X.; Zhang, H.; Wang, Y.; Li, Y.; He, C.; Zhu, J.; Xiong, Y.; Lin, Y. RNA-seq analysis reveals the positive role of KLF5 in the differentiation of subcutaneous adipocyte in goats. Gene 2022, 808, 145969. [Google Scholar] [CrossRef]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, 1–11. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Lin, S.; Zhu, J.; Wang, Y.; Lin, Y. The expression stability analysis of reference genes in the process of goat intramuscular preadipocytes differentiation in goat. Acta Vet. Zootech. Sin. 2018, 49, 907–918. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Sample | Raw_Reads(M) | Clean_Reads(M) | Clean_Bases(G) | Valid_Bases(%) | Q30(%) | GC(%) |
|---|---|---|---|---|---|---|
| IMA1 | 50.38 | 49.57 | 14.17 | 93.75 | 94.08 | 60.94 |
| IMA2 | 50.11 | 49.27 | 14.05 | 93.47 | 93.83 | 60.63 |
| IMA3 | 50.3 | 49.49 | 14.22 | 94.24 | 93.82 | 60.74 |
| IMPA1 | 49.83 | 49.01 | 13.99 | 93.58 | 94.03 | 61.13 |
| IMPA2 | 49.37 | 48.62 | 14.02 | 94.66 | 94.02 | 60.68 |
| IMPA3 | 50.31 | 49.49 | 14.11 | 93.49 | 93.89 | 61.01 |
| IMA1_input | 47.97 | 47.05 | 13.23 | 91.94 | 95.93 | 57.21 |
| IMA2_input | 49.37 | 48.39 | 13.51 | 91.21 | 96.04 | 56.96 |
| IMA3_input | 47.29 | 46.34 | 12.98 | 91.49 | 96.1 | 57.41 |
| IMPA1_input | 47.73 | 46.82 | 13.16 | 91.9 | 95.94 | 57.5 |
| IMPA2_input | 48.35 | 47.44 | 13.31 | 91.77 | 95.99 | 56.77 |
| IMPA3_input | 47.88 | 46.89 | 13.07 | 90.99 | 95.87 | 57.06 |
| Sample | Total_Reads | Total_Mapped | Multiple_Mapped | Uniquely_Mapped |
|---|---|---|---|---|
| IMA1 | 99,139,058 | 95,401,484 (96.22%) | 4,848,347 (4.89%) | 90,553,137 (91.33%) |
| IMA2 | 98,545,924 | 94,865,327 (96.26%) | 4,698,299 (4.76%) | 90,167,028 (91.49%) |
| IMA3 | 98,984,968 | 95,084,400 (96.05%) | 5,016,142 (5.06%) | 90,068,258 (90.99%) |
| IMPA1 | 98,018,414 | 94,268,208 (96.17%) | 5,241,506 (5.34%) | 89,026,702 (90.82%) |
| IMPA2 | 97,232,168 | 93,538,409 (96.20%) | 4,702,985 (4.83%) | 88,835,424 (91.36%) |
| IMPA3 | 98,974,940 | 95,109,577 (96.09%) | 5,017,553 (5.06%) | 90,092,024 (91.02%) |
| IMA1_input | 94,096,060 | 89,868,600 (95.50%) | 10,729,437 (11.40%) | 79,139,163 (84.10%) |
| IMA2_input | 96,775,848 | 92,699,532 (95.78%) | 10,800,206 (11.16%) | 81,899,326 (84.62%) |
| IMA3_input | 92,682,072 | 88,584,369 (95.57%) | 10,356,897 (11.17%) | 78,227,472 (84.40%) |
| IMPA1_input | 93,638,328 | 89376,862 (95.44%) | 10,923,483 (11.66%) | 78,453,379 (83.78%) |
| IMPA2_input | 94,873,142 | 90,865,112 (95.77%) | 10,783,550 (11.36%) | 80,081,562 (84.40%) |
| IMPA3_input | 93,789,268 | 89,386,330 (95.30%) | 10,808,685 (11.52%) | 78,577,645 (83.78%) |
| Chr | Start | End | circRNA | Log2FC | p-Value | Regulation | Gene Name |
|---|---|---|---|---|---|---|---|
| Chr12 | 35,549,676 | 35,551,085 | circRNA_1055 | 3.32478924 | 3.12E-87 | Up | LMO7 |
| Chr18 | 56,511,885 | 56,517,614 | circRNA_1438 | 9.255672903 | 1.11E-62 | Up | NUCB1 |
| Chr13 | 33,128,553 | 33,129,648 | circRNA_1119 | 2.485573285 | 5.46E-55 | Up | ZEB1 |
| Chr10 | 21,218,634 | 21,218,882 | circRNA_0873 | 8.626127066 | 2.59E-44 | Up | SLC8A3 |
| Chr10 | 16,161,014 | 16,161,210 | circRNA_0866 | 2.253712935 | 1.31E-42 | Up | LOC102187597 |
| Chr3 | 10,437,366 | 10,437,666 | circRNA_0238 | 8.487830648 | 4.97E-41 | Up | ZMYM4 |
| Chr11 | 74,783,037 | 74,799,539 | circRNA_1012 | 8.339699789 | 9.02E-38 | Up | ATAD2B |
| Chr27 | 12,234,705 | 12,235,515 | circRNA_1944 | 8.329999645 | 1.44E-37 | Up | WHSC1L1 |
| Chr13 | 61,360,331 | 61,366,664 | circRNA_1079 | 8.30930326 | 3.91E-37 | Up | DCLK1 |
| Chr20 | 66,412,450 | 66,418,587 | circRNA_1583 | 1.733511924 | 3.03E-35 | Up | PAPD7 |
| Chr18 | 23,020,758 | 23,021,057 | circRNA_1399 | 2.578567264 | 1.12E-51 | Down | CHD9 |
| Chr13 | 75,314,614 | 75,354,619 | circRNA_1149 | 9.125673331 | 3.81E-49 | Down | ZMYND8 |
| Chr19 | 53,784,636 | 53,812,138 | circRNA_1531 | 8.986718904 | 1.48E-45 | Down | SEPTIN9 |
| Chr18 | 3,010,918 | 3,011,590 | circRNA_1385 | 2.205131135 | 2.40E-43 | Down | DDX19A |
| Chr2 | 113,508,380 | 113,508,876 | circRNA_0178 | 2.756039782 | 4.60E-41 | Down | SP3 |
| Chr26 | 11,981,485 | 11,988,759 | circRNA_1905 | 8.661060515 | 4.30E-38 | Down | SEC23IP |
| Chr8 | 40,220,320 | 40,221,369 | circRNA_0743 | 2.262451821 | 3.39E-36 | Down | GLIS3 |
| Chr8 | 76,948,426 | 76,949,174 | circRNA_0762 | 8.473007382 | 2.47E-34 | Down | KIF27 |
| Chr20 | 13,792,302 | 13,792,500 | circRNA_1563 | 8.471672577 | 2.62E-34 | Down | NLN |
| Chr17 | 9,726,962 | 9,729,487 | circRNA_1343 | 8.469304477 | 2.91E-34 | Down | LOC102190983 |
| Gene Name | Primer Sequence | TM/°C | Product Length |
|---|---|---|---|
| UXT | GCAAGTGGATTTGGGCTGTAAC | 60 | 180 |
| ATGGAGTCCTTGGTGAGGTTGT | 60 | ||
| circRNA_LMO7 | TCAAAGTTAGTGTCTGGCAATG | 55.7 | 227 |
| GTGCTGGACTTTTGTGGG | 55.6 | ||
| circRNA_PAPD7 | TACATCCCAGCACCTAACC | 52.9 | 180 |
| CATCGGTCTGTTCCATCC | 53 | ||
| circRNA_CHD9 | ACTGCCAGTTCCCGTGACA | 59 | 235 |
| GAAGGGGTTCGTAGCAGCG | 60.6 | ||
| circRNA_SP3 | GGGTCCTTGTGGGGCTTAC | 59 | 237 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Li, X.; Qubi, W.; Li, Y.; Wang, Y.; Wang, Y.; Lin, Y. The Important Role of m6A-Modified circRNAs in the Differentiation of Intramuscular Adipocytes in Goats Based on MeRIP Sequencing Analysis. Int. J. Mol. Sci. 2023, 24, 4817. https://doi.org/10.3390/ijms24054817
Wang J, Li X, Qubi W, Li Y, Wang Y, Wang Y, Lin Y. The Important Role of m6A-Modified circRNAs in the Differentiation of Intramuscular Adipocytes in Goats Based on MeRIP Sequencing Analysis. International Journal of Molecular Sciences. 2023; 24(5):4817. https://doi.org/10.3390/ijms24054817
Chicago/Turabian StyleWang, Jianmei, Xin Li, Wuqie Qubi, Yanyan Li, Yong Wang, Youli Wang, and Yaqiu Lin. 2023. "The Important Role of m6A-Modified circRNAs in the Differentiation of Intramuscular Adipocytes in Goats Based on MeRIP Sequencing Analysis" International Journal of Molecular Sciences 24, no. 5: 4817. https://doi.org/10.3390/ijms24054817
APA StyleWang, J., Li, X., Qubi, W., Li, Y., Wang, Y., Wang, Y., & Lin, Y. (2023). The Important Role of m6A-Modified circRNAs in the Differentiation of Intramuscular Adipocytes in Goats Based on MeRIP Sequencing Analysis. International Journal of Molecular Sciences, 24(5), 4817. https://doi.org/10.3390/ijms24054817






