Blocking the Hormone Receptors Modulates NLRP3 in LPS-Primed Breast Cancer Cells
Abstract
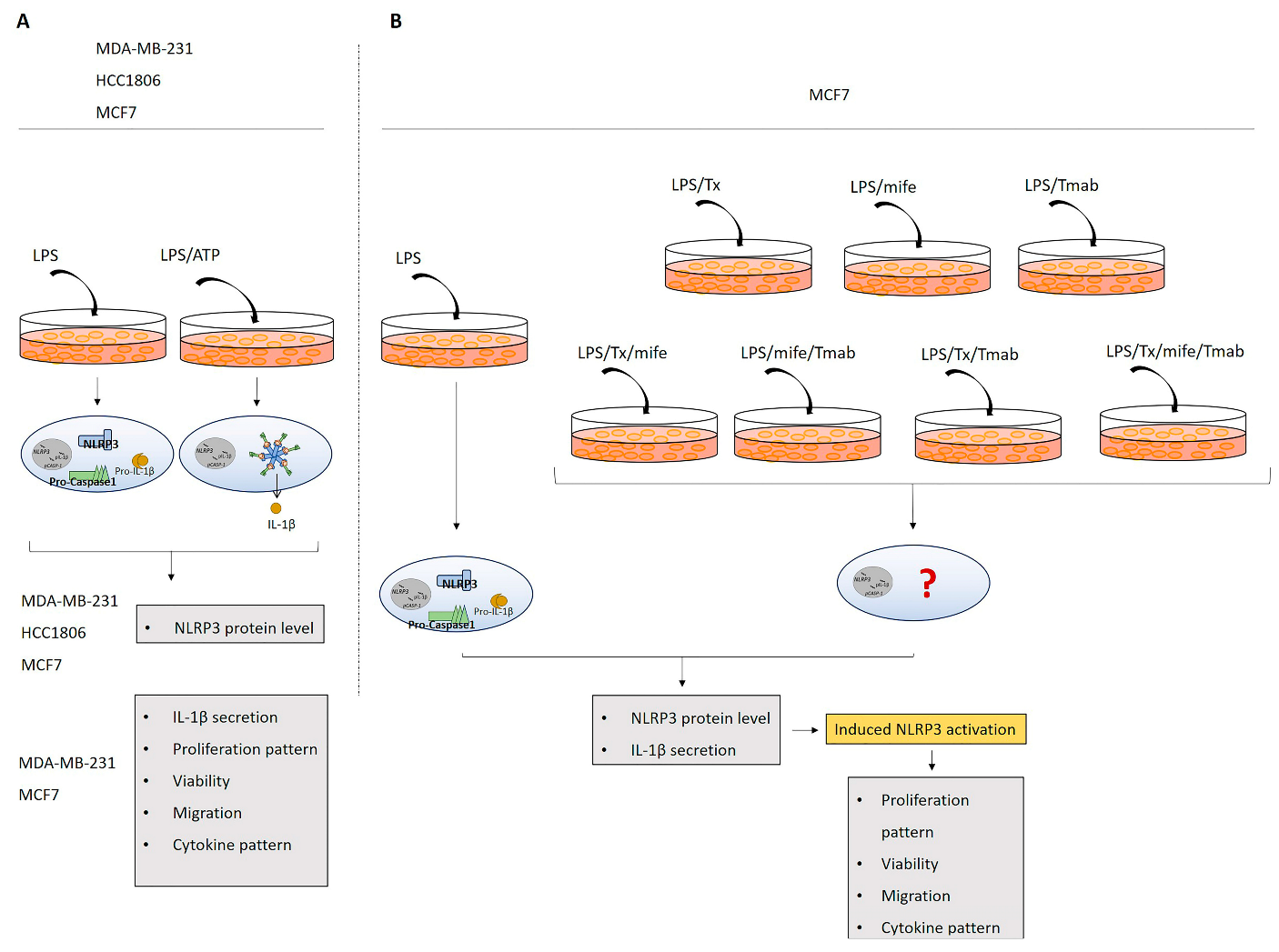
:1. Introduction
2. Results
2.1. Transcriptional Levels of NLRP3 in BC Tumors and Cell Lines
2.2. NLRP3 Protein Levels Vary in TNBC and Luminal A Type Cells
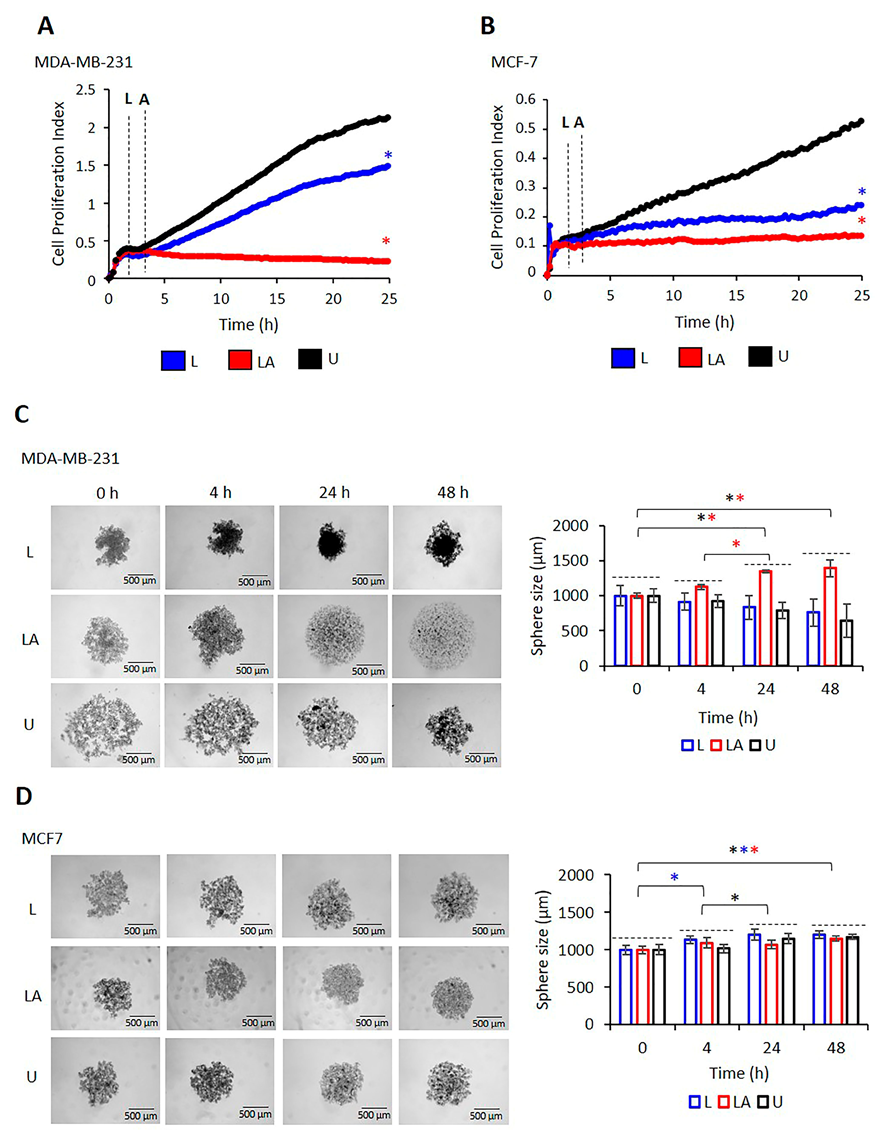
2.3. The Proliferation of MDA-MB-231 and MCF-7 Cells Differ after NLRP3 Activation
2.4. NLRP3 Expression Reduces the Migration Capacity of BC Cells
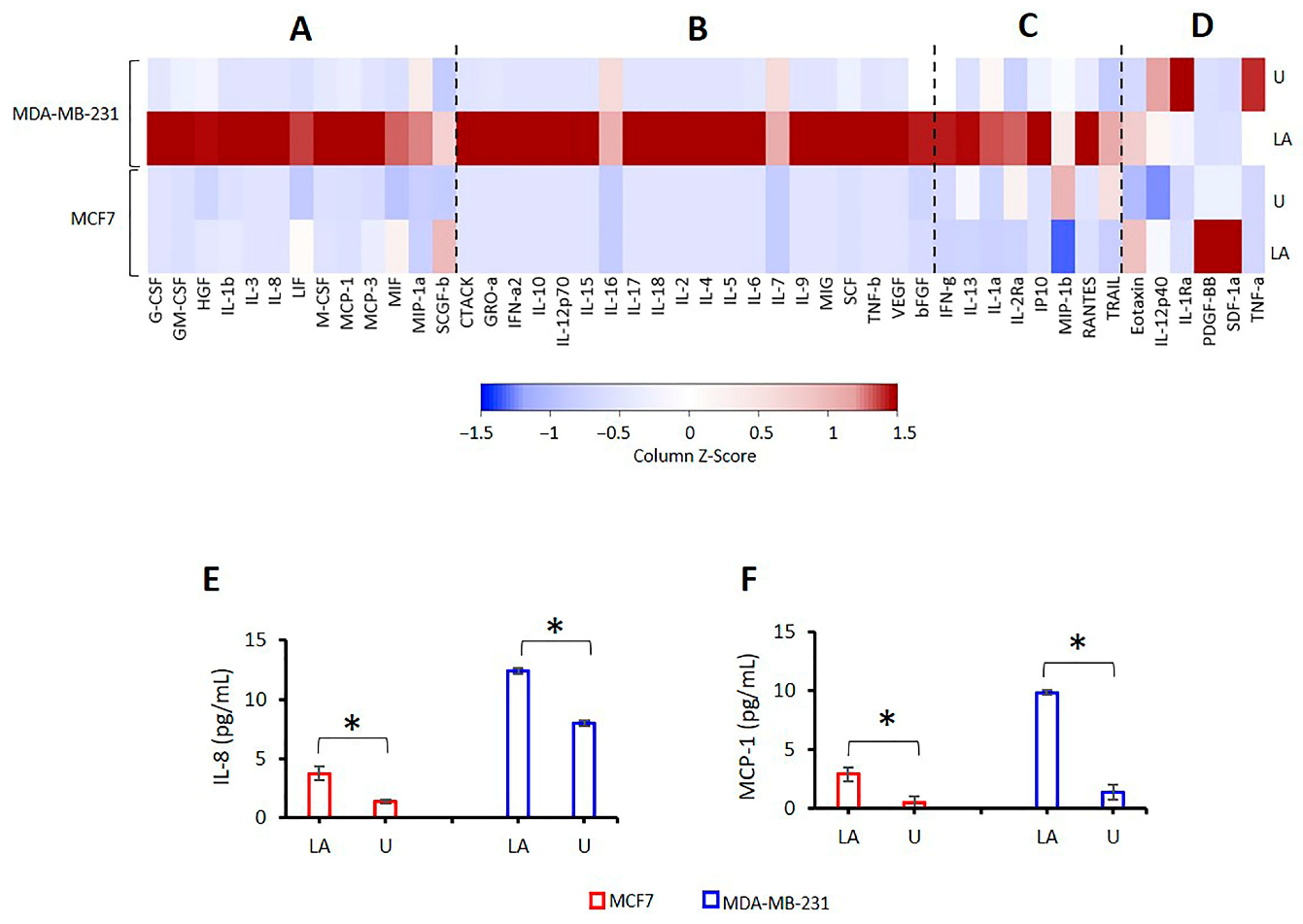
2.5. Effect of NLRP3 Activation on Cytokine Release in BC Cells
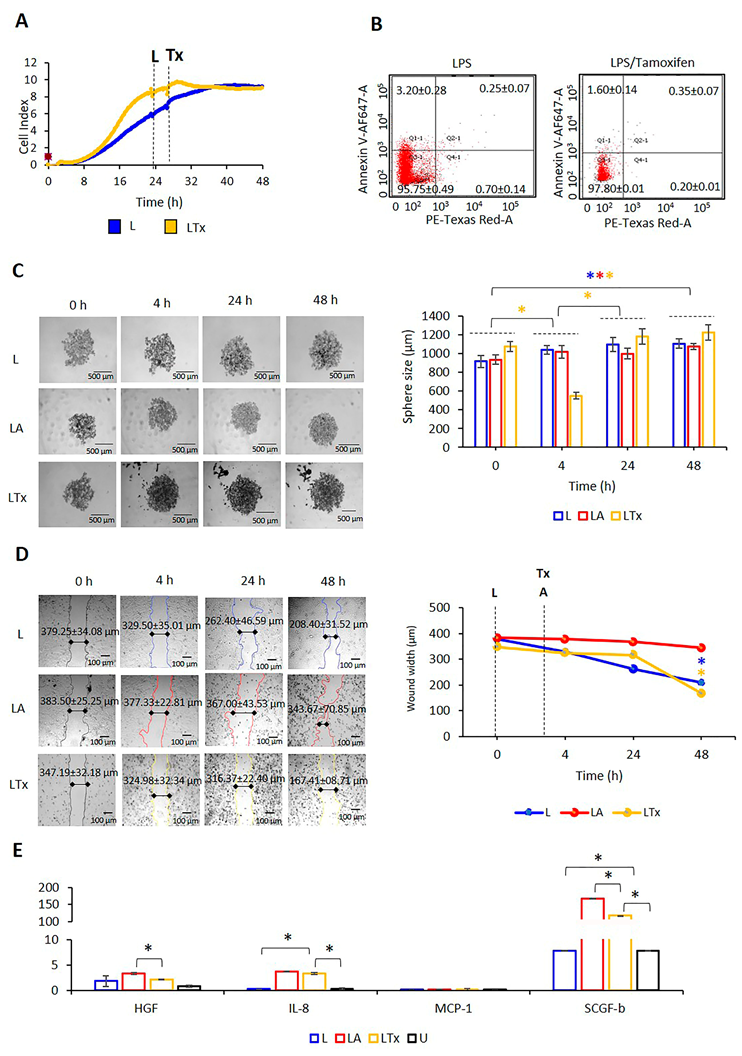
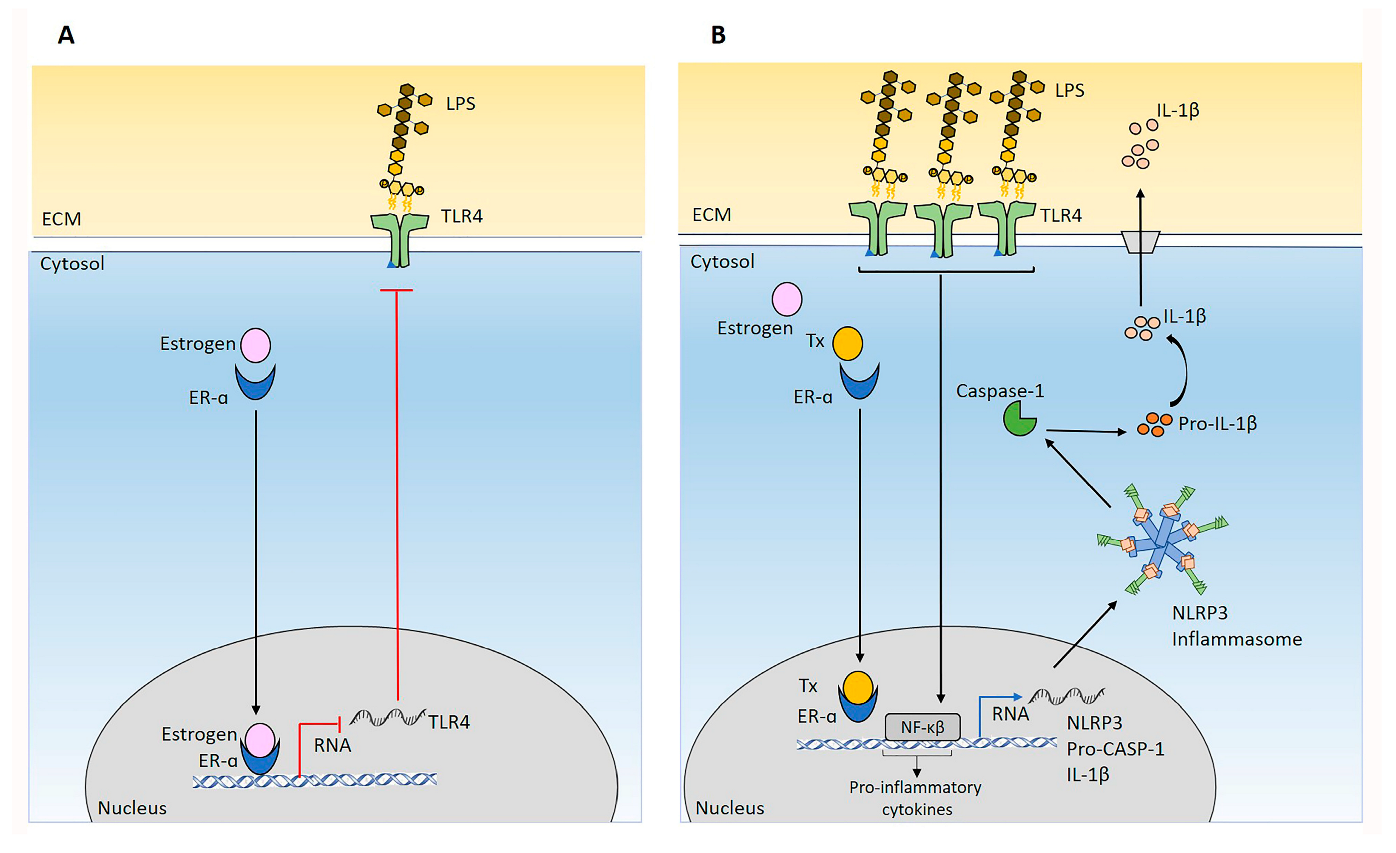
2.6. Targeting Hormone Receptors affect NLRP3 Status in MCF7 Cells
2.7. NLRP3 Activation Combined with Tx Blocking ER-α Promotes MCF7 Cells Migration and Proliferation
3. Discussion
4. Material and Methods
4.1. The Transcriptomic Profiling of NLRP3 in BC
4.2. Cell Lines and Reagents
4.3. Immunoblotting
4.4. Enzyme-Linked Immunosorbent Assay (ELISA)
4.5. Live-Cell Monitoring
4.6. Annexin V Assay
4.7. Scratch Wound Healing Assay
4.8. Sphere Formation Analysis
4.9. Cytokine and Chemokine Analysis
4.10. In Vitro Experimental Design
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bauernfeind, F.G.; Horvath, G.; Stutz, A.; Alnemri, E.S.; MacDonald, K.; Speert, D.; Fernandes-Alnemri, T.; Wu, J.; Monks, B.G.; Fitzgerald, K.A.; et al. Cutting Edge: NF-κB Activating Pattern Recognition and Cytokine Receptors License NLRP3 Inflammasome Activation by Regulating NLRP3 Expression. J. Immunol. 2009, 183, 787–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dostert, C.; Pétrilli, V.; van Bruggen, R.; Steele, C.; Mossman, B.T.; Tschopp, J. Innate Immune Activation through Nalp3 Inflammasome Sensing of Asbestos and Silica. Science 2008, 320, 674–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piccini, A.; Carta, S.; Tassi, S.; Lasiglié, D.; Fossati, G.; Rubartelli, A. ATP is released by monocytes stimulated with pathogen-sensing receptor ligands and induces IL-1β and IL-18 secretion in an autocrine way. Proc. Natl. Acad. Sci. USA 2008, 105, 8067–8072. [Google Scholar] [CrossRef] [Green Version]
- Kantono, M.; Guo, B. Inflammasomes and Cancer: The Dynamic Role of the Inflammasome in Tumor Development. Front. Immunol. 2017, 8, 1132. [Google Scholar] [CrossRef] [Green Version]
- Karki, R.; Man, S.M.; Kanneganti, T.-D. Inflammasomes and Cancer. Cancer Immunol. Res. 2017, 5, 94–99. [Google Scholar] [CrossRef] [Green Version]
- Dupaul-Chicoine, J.; Arabzadeh, A.; Dagenais, M.; Douglas, T.; Champagne, C.; Morizot, A.; Rodrigue-Gervais, I.G.; Breton, V.; Colpitts, S.L.; Beauchemin, N.; et al. The Nlrp3 Inflammasome Suppresses Colorectal Cancer Metastatic Growth in the Liver by Promoting Natural Killer Cell Tumoricidal Activity. Immunity 2015, 43, 751–763. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Li, S.; Qi, J.; Chen, Z.; Wu, Y.; Guo, J.; Wang, K.; Sun, X.; Zheng, J. Cleavage of GSDME by caspase-3 determines lobaplatin-induced pyroptosis in colon cancer cells. Cell Death Dis. 2019, 10, 193. [Google Scholar] [CrossRef] [Green Version]
- Tezcan, G.; Garanina, E.E.; Alsaadi, M.; Gilazieva, Z.E.; Martinova, E.V.; Markelova, M.I.; Arkhipova, S.S.; Hamza, S.; McIntyre, A.; Rizvanov, A.A.; et al. Therapeutic Potential of Pharmacological Targeting NLRP3 Inflammasome Complex in Cancer. Front. Immunol. 2021, 11, 607881. [Google Scholar] [CrossRef]
- Zheng, Q.; Yao, D.; Cai, Y.; Zhou, T. NLRP3 augmented resistance to gemcitabine in triple-negative breast cancer cells via EMT/IL-1β/Wnt/β-catenin signaling pathway. Biosci. Rep. 2020, 40, BSR20200730. [Google Scholar] [CrossRef]
- Feng, X.; Luo, Q.; Zhang, H.; Wang, H.; Chen, W.; Meng, G.; Chen, F. The role of NLRP3 inflammasome in 5-fluorouracil resistance of oral squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 81. [Google Scholar] [CrossRef] [Green Version]
- Bruchard, M.; Mignot, G.; Derangère, V.; Chalmin, F.; Chevriaux, A.; Vegran, F.; Boireau, W.; Simon, B.; Ryffel, B.; Connat, J.L.; et al. Chemotherapy-triggered cathepsin B release in myeloid-derived suppressor cells activates the Nlrp3 inflammasome and promotes tumor growth. Nat. Med. 2012, 19, 57–64. [Google Scholar] [CrossRef]
- Mørch, L.S.; Løkkegaard, E.; Andreasen, A.H.; Krüger-Kjær, S.; Lidegaard, O. Hormone Therapy and Ovarian Cancer. JAMA 2009, 302, 298–305. [Google Scholar] [CrossRef] [Green Version]
- Drăgănescu, M.; Carmocan, C. Hormone Therapy in Breast Cancer. Chirurgia 2017, 112, 413–417. [Google Scholar] [CrossRef]
- Whitmore, W.F. Hormone therapy in prostatic cancer. Am. J. Med. 1956, 21, 697–713. [Google Scholar] [CrossRef]
- Fisher, B.; Dignam, J.; Emir, B.; Bryant, J.; DeCillis, A.; Wolmark, N.; Wickerham, D.L.; Dimitrov, N.V.; Abramson, N.; Atkins, J.N.; et al. Tamoxifen and Chemotherapy for Lymph Node-Negative, Estrogen Receptor-Positive Breast Cancer. Gynecol. Oncol. 1997, 89, 1673–1682. [Google Scholar] [CrossRef]
- Farrar, M.C.; Jacobs, T.F. Tamoxifen; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Goyeneche, A.A.; Carón, R.W.; Telleria, C.M. Mifepristone Inhibits Ovarian Cancer Cell Growth in vitro and in vivo. Clin. Cancer Res. 2007, 13, 3370–3379. [Google Scholar] [CrossRef] [Green Version]
- Elía, A.; Saldain, L.; Vanzulli, S.I.; Helguero, L.A.; Lamb, C.A.; Fabris, V.; Pataccini, G.; Martínez-Vazquez, P.; Burruchaga, J.; Caillet-Bois, I.; et al. Beneficial Effects of Mifepristone Treatment in Patients with Breast Cancer Selected by the Progesterone Receptor Isoform Ratio: Results from the MIPRA Trial. Clin. Cancer Res. 2022, OF1–OF12. [Google Scholar] [CrossRef] [PubMed]
- Zahavi, D.; Weiner, L. Monoclonal Antibodies in Cancer Therapy. Antibodies 2020, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Emens, L.A.; Davidson, N.E. Trastuzumab in breast cancer. Oncology 2004, 18, 1117–1128. [Google Scholar] [PubMed]
- Davidson, M.; Starling, N. Trastuzumab in the management of gastroesophageal cancer: Patient selection and perspectives. OncoTargets Ther. 2016, 9, 7235–7245. [Google Scholar] [CrossRef] [Green Version]
- Meza-Junco, J.; Au, H.-J.; Sawyer, M.B. Trastuzumab for gastric cancer. Expert Opin. Biol. Ther. 2009, 9, 1543–1551. [Google Scholar] [CrossRef] [PubMed]
- Shou, J.; Massarweh, S.; Osborne, C.K.; Wakeling, A.E.; Ali, S.; Weiss, H.; Schiff, R. Mechanisms of Tamoxifen Resistance: Increased Estrogen Receptor-HER2/neu Cross-Talk in ER/HER2-Positive Breast Cancer. Gynecol. Oncol. 2004, 96, 926–935. [Google Scholar] [CrossRef] [Green Version]
- Onitilo, A.A.; Engel, J.M.; Greenlee, R.T.; Mukesh, B.N. Breast Cancer Subtypes Based on ER/PR and Her2 Expression: Comparison of Clinicopathologic Features and Survival. Clin. Med. Res. 2009, 7, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.D.; Warren, R.L.; Gibb, E.A.; Martin, S.D.; Spinelli, J.J.; Nelson, B.H.; Holt, R.A. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014, 24, 743–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, B.; Chin, S.-F.; Rueda, O.M.; Vollan, H.-K.M.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.-J.; et al. The somatic mutation profiles of 2433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef] [Green Version]
- Mitri, Z.; Constantine, T.; O’Regan, R. The HER2 Receptor in Breast Cancer: Pathophysiology, Clinical Use, and New Advances in Therapy. Chemother. Res. Pract. 2012, 2012, 743193. [Google Scholar] [CrossRef] [Green Version]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-Negative Breast Cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [Green Version]
- Heldring, N.; Nilsson, M.; Buehrer, B.; Treuter, E.; Gustafsson, J.-A. Identification of Tamoxifen-Induced Coregulator Interaction Surfaces within the Ligand-Binding Domain of Estrogen Receptors. Mol. Cell. Biol. 2004, 24, 3445–3459. [Google Scholar] [CrossRef] [Green Version]
- Perrault, D.; Eisenhauer, E.A.; Pritchard, K.I.; Panasci, L.; Norris, B.; Vandenberg, T.; Fisher, B. Phase II study of the progesterone antagonist mifepristone in patients with untreated metastatic breast carcinoma: A National Cancer Institute of Canada Clinical Trials Group study. J. Clin. Oncol. 1996, 14, 2709–2712. [Google Scholar] [CrossRef]
- Rossi, M.; Carioli, G.; Bonifazi, M.; Zambelli, A.; Franchi, M.; Moja, L.; Zambon, A.; Corrao, G.; La Vecchia, C.; Zocchetti, C.; et al. Trastuzumab for HER2+ metastatic breast cancer in clinical practice: Cardiotoxicity and overall survival. Eur. J. Cancer 2015, 52, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.J.; Jung, S.J.; Kim, T.H.; Oh, M.K.; Yoon, H.-K. Differences in Clinical Outcomes between Luminal A and B Type Breast Cancers according to the St. Gallen Consensus 2013. J. Breast Cancer 2015, 18, 149–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, G.; Zhang, S.; Yazdanparast, A.; Li, M.; Pawar, A.V.; Liu, Y.; Inavolu, S.M.; Cheng, L. Comprehensive comparison of molecular portraits between cell lines and tumors in breast cancer. BMC Genom. 2016, 17, 281–301. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [Green Version]
- Landman, K.A.; Cai, A.Q.; Hughes, B.D. Travelling Waves of Attached and Detached Cells in a Wound-Healing Cell Migration Assay. Bull. Math. Biol. 2007, 69, 2119–2138. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Manegold, P.C.; Hong, Y.K.; Zhang, W.; Pohl, A.; Lurje, G.; Winder, T.; Yang, D.; LaBonte, M.J.; Wilson, M.L.; et al. Interleukin-8 is associated with proliferation, migration, angiogenesis and chemosensitivity in vitro and in vivo in colon cancer cell line models. Int. J. Cancer 2010, 128, 2038–2049. [Google Scholar] [CrossRef] [Green Version]
- Levina, V.; Marrangoni, A.M.; Demarco, R.; Gorelik, E.; Lokshin, A.E. Drug-Selected Human Lung Cancer Stem Cells: Cytokine Network, Tumorigenic and Metastatic Properties. PLoS ONE 2008, 3, e3077. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Huang, C.-F.; Li, Y.-C.; Deng, W.-W.; Mao, L.; Wu, L.; Zhang, W.-F.; Zhang, L.; Sun, Z.-J. Blockage of the NLRP3 inflammasome by MCC950 improves anti-tumor immune responses in head and neck squamous cell carcinoma. Cell. Mol. Life Sci. 2017, 75, 2045–2058. [Google Scholar] [CrossRef]
- Xue, Y.; Du, H.-D.; Tang, D.; Zhang, D.; Zhou, J.; Zhai, C.-W.; Yuan, C.-C.; Hsueh, C.-Y.; Li, S.-J.; Heng, Y.; et al. Correlation between the NLRP3 Inflammasome and the Prognosis of Patients with LSCC. Front. Oncol. 2019, 9, 588. [Google Scholar] [CrossRef]
- Poli, G.; Brancorsini, S.; Cochetti, G.; Barillaro, F.; Egidi, M.G.; Mearini, E. Expression of inflammasome-related genes in bladder cancer and their association with cytokeratin 20 messenger RNA. Urol. Oncol. Semin. Orig. Investig. 2015, 33, 505.e1–505.e7. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, H.; Qin, Z.; Zhao, F.; Zhou, L.; Xu, L.; Jia, R. NLRP3 inflammasome promoted the malignant progression of prostate cancer via the activation of caspase-1. Cell Death Discov. 2021, 7, 399. [Google Scholar] [CrossRef]
- Zhang, L.; Li, H.; Zang, Y.; Wang, F. NLRP3 inflammasome inactivation driven by miR-223-3p reduces tumor growth and increases anticancer immunity in breast cancer. Mol. Med. Rep. 2019, 19, 2180–2188. [Google Scholar] [CrossRef] [Green Version]
- Guo, B.; Fu, S.; Zhang, J.; Liu, B.; Li, Z. Targeting inflammasome/IL-1 pathways for cancer immunotherapy. Sci. Rep. 2016, 6, 36107. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Hara, H.; Núñez, G. Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends Biochem. Sci. 2016, 41, 1012–1021. [Google Scholar] [CrossRef] [Green Version]
- Martinon, F.; Burns, K.; Tschopp, J. The Inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef]
- Kayagaki, N.; Wong, M.T.; Stowe, I.B.; Ramani, S.R.; Gonzalez, L.C.; Akashi-Takamura, S.; Miyake, K.; Zhang, J.; Lee, W.P.; Muszyński, A.; et al. Noncanonical Inflammasome Activation by Intracellular LPS Independent of TLR4. Science 2013, 341, 1246–1249. [Google Scholar] [CrossRef]
- Franchi, L.; Eigenbrod, T.; Muñoz-Planillo, R.; Nuñez, G. The inflammasome: A caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 2009, 10, 241–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, E.-K.; Kim, J.K.; Shin, D.-M.; Sasakawa, C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell. Mol. Immunol. 2015, 13, 148–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cullen, S.P.; Kearney, C.J.; Clancy, D.M.; Martin, S.J. Diverse Activators of the NLRP3 Inflammasome Promote IL-1β Secretion by Triggering Necrosis. Cell Rep. 2015, 11, 1535–1548. [Google Scholar] [CrossRef] [Green Version]
- Boichuk, S.; Galembikova, A.; Sitenkov, A.; Khusnutdinov, R.; Dunaev, P.; Valeeva, E.; Usolova, N. Establishment and characterization of a triple negative basal-like breast cancer cell line with multi-drug resistance. Oncol. Lett. 2017, 14, 5039–5045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.V.; Oesterreich, S.; Davidson, N.E. MCF-7 Cells—Changing the Course of Breast Research and Care for 45 Years. J. Natl. Cancer Inst. 2015, 107, djv073. [Google Scholar] [CrossRef] [Green Version]
- Hui, L.; Zheng, Y.; Yan, Y.; Bargonetti, J.; Foster, D.A. Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D. Oncogene 2006, 25, 7305–7310. [Google Scholar] [CrossRef] [Green Version]
- Patra, S.; Young, V.; Llewellyn, L.; Senapati, J.; Mathew, J. BRAF, KRAS and PIK3CA Mutation and Sensitivity to Trastuzumab in Breast Cancer Cell Line Model. Asian Pac. J. Cancer Prev. 2017, 18, 2209–2213. [Google Scholar] [CrossRef]
- Eng, C.H.; Wang, Z.; Tkach, D.; Toral-Barza, L.; Ugwonali, S.; Liu, S.; Fitzgerald, S.L.; George, E.; Frias, E.; Cochran, N.; et al. Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc. Natl. Acad. Sci. USA 2015, 113, 182–187. [Google Scholar] [CrossRef] [Green Version]
- Hamarsheh, S.; Osswald, L.; Saller, B.S.; Unger, S.; de Feo, D.; Vinnakota, J.M.; Konantz, M.; Uhl, F.M.; Becker, H.; Lübbert, M.; et al. Oncogenic KrasG12D causes myeloproliferation via NLRP3 inflammasome activation. Nat. Commun. 2020, 11, 1659. [Google Scholar] [CrossRef] [Green Version]
- Hamarsheh, S.; Groß, O.; Brummer, T.; Zeiser, R. Immune modulatory effects of oncogenic KRAS in cancer. Nat. Commun. 2020, 11, 5439. [Google Scholar] [CrossRef]
- Miller, L.J.; Kurtzman, S.H.; Anderson, K.; Wang, Y.; Stankus, M.; Renna, M.; Lindquist, R.; Barrows, G.; Kreutzer, D.L. Interleukin-1 Family Expression in Human Breast Cancer: Interleukin-1 Receptor Antagonist. Cancer Investig. 2000, 18, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Chavey, C.; Bibeau, F.; Gourgou-Bourgade, S.; Burlinchon, S.; Boissière, F.; Laune, D.; Roques, S.; Lazennec, G. Oestrogen receptor negative breast cancers exhibit high cytokine content. Breast Cancer Res. 2007, 9, R15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucero, M.; Thind, J.; Sandoval, J.; Senaati, S.; Jimenez, B.; Kandpal, R.P. Stem-like Cells from Invasive Breast Carcinoma Cell Line MDA-MB-231 Express a Distinct Set of Eph Receptors and Ephrin Ligands. Cancer Genom. Proteom. 2020, 17, 729–738. [Google Scholar] [CrossRef]
- Fillmore, C.M.; Kuperwasser, C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008, 10, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Chaudhry, A.; Lee, I.; Frank, J.A. Effects of Long-Term Hypoxia and Pro-Survival Cocktail in Bone Marrow-Derived Stromal Cell Survival. Stem Cells Dev. 2014, 23, 530–540. [Google Scholar] [CrossRef] [Green Version]
- Mego, M.; Cholujova, D.; Gronesova, P.; Pastorek, M.; Miskovska’, V.; Obertova, J.; Usakova, V.; Ondrus, D.; Spanik, S.; Mardiak, J. A Cytokine and Angiogenic Factor (CAF) Analysis in Plasma in Testicular Germ Cell Tumor Patients. Ann. Oncol. 2012, 23, ix286. [Google Scholar] [CrossRef]
- Jiang, J.; Ye, F.; Yang, X.; Zong, C.; Gao, L.; Yang, Y.; Zhao, Q.; Han, Z.; Wei, L. Peri-tumor associated fibroblasts promote intrahepatic metastasis of hepatocellular carcinoma by recruiting cancer stem cells. Cancer Lett. 2017, 404, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Sukowati, C.H.C.; Patti, R.; Pascut, D.; Ladju, R.B.; Tarchi, P.; Zanotta, N.; Comar, M.; Tiribelli, C.; Crocè, L.S. Serum Stem Cell Growth Factor Beta for the Prediction of Therapy Response in Hepatocellular Carcinoma. BioMed Res. Int. 2018, 2018, 6435482. [Google Scholar] [CrossRef] [PubMed]
- Tanno, T.; Matsui, W. Development and Maintenance of Cancer Stem Cells under Chronic Inflammation. J. Nippon. Med. Sch. 2011, 78, 138–145. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.-Y.; Tsai, M.-C.; Tu, W.; Yeh, H.-C.; Wang, S.-C.; Huang, S.-P.; Li, C.-Y. Role of the NLRP3 Inflammasome: Insights into Cancer Hallmarks. Front. Immunol. 2021, 11, 610492. [Google Scholar] [CrossRef]
- Schwab, L.P.; Peacock, D.L.; Majumdar, D.; Ingels, J.F.; Jensen, L.C.; Smith, K.D.; Cushing, R.C.; Seagroves, T.N. Hypoxia-inducible factor 1α promotes primary tumor growth and tumor-initiating cell activity in breast cancer. Breast Cancer Res. 2012, 14, R6. [Google Scholar] [CrossRef] [Green Version]
- Yun, Z.; Lin, Q. Hypoxia and Regulation of Cancer Cell Stemness. Adv. Exp. Med. Biol. 2013, 772, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, H.; Wyckoff, J.; Condeelis, J. Cell migration in tumors. Curr. Opin. Cell Biol. 2005, 17, 559–564. [Google Scholar] [CrossRef]
- Kiso, M.; Tanaka, S.; Saji, S.; Toi, M.; Sato, F. Long isoform of VEGF stimulates cell migration of breast cancer by filopodia formation via NRP1/ARHGAP17/Cdc42 regulatory network. Int. J. Cancer 2018, 143, 2905–2918. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Wu, Y.; Helman, J.I.; Wen, Y.; Wang, C.; Li, L. CXCR4 Promotes Oral Squamous Cell Carcinoma Migration and Invasion through Inducing Expression of MMP-9 and MMP-13 via the ERK Signaling Pathway. Mol. Cancer Res. 2011, 9, 161–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dulak, A.M.; Gubish, C.T.; Stabile, L.P.; Henry, C.; Siegfried, J.M. HGF-independent potentiation of EGFR action by c-Met. Oncogene 2011, 30, 3625–3635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Liu, P.; Wang, Y.; Li, H.; Wei, F.; Cheng, Y.; Han, L.; Yu, J. Neurotensin, a Novel Messenger to Cross-Link Inflammation and Tumor Invasion via Epithelial-Mesenchymal Transition Pathway. Int. Rev. Immunol. 2016, 35, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, S.; Lin, Y.; Miao, Y.; Zeng, Y.; Nie, Y.; Guo, P.; Jiang, G.; Wu, J. Epithelial-mesenchymal transition in cancer: Role of the IL-8/IL-8R axis. Oncol. Lett. 2017, 13, 4577–4584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.-C.; Ho, H.-C.; Su, Y.-C.; Lee, M.-S.; Hung, S.-K.; Lin, C.-H. MCP1-Induced Epithelial-Mesenchymal Transition in Head and Neck Cancer by AKT Activation. Anticancer Res. 2015, 35, 3299–3306. [Google Scholar]
- Li, S.; Lu, J.; Chen, Y.; Xiong, N.; Li, L.; Zhang, J.; Yang, H.; Wu, C.; Zeng, H.; Liu, Y. MCP-1-induced ERK/GSK-3β/Snail signaling facilitates the epithelial–mesenchymal transition and promotes the migration of MCF-7 human breast carcinoma cells. Cell. Mol. Immunol. 2017, 14, 621–630. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Wang, B.; Wang, T.; Xu, L.; He, C.; Wen, H.; Chunyan, H.; Su, H.; Zhu, X. Toll-Like Receptor 4 Prompts Human Breast Cancer Cells Invasiveness via Lipopolysaccharide Stimulation and Is Overexpressed in Patients with Lymph Node Metastasis. PLoS ONE 2014, 9, e109980. [Google Scholar] [CrossRef]
- Wang, P.-P.; Xie, D.-Y.; Liang, X.-J.; Peng, L.; Zhang, G.-L.; Ye, Y.-N.; Xie, C.; Gao, Z.-L. HGF and Direct Mesenchymal Stem Cells Contact Synergize to Inhibit Hepatic Stellate Cells Activation through TLR4/NF-kB Pathway. PLoS ONE 2012, 7, e43408. [Google Scholar] [CrossRef]
- He, W.; Qu, T.; Yu, Q.; Wang, Z.; Lv, H.; Zhang, J.; Zhao, X.; Wang, P. LPS induces IL-8 expression through TLR4, MyD88, NF-kappaB and MAPK pathways in human dental pulp stem cells. Int. Endod. J. 2012, 46, 128–136. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, X.; Deng, B.; Xiao, J.; Jin, J.; Huang, Z. Lipopolysaccharide and palmitic acid synergistically induced MCP-1 production via MAPK-meditated TLR4 signaling pathway in RAW264.7 cells. Lipids Health Dis. 2019, 18, 71. [Google Scholar] [CrossRef] [Green Version]
- Seo, S.-W.; Park, S.-K.; Oh, S.-J.; Shin, O.S. TLR4-mediated activation of the ERK pathway following UVA irradiation contributes to increased cytokine and MMP expression in senescent human dermal fibroblasts. PLoS ONE 2018, 13, e0202323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Z.; Gu, X.; Chen, Y.; Liu, J.; Hou, C.; Lin, S.; Hao, N.; Liang, Y.; Chen, W.; Meng, H. Toll-like receptor 4 activates the NLRP3 inflammasome pathway and periodontal inflammaging by inhibiting Bmi-1 expression. Int. J. Mol. Med. 2020, 47, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wise, L.; Fukuchi, K.-I. TLR4 Cross-Talk with NLRP3 Inflammasome and Complement Signaling Pathways in Alzheimer’s Disease. Front. Immunol. 2020, 11, 724. [Google Scholar] [CrossRef]
- Mehmeti, M.; Allaoui, R.; Bergenfelz, C.; Saal, L.H.; Ethier, S.P.; Johansson, M.E.; Jirström, K.; Leandersson, K. Expression of functional toll like receptor 4 in estrogen receptor/progesterone receptor-negative breast cancer. Breast Cancer Res. 2015, 17, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, T.; Zhang, Z.; Yu, H.; Ren, S.; Wang, J.; Li, S.; Sun, L. Tamoxifen Exerts Anticancer Effects on Pituitary Adenoma Progression via Inducing Cell Apoptosis and Inhibiting Cell Migration. Int. J. Mol. Sci. 2022, 23, 2664. [Google Scholar] [CrossRef]
- Vegeto, E.; Bonincontro, C.; Pollio, G.; Sala, A.; Viappiani, S.; Nardi, F.; Brusadelli, A.; Viviani, B.; Ciana, P.; Maggi, A. Estrogen Prevents the Lipopolysaccharide-Induced Inflammatory Response in Microglia. J. Neurosci. 2001, 21, 1809–1818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawas, A.F.; Mistry, R.; Narayanan, S.; Thomas-Jardin, S.E.; Ramachandran, J.; Ravichandran, J.; Neduvelil, E.; Luangpanh, K.; Delk, N.A. IL-1 induces p62/SQSTM1 and autophagy in ERα+/PR+ BCa cell lines concomitant with ERα and PR repression, conferring an ERα−/PR− BCa-like phenotype. J. Cell. Biochem. 2018, 120, 1477–1491. [Google Scholar] [CrossRef] [PubMed]
- Escobar, P.; Bouclier, C.; Serret, J.; Bièche, I.; Brigitte, M.; Caicedo, A.; Sanchez, E.; Vacher, S.; Vignais, M.-L.; Bourin, P.; et al. IL-1β produced by aggressive breast cancer cells is one of the factors that dictate their interactions with mesenchymal stem cells through chemokine production. Oncotarget 2015, 6, 29034–29047. [Google Scholar] [CrossRef] [Green Version]
- Mon, N.N.; Senga, T.; Ito, S. Interleukin-1β activates focal adhesion kinase and Src to induce matrix metalloproteinase-9 production and invasion of MCF-7 breast cancer cells. Oncol. Lett. 2016, 13, 955–960. [Google Scholar] [CrossRef] [Green Version]
- Holen, I.; Lefley, D.V.; Francis, S.E.; Rennicks, S.; Bradbury, S.; Coleman, R.E.; Ottewell, P. IL-1 drives breast cancer growth and bone metastasis in vivo. Oncotarget 2016, 7, 75571–75584. [Google Scholar] [CrossRef] [Green Version]
- Tulotta, C.; Ottewell, P.D. The role of IL-1B in breast cancer bone metastasis. Endocr.-Relat. Cancer 2018, 25, R421–R434. [Google Scholar] [CrossRef] [Green Version]
- Sobesky, J.L.; D’Angelo, H.M.; Weber, M.D.; Anderson, N.D.; Frank, M.G.; Watkins, L.R.; Maier, S.F.; Barrientos, R.M. Glucocorticoids Mediate Short-Term High-Fat Diet Induction of Neuroinflammatory Priming, the NLRP3 Inflammasome, and the Danger Signal HMGB1. Eneuro 2016, 3. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Wei, H.; Li, S.; Wu, P.; Mao, X. The Role of Progesterone Receptors in Breast Cancer. Drug Des. Dev. Ther. 2022, 16, 305–314. [Google Scholar] [CrossRef]
- Narvekar, N.; Cameron, S.; Critchley, H.O.D.; Lin, S.; Cheng, L.; Baird, D.T. Low-Dose Mifepristone Inhibits Endometrial Proliferation and Up-Regulates Androgen Receptor. J. Clin. Endocrinol. Metab. 2004, 89, 2491–2497. [Google Scholar] [CrossRef] [Green Version]
- Maurea, N.; Quagliariello, V.; Iaffaioli, R.V.; de Laurentiis, M.; Botti, G. The sodium-glucose cotransporter-2 inhibitor dapagliflozin exerts cardioprotective effects against doxorubicin and trastuzumab toxicity through tlr4/MYD88/NF-KB signaling and NLRP3 inflam-masome pathway. J. Am. Coll. Cardiol. 2020, 75, 1192. [Google Scholar] [CrossRef]
- Larsen, S.S.; Egeblad, M.; Jäättelä, M.; Lykkesfeldt, A.E. Acquired antiestrogen resistance in MCF-7 human breast cancer sublines is not accomplished by altered expression of receptors in the ErbB-family. Breast Cancer Res. Treat. 1999, 58, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Emlet, D.R.; Brown, K.A.; Kociban, D.L.; Pollice, A.A.; Smith, C.A.; Ong, B.B.L.; Shackney, S.E. Response to trastuzumab, erlotinib, and bevacizumab, alone and in combination, is correlated with the level of human epidermal growth factor receptor-2 expression in human breast cancer cell lines. Mol. Cancer Ther. 2007, 6, 2664–2674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigelt, B.; Lo, A.T.; Park, C.C.; Gray, J.W.; Bissell, M.J. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res. Treat. 2009, 122, 35–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paik, S.; Kim, C.; Wolmark, N. HER2 Status and Benefit from Adjuvant Trastuzumab in Breast Cancer. N. Engl. J. Med. 2008, 358, 1409–1411. [Google Scholar] [CrossRef] [PubMed]
- Burguin, A.; Furrer, D.; Ouellette, G.; Jacob, S.; Diorio, C.; Durocher, F. Trastuzumab effects depend on HER2 phosphorylation in HER2-negative breast cancer cell lines. PLoS ONE 2020, 15, e0234991. [Google Scholar] [CrossRef]
- Rubartelli, A.; Bajetto, A.; Allavena, G.; Cozzolino, F.; Sitia, R. Post-translational regulation of interleukin 1β secretion. Cytokine 1993, 5, 117–124. [Google Scholar] [CrossRef]
- Tassi, S.; Carta, S.; Delfino, L.; Caorsi, R.; Martini, A.; Gattorno, M.; Rubartelli, A. Altered redox state of monocytes from cryopyrin-associated periodic syndromes causes accelerated IL-1β secretion. Proc. Natl. Acad. Sci. USA 2010, 107, 9789–9794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carta, S.; Tassi, S.; Pettinati, I.; Delfino, L.; Dinarello, C.A.; Rubartelli, A. The Rate of Interleukin-1β Secretion in Different Myeloid Cells Varies with the Extent of Redox Response to Toll-like Receptor Triggering. J. Biol. Chem. 2011, 286, 27069–27080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perregaux, D.G.; Laliberte, R.E.; Gabel, C.A. Human Monocyte Interleukin-1β Posttranslational Processing. J. Biol. Chem. 1996, 271, 29830–29838. [Google Scholar] [CrossRef] [Green Version]
- Lagarkova, M.A.; Shutova, M.V.; Bogomazova, A.; Vassina, E.M.; Glazov, E.A.; Zhang, P.; Rizvanov, A.; Chestkov, I.V.; Kiselev, S. Induction of pluripotency in human endothelial cells resets epigenetic profile on genome scale. Cell Cycle 2010, 9, 937–946. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.-C.; Park, A.Y.; Guan, J.-L. In Vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badea, M.A.; Balas, M.; Hermenean, A.; Ciceu, A.; Herman, H.; Ionita, D.; Dinischiotu, A. Influence of Matrigel on Single- and Multiple-Spheroid Cultures in Breast Cancer Research. SLAS Discov. Adv. Sci. Drug Discov. 2019, 24, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Sagulenko, V.; Zamoshnikova, A.; Richards, A.A.; Cridland, J.A.; Irvine, K.M.; Stacey, K.J.; Sweet, M.J. Acute lipopolysaccharide priming boosts inflammasome activation independently of inflammasome sensor induction. Immunobiology 2012, 217, 1325–1329. [Google Scholar] [CrossRef] [Green Version]
- Juliana, C.; Fernandes-Alnemri, T.; Kang, S.; Farias, A.; Qin, F.; Alnemri, E.S. Non-transcriptional Priming and Deubiquitination Regulate NLRP3 Inflammasome Activation. J. Biol. Chem. 2012, 287, 36617–36622. [Google Scholar] [CrossRef] [Green Version]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamza, S.; Garanina, E.E.; Alsaadi, M.; Khaiboullina, S.F.; Tezcan, G. Blocking the Hormone Receptors Modulates NLRP3 in LPS-Primed Breast Cancer Cells. Int. J. Mol. Sci. 2023, 24, 4846. https://doi.org/10.3390/ijms24054846
Hamza S, Garanina EE, Alsaadi M, Khaiboullina SF, Tezcan G. Blocking the Hormone Receptors Modulates NLRP3 in LPS-Primed Breast Cancer Cells. International Journal of Molecular Sciences. 2023; 24(5):4846. https://doi.org/10.3390/ijms24054846
Chicago/Turabian StyleHamza, Shaimaa, Ekaterina E. Garanina, Mohammad Alsaadi, Svetlana F. Khaiboullina, and Gulcin Tezcan. 2023. "Blocking the Hormone Receptors Modulates NLRP3 in LPS-Primed Breast Cancer Cells" International Journal of Molecular Sciences 24, no. 5: 4846. https://doi.org/10.3390/ijms24054846





